Abstract
Small interfering RNAs (siRNAs) represent a new class of antigene agents, which has emerged as a powerful tool for functional genomics and might serve as a potent therapeutic approach. However, several studies have showed that they could trigger several bystander effects, including immune activation and inhibition of unintended target genes. Although activation of innate immunity by siRNAs might be beneficial for therapy in some instances, uncontrolled activation can be toxic, and is therefore a major challenging problem. Interestingly, replacement of uridines in siRNA sequences with their 2′-modified counterparts abrogated siRNA bystander effects. Here we highlight these important findings that are expected to facilitate the rational design of siRNAs that avoid the induction of bystander effects.
INTRODUCTION
A variety of well-documented approaches have been developed for inhibiting gene expression. These include the use of antisense RNAs, oligodeoxynucleotides (ODNs), ribozymes, and RNA interference (RNAi) [1–3]. Among these approaches, RNAi-mediated mRNA degradation is emerging as the most highly effective antigene strategies [2, 3]. Although the same phenomenon was described in transgenic plants in the late 1980s [4], only in 1998 Fire and colleagues have demonstrated that ds RNA introduced into Caenorhabditis elegans can silence the expression of homologous target gene by directing degradation of its mRNA [5]. During RNAi, the precursor double-stranded (ds) RNA is processed by the Dicer endonuclease into short 21–24 nucleotides ds siRNAs containing 2-base 3′-overhangs. Subsequently, the siRNA duplexes are then incorporated into a multiprotein complex, the RNA-induced silencing complex (RISC), which mediates the degradation of mRNAs with sequence homologous to the siRNA. Recent studies have showed that nucleotides 2–8 of the siRNA antisense strand form a seed sequence that directs target mRNA recognition [6]. Some of the protein components of RISC have been identified and assigned functions. Argonaute 2 (Ago2) is the RNA endonuclease that cleaves target mRNA [7].
Despite the demonstration of RNAi in plants and worms, researchers have been pessimistic about using RNAi in mammalian cells due to the induction of the interferon (INF) pathway by long ds RNAs (> 30 nucleotides), leading to nonspecific inhibition of protein synthesis and degradation of mRNAs [8]. However, Tuschl and colleagues found that siRNA duplexes mimicking the siRNAs generated by Dicer cleavage in lower organisms could trigger RNAi without inducing the IFN pathway, which is observed with long double-stranded RNA in most cell types [9]. This discovery has paved the way for the study of gene function in mammalian cells. In contrast to invertebrates, mammalian cells do not have transitive RNAi because of the absence of RNA-dependent RNA polymerase [2, 3]. Consequently, the number of siRNA molecules introduced into a cell limits gene silencing induced by synthetic siRNAs. To circumvent this potential problem, several groups have developed expression vectors that direct the synthesis of RNA duplexes or short hairpin RNAs (shRNAs) in mammalian cells, leading to sustained production of siRNAs [3]. One approach for silencing relies on the expression of shRNAs under the control of H1 or U6 pol III promoter, which is active in most mammalian cell types.
During the last 5 years, siRNA-mediated gene knockdown has become a standard method for studying gene function and drug target validation. However, despite the success that has been achieved, some serious concerns such as the activation of innate immunity, inhibition of unintended target mRNAs, and potential interference with regulatory functions of endogenous microRNAs need to be resolved prior therapeutic applications in humans [2, 10–12].
CELLULAR RESPONSE TO siRNAs
The gradual maturation of RNAi technology from the laboratory to the clinic involves several major challenges, many of which still need to be resolved. These include delivery to target cells, intracellular stability, and specificity [2, 3]. It should be noted that the introduction of synthetic siRNAs into mammalian cells in culture is relatively simple by transfection reagents [9]. siRNAs can be chemically synthesized, or produced by in vitro transcription or by digestion of long dsRNAs by recombinant RNase III or Dicer [2, 3]. Most of the techniques that have been used for antisense delivery can be applied to synthetic siRNAs and to DNA constructs engineered to express shRNAs. These include electroporation, cell microinjection, and lipophilic transfection [13].
Although siRNAs were initially thought to be small enough to avoid the activation of the IFN pathway [9], recent studies showed they could activate innate immunity in mammalian cells [10, 14–16]. In this respect, Sledz and colleagues reported that siRNA could activate PKR, and the effects were sequence independent and do not occur with the sense or the antisense RNA used to prepare the siRNA duplexes [15]. In contrast, Kariko and colleagues found that siRNA could activate cytokine and interferon production via TLR3, a receptor known to bind viral ds RNAs [16]. TLR3 is mainly expressed on the cell surface. However, we and others recently have demonstrated that PKR and TLR-3 do not represent the major mechanism by which chemically synthesized siRNA activate innate immunity [17–19]. In addition, internalization of ds siRNAs or ss siRNAs is required for immune activation. Indeed, inhibitors of endosomal maturation/acidification like bafilomycin A1, a drug that inhibit endosomal H+-proton pumps, blocked immune activation [17], indicating the involvement of endosomal TLR7 and TLR8. Consistent with the role of endosomal TLRs in siRNA signaling, TLR7 knockout mice did not mount immune activation in response to siRNAs [18]. Taken together, the available data indicate that immune cells such as monocytes and dendritic cells recognize ds siRNAs and ss siRNAs through TLR7 and TLR8, leading to the production of inflammatory cytokines and type I interferons [20]. We also found that ss siRNAs are more effective than ds siRNA in triggering TLR7 and TLR8 responses [17, 21]. In addition, we have found that human bone marrow hematopoietic CD34+ progenitor cells express TLR7 and TLR8. Interestingly, incubation of these cells with either immunostimulatory synthetic siRNAs or R848, a specific ligand for TLR7 and TLR8, induced their differentiation into the myeloid lineage (Sioud et al in preparation). Thus, the interaction of viral RNAs with CD34+ progenitor cells may increase the pool of innate immune cells.
As mentioned above mammalian cells respond to siRNAs, provided they are delivered to the endosomes. Notably, the immune system uses a set of germ line encoded receptors called pattern recognition receptors (PRRs) to recognize common microbial structures known as pathogen associated molecular patterns [22]. Whereas several TLRs are expressed in the cell surface, TLR3, TLR7, TLR8, and TLR9, traffic between the endoplasmic reticulum and intracellular compartment such as the endosomes and the lysosomes [22]. An obvious function of these trafficking pathways is to scan for viral and/or bacterial nucleic acids, thus playing a central role in innate antiviral responses. Therefore siRNAs internalized via endocytosis are more likely to activate endosomal TLRs. It should be noted that cytoplasmic delivery of immunostimulatoty synthetic siRNAs or shRNA did not induce immune response in human blood cells [17, 21], suggesting that they are not recognized by cytolasmic sensors of ds RNAs. In this respect, a recent study has showed that endogenously expressed shRNAs are not immunostimulatory in human cells [23]. Interestingly, Williams and colleagues demonstrated that the presence of 2-base 3′-overhangs in synthetic siRNAs or in Dicer processed shRNAs blocks the activation of RIG-1, a major cytolasmic sensor for viral ds RNAs [24].
STRUCTURE FEATURES THAT ACTIVATED TLR7 AND TLR8
Initial experiments indicate that some types of secondary structures and/or specific nucleotides are responsible for the activation of NF-κB signaling pathway by siRNAs in adherent PBMC, an enriched monocyte population [10]. Monocytes are circulating peripheral blood cells that can be differentiated by cytokines into macrophages of different phenotypes as well as into dendritic cells. As mentioned above, siRNA effects are sequence dependent and can occur with ds siRNAs and ss siRNAs [20]. Thus, what is the nature of IFN-inducing motif present in one sequence but absent in another? Although GU dinucleotides were found to trigger TLR7 and TLR8 activation [25], their absolute requirement in siRNA activation of innate immunity is still not clear [21]. Judge and colleagues identified one RNA motif and its immunostimulatory effect seems to depend on the GU content [19]. However, Hornung and colleagues identified a second RNA motif that is recognized by TLR7 in the context of siRNA duplexes and the activity does not depend on GU content [18]. It is worth noting that several siRNA sequences without GU nucleotides activated the immune system [21, 26]. Thus, it is likely that in addition to GU dinucleotides other characteristics such as RNA structure, base position, and base composition of the siRNA flanking sequences may be involved.
2′-RIBOSE MODIFICATIONS OF URIDINES BLOCKS IMMUNE ACTIVATION
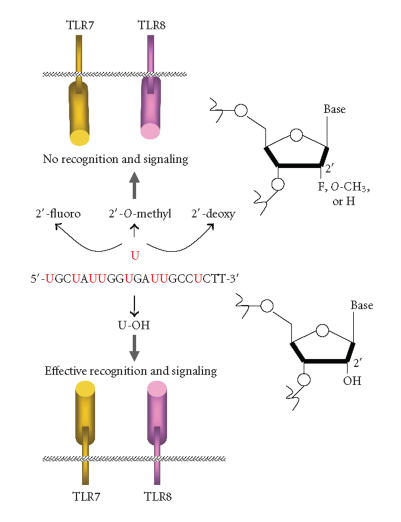
The identification of the immunostimulatory sequence that can activate innate immunity will allow the design of siRNAs to minimize activation or to increase the immune response for combating infections and tumor cells. Considering the simplicity of the immunostimulatory motifs and their high frequency in human RNAs, it is desirable to find strategies that evade immune recognition of siRNAs. At least two basic strategies can be used to block immune recognition of siRNA. One is to use delivery agents that avoid the delivery and/or retention of siRNA within the endosomes. The other is to apply chemical modifications. Regarding the first strategy, Song and colleagues described an antibody-based delivery strategy that can result in gene silencing without immune activation [27]. The second strategy relies in the use of modified nucleotides, which are essential to protect siRNAs from nuclease degradation and ameliorate their pharmacokinetic parameters in vivo [13]. However, the chemical modification that blocks immune activation must be chosen carefully so as not to inhibit siRNA silencing activity. Thus, finding the appropriate chemical modifications for inhibiting siRNA immune activation will be important for exploring their therapeutic applications. In this respect, replacement of the 2′-hydroxyl uridines with either 2′-fluoro, 2′-deoxy, or 2′-O-methyl uridines abrogated immune recognition of siRNAs by TLRs [21]. Thus, endosomal TLRs can distinguish between modified and unmodified RNAs (Figure 1). In order to distinguish between self and nonself RNAs, the immune system may use both endosomal compartmentalization and RNA modification strategies [20]. Collectively, these recent findings offer the possibility of choosing the appropriate modifications that evade immune activation without reducing siRNA-silencing potency. Of note, siRNA with either 2′-fluoro uridines or 2′-deoxy uridines maintained silencing activity [26, 28].
Figure 1.
Unmodified siRNA duplexes and single-stranded siRNAs (sense or antisense strand) activate innate immunity through TLR7 and TLR8 resulting in cytokines and type I interferon production. 2′-ribose modifications of uridines abrogate TLR recognition of RNAs and therefore signaling [21]. Modified ss siRNAs did not compete with unmodified siRNAs to activate TLR signaling, indicating that 2′ modifications of uridines block the binding to TLRs (unpublished data).
Another potential source of toxicity is the destruction of cellular mRNAs that share partial homology to the siRNA sequences. Indeed, recent studies demonstrated that both siRNAs and microRNAs could interact with undesired target mRNAs via base pairing of only few nucleotides, leading to inhibition of gene expression [12, 29]. Interestingly, we have found that chemical modifications of siRNAs not only evade immune activation but also reduce the ss siRNAs and ds siRNA “off-target effects” [30]. Although the evading mechanism is not known, it is probable that the interaction of ss siRNAs or ds siRNAs with unintended cellular mRNAs is affected by chemical modifications.
IMMUNOSTIMULATORY siRNAs AS VACCINE ADJUVANTS
It is generally accepted that initiation of a specific immune response requires activation of innate immunity resulting in a proinflammatory response. The produced cytokines and chemokines assist in activating and directing the adaptive immune responses. Therefore, a vaccine has to induce a proinflammatory response to be effective. As discussed above, siRNA sequences containing immunostimulatory motifs induced interferons, chemokines, proinflammatory cytokines, monocyte differentiation, and dendritic cell maturation [20]. Activated DCs produce high levels of IL-12, INF-α, and proinflammatory cytokines such as IL-6 and TNF-α. IFN-α triggers not only innate immune defense such as the activation of NK cells, but also adaptive Th-1 responses, which are important for killing tumor cells and virus infected cells [20]. Engagement of endosomal TLR7 and/or TLR8 with siRNAs causes activation of at least three key transcription factors, NF-κB, IFN regulatory factor (IRF)-3, and IRF-7, which are important immune responses [30]. Previous studies have shown that the activation of TLR9 can improve both cell-mediated and humoral responses to antigens [31]. Notably, endosomal TLR9 recognize unmethylated 2′-deoxyribo (cytidine-phosphate-guanosine) CpG motifs that are more commonly found in bacterial and viral genomes [32]. Most, but not all, CpG dinucleotides are methylated in the genomic DNA of vertebrates. Although much remains to be revealed regarding the adjuvant potency of RNA oligonucleotides, the reported data indicate that ss siRNA- and ds siRNAs-containing immunostimulatory RNA motifs might improve cancer and viral vaccines.
CONCLUDING REMARKS
As with any new agent, issues of delivery and specificity are major obstacles before siRNAs can be used in patients. Similar to antisense oligonucleotides, certain synthetic siRNAs activated innate immunity via TLRs, in particular TLR7 and TLR8. Therefore, there is a need to examine the immunostimulatory effects of any potential therapeutic siRNA in human immune cells prior to clinical applications. The recent findings clearly show that the 2′-hydroxyl uridines are absolutely essential for TLR7 and/or TLR8 recognition and signaling. Indeed, replacement of uridines with 2′-fluoro, 2′-deoxy, or 2′-O-methyl modified counterparts abrogated immune activation by ss siRNA and siRNA duplexes. Alternatively, the presence of 2′-modified uridines might protect siRNAs for being sensed by the immune system. Also, most of the other bystander effects not related to immune activation were also inhibited by chemical modifications. These findings will enable the rational design of siRNAs that avoid the induction of bystander effects.
ACKNOWLEDGMENT
We thank Dr. Anne Dybwad for critical reading of the manuscript.
References
- 1.Sioud M. Nucleic acid enzymes as a novel generation of anti-gene agents. Current Molecular Medicine. 2001;1(5):575–588. doi: 10.2174/1566524013363366. [DOI] [PubMed] [Google Scholar]
- 2.Sioud M. Therapeutic siRNAs. Trends in Pharmacological Sciences. 2004;25(1):22–28. doi: 10.1016/j.tips.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 3.Hannon GJ, Rossi JJ. Unlocking the potential of the human genome with RNA interference. Nature. 2004;431(7006):371–378. doi: 10.1038/nature02870. [DOI] [PubMed] [Google Scholar]
- 4.Jorgensen R. Altered gene expression in plants due to trans interactions between homologous genes. Trends in Biotechnology. 1990;8(12):340–344. doi: 10.1016/0167-7799(90)90220-r. [DOI] [PubMed] [Google Scholar]
- 5.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by doublestranded RNA in caenorhabditis elegans. Nature. 1998;391(6669):806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 6.Ma J-B, Yuan Y-R, Meister G, Pei Y, Tuschl T, Patel DJ. Structural basis for 5′-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature. 2005;434(7033):666–670. doi: 10.1038/nature03514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu J, Carmell MA, Rivas FV, et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305(5689):1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 8.Sen GC. Viruses and interferons. Annual Review of Microbiology. 2001;55:255–281. doi: 10.1146/annurev.micro.55.1.255. [DOI] [PubMed] [Google Scholar]
- 9.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411(6836):494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 10.Sioud M, Sørensen DR. Cationic liposome-mediated delivery of siRNAs in adult mice. Biochemical and Biophysical Research Communications. 2003;312(4):1220–1225. doi: 10.1016/j.bbrc.2003.11.057. [DOI] [PubMed] [Google Scholar]
- 11.Jackson AL, Bartz SR, Schelter J, et al. Expression profiling reveals off-target gene regulation by RNAi. Nature Biotechnology. 2003;21(6):635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- 12.Semizarov D, Frost L, Sarthy A, Kroeger P, Halbert DN, Fesik SW. Specificity of short interfering RNA determined through gene expression signatures. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(11):6347–6352. doi: 10.1073/pnas.1131959100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sioud M. On the delivery of small interfering RNAs into mammalian cells. Expert Opinion on Drug Delivery. 2005;2(4):639–651. doi: 10.1517/17425247.2.4.639. [DOI] [PubMed] [Google Scholar]
- 14.Persengiev SP, Zhu X, Green MR. Nonspecific, concentration-dependent stimulation and repression of mammalian gene expression by small interfering RNAs (siRNAs) RNA. 2004;10(1):12–18. doi: 10.1261/rna5160904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sledz CA, Holko M, De Veer MJ, Silverman RH, Williams BRG. Activation of the interferon system by short-interfering RNAs. Nature Cell Biology. 2003;5(9):834–839. doi: 10.1038/ncb1038. [DOI] [PubMed] [Google Scholar]
- 16.Karikó K, Bhuyan P, Capodici J, Weissman D. Small interfering RNAs mediate sequence-independent gene suppression and induce immune activation by signaling through toll-like receptor 3. Journal of Immunology. 2004;172(11):6545–6549. doi: 10.4049/jimmunol.172.11.6545. [DOI] [PubMed] [Google Scholar]
- 17.Sioud M. Induction of inflammatory cytokines and interferon responses by double-stranded and single-stranded siRNAs is sequence-dependent and requires endosomal localization. Journal of Molecular Biology. 2005;348(5):1079–1090. doi: 10.1016/j.jmb.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 18.Hornung V, Guenthner-Biller M, Bourquin C, et al. Sequence-specific potent induction of IFN-α by short interfering RNA in plasmacytoid dendritic cells through TLR7. Nature Medicine. 2005;11(3):263–270. doi: 10.1038/nm1191. [DOI] [PubMed] [Google Scholar]
- 19.Judge AD, Sood V, Shaw JR, Fang D, McClintock K, MacLachlan I. Sequence-dependent stimulation of the mammalian innate immune response by synthetic siRNA. Nature Biotechnology. 2005;23(4):457–462. doi: 10.1038/nbt1081. [DOI] [PubMed] [Google Scholar]
- 20.Sioud M. Innate sensing of self and non-self RNAs by toll-like receptors. Trends in Molecular Medicine. 2006;12(4):167–176. doi: 10.1016/j.molmed.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 21.Sioud M. Single-stranded small interfering RNA are more immunostimulatory than their double-stranded counterparts: a central role for 2′-hyroxyl uridines in immune responses. European Journal of Immunology. 2006;36(5):1222–1230. doi: 10.1002/eji.200535708. [DOI] [PubMed] [Google Scholar]
- 22.Takeda K, Akira S. Toll-like receptors in innate immunity. International Immunology. 2005;17(1):1–14. doi: 10.1093/intimm/dxh186. [DOI] [PubMed] [Google Scholar]
- 23.Robbins MA, Li M, Leung I, et al. Stable expression of shRNAs in human CD34+ progenitor cells can avoid induction of interferon responses to siRNAs in vitro. Nature Biotechnology. 2006;24(5):566–571. doi: 10.1038/nbt1206. [DOI] [PubMed] [Google Scholar]
- 24.Marques JT, Devosse T, Wang D, et al. A structural basis for discriminating between self and nonself double-stranded RNAs in mammalian cells. Nature Biotechnology. 2006;24(5):559–565. doi: 10.1038/nbt1205. [DOI] [PubMed] [Google Scholar]
- 25.Heil F, Hemmi H, Hochrein H, et al. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science. 2004;303(5663):1526–1529. doi: 10.1126/science.1093620. [DOI] [PubMed] [Google Scholar]
- 26.Layzer JM, McCaffrey AP, Tanner AK, Huang Z, Kay MA, Sullenger BA. In vivo activity of nuclease-resistant siRNAs. RNA. 2004;10(5):766–771. doi: 10.1261/rna.5239604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song E, Zhu P, Lee S-K, et al. Antibody mediated in vivo delivery of small interfering RNAs via cell-surface receptors. Nature Biotechnology. 2005;23(6):709–717. doi: 10.1038/nbt1101. [DOI] [PubMed] [Google Scholar]
- 28.Chiu Y-L, Rana TM. siRNA function in RNAi: a chemical modification analysis. RNA. 2003;9(9):1034–1048. doi: 10.1261/rna.5103703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lim LP, Lau NC, Garrett-Engele P, et al. Microarray analysis shows that some microRNAs downregulate large numbers of-target mRNAs. Nature. 2005;433(7027):769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 30.Cekaite L, Furset G, Hovig E, Sioud M. Gene expression analysis in blood cells in response to unmodified and 2′-modified ss siRNAs reveals TLR-dependent and independent effects. doi: 10.1016/j.jmb.2006.09.034. submitted. [DOI] [PubMed] [Google Scholar]
- 31.Krieg AM. CpG motifs: the active ingredient in bacterial extracts? Nature Medicine. 2003;9(7):831–835. doi: 10.1038/nm0703-831. [DOI] [PubMed] [Google Scholar]
- 32.Krieg AM. CpG motifs in bacterial DNA and their immune effects. Annual Review Immunology. 2002;20:709–760. doi: 10.1146/annurev.immunol.20.100301.064842. [DOI] [PubMed] [Google Scholar]