Abstract
Single nucleotide polymorphisms (SNPs) in five genes have been used to identify four major subtypes of wild-type varicella-zoster virus (VZV) A, B, C, and J. Additional SNPs, located in the IE62 major transactivating gene can be used to differentiate the Oka vaccine strain (vOka) from wild-type VZV. Primer-probe sets for the detection of the five polymorphic loci were designed by Applied Biosystems for the ABI 7900HT platform. Probes for each allele were labeled with VIC or 6-carboxyfluorescein fluorogenic markers. Each primer-probe set was validated to establish assay sensitivity and specificity using VZV DNA of predetermined copy number and genotype. Further evaluation was carried out using DNA samples from the vesicle fluid or skin swab of the rash of adult patients with herpes zoster or rashes due to vOka. Assay sensitivity ranged from 10 and 108 copies/ml of VZV DNA (equivalent to 2 to 20 copies per reaction). Statistical analyses showed that for each genotype, a set of two probes clearly differentiated the nucleotide present (allele) at that locus (P < 0.0001). It was possible to determine the genotype of wild-type VZV using one of four SNP assays and also to differentiate wild type from vOka using a single SNP assay. The assay can be used for diagnostic and epidemiological studies of VZV, including the differentiation of vOka from wild-type strains, investigation of breakthrough infections, and varicella outbreaks following immunization.
Varicella-zoster virus (VZV) is the etiological agent of chickenpox in childhood and shingles (herpes zoster) following the reactivation of latent virus dormant in the dorsal root ganglia (2). VZV is genetically stable, and one major serotype has been described previously (2). Using single nucleotide polymorphisms (SNPs) located in five open reading frames (ORFs) of VZV, we have identified four major subtypes of wild-type VZV: A (African/Asian), B, C (European), and J (Japanese) (1). Similar results have been described by other groups of investigators, although nomenclature remains to be agreed (5, 13, 18). A live attenuated VZV vaccine, the vOka strain, is derived from a Japanese wild-type genotype J virus. Since 1995 vOka has been licensed for toddler immunization in the United States (4). The vaccine is effective in healthy and immunocompromised individuals, with 95% protection against severe infection and 75% against all infection (11). Complications are rare, the commonest being rashes which occur in 5% of children and 10% of adults (6, 19).
Establishing the vaccine origin of rashes occurring postvaccination is important for the purposes of surveillance (15). Initial methods exploited the geographical SNP variations between the Japanese vaccine strain and wild-type strains found in the United States (10). However, these methods did not distinguish between the vaccine and some wild-type Japanese strains (16).
An Sma1 and an Nae1 site in the ORF 62 gene are present in all preparations of the vaccine and in all vaccine virus rashes and are absent from all wild-type strains, whatever their geographical origin (7, 12, 14). In order to maximize detection and typing of low-copy-number vaccine virus in vaccine rashes, we report the development of a sensitive and discriminatory real-time PCR that will facilitate the epidemiological surveillance of vaccine rashes.
MATERIALS AND METHODS
VZV strains and DNA extraction.
VZV DNA was extracted (QIAamp DNA MiniKit; QIAGEN, Crawley, United Kingdom) from eight clinical isolates, 79 vesicle fluid samples of patients with shingles, vesicular swabs of the rashes of 14 patients with suspected zoster due to vaccine breakthrough, and commercially available VZV vaccine (MERCK and GSK). The extracted DNA was genotyped as A, B, C, or J using sequence analysis of four regions of the VZV genome, ORFs 1, 21, 50, and 54 (1). The identity of the vaccine strains of VZV was confirmed as vOka by PCR and digestion across the SmaI restriction endonuclease site in ORF 62 (14).
TaqMan SNP assays.
For each of the SNPs in Table 1, primer-probe sets (Table 2) were made using the Applied Biosystems design service (California). Two fluorogenic minor groove binder probes were used for each locus using the dyes 6-carboxyfluorescein (FAM; excitation, 494 nm) and VIC (excitation, 538 nm) which are easily differentiated in the Applied Biosystems Prism 7900HT PCR system. Real-time PCR was performed using 7.5 μl of TaqMan 2× universal master mix (Applied Biosystems, CA), 0.19 μl of primer-probe (470 nM and 100 μM, respectively), 5.31 μl of RNase- and DNase-free water (Sigma), and 2 μl of sample DNA, in a total volume of 15 μl per single tube reaction. Two wells of a 384 plate (Applied Biosystems, CA) were used for each sample or SNP. DNase-free water used as nontemplate control and DNA of known VZV genotype (appropriate to the SNP of interest) used as a positive control were included in each assay run. Assay conditions were 2 min at 50°, 10 min at 95°C, and 40 cycles of 95°C for 15 s and 60°C for 1 min. Initially, the SNP assay was set up using SDS, version 2.1, software (Applied Biosystems, CA) as an absolute quantification assay, but after assay completion the plate was read using the allelic discrimination settings. Postassay analysis was performed using the SDS software.
TABLE 1.
VZV SNP for TaqMan allelic discrimination assay
| Geographic location or source | Genotype | Residue of indicated SNP in ORF:
|
||||
|---|---|---|---|---|---|---|
| 1 | 21 | 37 | 60 | 62 | ||
| SNP 685 | SNP 33725 | SNP 66288 | SNP 101464 | SNP 106262 | ||
| Europe | C | G | T | G | C | T |
| Europe | B | G | C | A | C | T |
| Africa/Asia | A | G | C | G | A | T |
| Japan (pOka) | J (J1) | A | C | G | C | T |
| Japan (European) | J (J2) | A | C | G | C | T |
| Vaccine (vOka) | J | A | C | G | C | C |
TABLE 2.
TaqMan primers and probes evaluated for VZV genotyping
| ORF | SNP | Primer (5′-3′)a | Probeb | Identityc |
|---|---|---|---|---|
| 1 | 685 | CGTCGCCATCTGGAGTACTAC-F | VIC-ACCCAGTACGTTGCAT | Non-J |
| GCGCAACTGAAAATGCAAATGG-R | FAM-ACCCAGTACATTGCAT | GT J | ||
| 21 | 33725 | AGGAACATGGGAGAATGTAAATGC-F | VIC-ACGTTTTTACATGATAATG | GT C |
| TACGTCTCTGACCGTTGTCGTT-R | FAM-ACATGACAATGTTAAAAT | Non-C | ||
| 37 | 66288 | CCTACTGGTTTCGATGAAGAACTCA-F | VIC-AAAACGTGTTTTCTATCATT | Non-B |
| CTTCGTGTGTTGTAGGGTTAACCTT-R | FAM-AAAACGTGTTTTTTATCATT | GT B | ||
| 60 | 101464 | TCTTAACCACGATTCCCGATAGC-F | VIC-ACCCCCGGTATCAA | Non-A |
| GTATATGAGGCGTGGGACTATGC-R | FAM-CACCCCCTGTATCAA | GT A | ||
| 62 | 106262 | CGTACACGTGATACTGAGACAAAGC-F | VIC-ATCCCTGGGCCACC | Wild |
| GCCGGTTGCTGGTGTTG-R | FAM-ATCCCCGGGCCAC | vOka |
F, forward primer; R, reverse primer.
Allelic changes in the probe are shown in boldface.
GT, genotype.
SNP assay evaluation.
DNA from clinical isolates of known genotype and vaccine strains were tested to assess the ability of each SNP assay to differentiate one allele from another, as well as background and cross talk signals. SNP assays that produced good discrimination were further evaluated using the zoster patient samples, representative of the range of quality and quantity of VZV DNA expected in clinical specimens.
Assay sensitivity.
DNA from VZV isolates genotyped as A, B, C, or J by sequencing were quantified using a validated in-house TaqMan viral load assay (8), and serial dilutions from 10 to 108 copies/ml were used to assess the sensitivity of each SNP assay.
Statistical methods.
An unpaired t test (GraphPad QuickCalcs; Prism, California) was used to compare the mean and standard deviation of the reactivity for each probe at each locus.
RESULTS
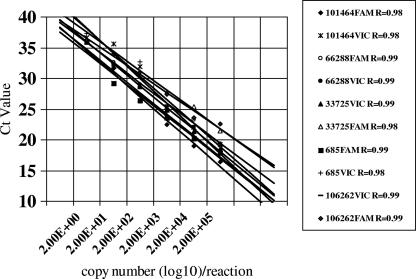
The results of an assessment of the sensitivity of the five probes for each particular genotype at each locus using serially diluted samples of known copy number are shown in Fig. 1. Assay reactivity was directly proportional to copy number (R2 of >0.95) for all probes. All but one probe was capable of detecting between 2 to 20 copies/reaction. One probe (33725FAM non-genotype C) was relatively insensitive and failed to detected DNA in samples with fewer than 2,000 copies/reaction.
FIG. 1.
Efficiency of each TaqMan SNP assay. CT, cycle threshold.
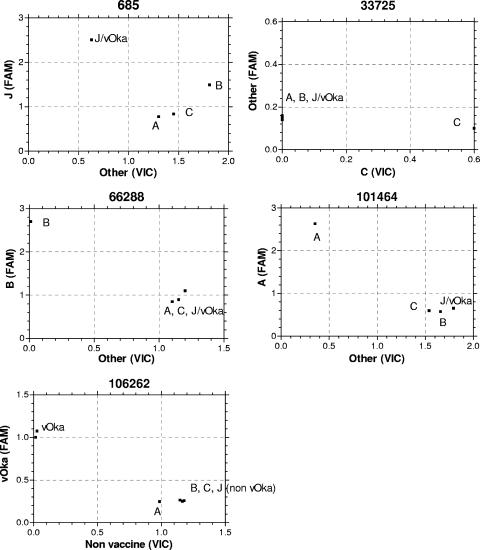
Each SNP assay showed excellent discrimination between genotypes using clinical isolates and vaccine (Fig. 2). The adjusted mean fluorescence (absolute value minus background template control) and standard deviation for each probe at each locus was calculated for all the zoster and vaccine strains tested (Table 3). Statistical comparison of these values (unpaired t test) showed highly significant discrimination (P < 0.0001) between probes at each locus. No false-positive results occurred, although for locus 33725 discrimination of genotype C and non-genotype C viral DNA was based solely on the level of reactivity of the VIC (genotype C specific) probe unless the sample contained high copy numbers of non-genotype C DNA owing to the lower sensitivity of the FAM probe.
FIG. 2.
Allelic discrimination for each genotype at each locus. Results are based on adjusted mean fluorescence of paired wells (absolute value minus background template control).
TABLE 3.
Analysis of the TaqMan genotype discrimination results of DNA samples derived from vesicle fluid taken from zoster patients
| Genotype or source of vesicle fluid sample | Adjusted mean fluorescence (± SD) of the indicated probe and genotype for:a
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| SNP 685
|
SNP 33725
|
SNP 66288
|
SNP 101464
|
SNP 106262
|
||||||
| VIC with non-GT J | FAM with GT J | VIC with GT C | FAM with non-GT C | VIC with non-GT B | FAM with GT B | VIC with non-GT A | FAM with GT A | VIC with WT | FAM with Vac | |
| GT A (n = 7) | 1.05 (0.29) | 0.33 (0.14) | 0.01 (0.01) | 0.48 (0.10) | 0.96 (0.17) | 0.25 (0.09) | 0.32 (0.17) | 2.27 (0.39) | 1.00(0.18) | 0.10 (0.09) |
| GT B (n = 18) | 0.78 (0.45) | 0.24 (0.17) | 0.00 (0.00) | 0.34 (0.12) | 0.03 (0.03) | 2.13 (0.43) | 1.49 (0.08) | 0.36 (0.02) | 0.90 (0.16) | 0.20 (0.12) |
| GT C (n= 37) | 0.98 (0.30) | 0.28 (0.10) | 0.50 (0.22) | 0.14 (0.11) | 0.91 (0.19) | 0.22 (0.06) | 1.65 (0.08) | 0.39 (0.04) | 1.10 (0.25) | 0.20 (0.09) |
| GT J (n = 17) | 0.07 (0.11) | 2.01 (0.64) | 0.01 (0.12) | 0.17 (0.08) | 0.86 (0.17) | 0.32 (0.09) | 1.68 (0.35) | 0.41 (0.13) | 1.20 (0.23) | 0.20 (0.15) |
| vOka (n = 14) | 0.08 (0.05) | 2.00 (0.57) | 0.01 (0.05) | 0.17 (0.05) | 0.80 (0.24) | 0.32 (0.11) | 1.58 (0.24) | 0.32 (0.18) | 0.10 (0.10) | 1.60 (0.35) |
Values are the mean fluorescence of the probe minus the background control. In all cases, a comparison (unpaired t test) of mean (± standard deviation) fluorescence values minus the background of each probe at each locus gave a P value of <0.0001. GT, genotype; WT, wild type; Vac, vaccine.
DISCUSSION
Real-time PCR assays are characterized by a wide dynamic range of quantification and offer a high degree of technical sensitivity and precision (9). No post-PCR steps are required, which avoids the possibility of cross-contamination, and these plate-based assays are easily adaptable for rapid, high-throughput screening. The use of fluorogenic probes for the detection of PCR product during the extension phase of the PCR provides empirical data yielding quantitative results that can be manipulated by statistical procedures to allow accurate quality assurance and control. We used TaqMan real-time allelic discrimination to genotype VZVs using specific primer and probe sets designed around informative polymorphic loci. The resulting assays are sensitive, detecting between 2 and 20 copies per reaction with good genotype specificity. The SNP 22725 assay was able to discriminate genotype C virus but only when used in conjunction with at least one other SNP assay owing to the lack of sensitivity of the non-genotype C (FAM) probe.
The sensitivity of the SNP assays compares with a previously published TaqMan-based assay method for which the limit of detection was 100 copies per reaction for the 107252 SNP of VZV (Oka versus wild type) (3). Five SNPs (SNP 101464 for genotype A, SNP 66288 for genotype B, SNP 33725 for genotype C, SNP 685 for genotype J, and SNP 106262) and the Sma1 restriction endonuclease site in ORF 62 of VZV can be used to distinguish vaccine and wild-type VZV. The sensitivity and discrimination of the assay allow genotyping even when copy number is low, as in the case of vaccine virus rashes.
Two groups have described melting curve assays based on real-time PCR using a LightCycler (Roche Applied Science) for the differentiation of vaccine from wild type using SNP 69649 in ORF 38 (17) and SNP 106262 in ORF 62 (12). The geographical SNPs used in our assay provide additional genotype information that will be especially important for monitoring the origin of varicella outbreaks in countries that have good vaccine coverage and low levels of endogenous varicella. In addition, the method provides proof that recombination of vaccine and wild type is not occurring as vaccine is introduced.
In summary, we have described a highly discriminatory and sensitive method for the detection and genotyping of virus from post-Oka vaccine rashes. The method is simple and reproducible and will be useful for postvaccination surveillance.
Acknowledgments
We acknowledge the help of Fiona Scott in collecting the viral strains and Anne Gershon for the gift of the vaccine rash strains. We also thank Rachael Nugent and Mahmoud Al-Bassam for technical help.
This work was supported by grants from Barts and the London Special Trustees and by Sanofi Pasteur.
REFERENCES
- 1. Barrett-Muir, W., R. A. Nichols, and J. Breuer. 2002. Phylogenetic analysis of varicella-zoster virus: evidence of intercontinental spread of genotypes and recombination. J. Virol. 76:1971-1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Breuer, J. 2004. Varicella zoster virus, p53-83. In A. J. Zuckerman, J. E. Banatvala, J. R. Pattison, P. D. Griffiths, and B. D. Schoub (ed.), Principles and practice of clinical virology, 5th edition. John Wiley, Chichester, United Kingdom.
- 3.Campsall, P. A., N. H. Au, J. S. Prendiville, D. P. Speert, R. Tan, and E. E. Thomas. 2004. Detection and genotyping of varicella-zoster virus by TaqMan allelic discrimination real-time PCR. J. Clin. Microbiol. 42:1409-1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. 1996. Prevention of varicella: recommendations of the Advisory Committee on Immunization Practices (ACIP). Morb. Mortal. Wkly. Rep. 45:1-25. [PubMed] [Google Scholar]
- 5.Faga, B., W. Maury, D. A. Bruckner, and C. Grosse. 2001. Identification and mapping of single nucleotide polymorphisms in the varicella-zoster virus genome. Virology 280:1-6. [DOI] [PubMed] [Google Scholar]
- 6.Gershon, A. A. 1995. Varicella-zoster virus: Prospects for control. Adv. Ped. Infect. Dis. 10:93-123. [PubMed] [Google Scholar]
- 7.Gomi, Y., T. Magana, M. Takahashi, and K. Yamanishi. 2000. Oka varicella vaccine is distinguishable from its parental virus in DNA sequence of open reading frame 62 and its transactivation activity. J. Med. Virol. 61:497-503. [DOI] [PubMed] [Google Scholar]
- 8.Hawrami, K., and J. Breuer. 1999. Development of a fluorogenic polymerase chain reaction assay (TaqMan) for the detection and quantitation of VZV. J. Virol. Methods 79:33-40. [DOI] [PubMed] [Google Scholar]
- 9.Klein, D. 2002. Quantification using real-time PCR technology: applications and limitations. Trends Mol. Med. 8:257-260. [DOI] [PubMed] [Google Scholar]
- 10.LaRussa, P., O. Lungu, I. Hardy, A. Gershon, S. P. Steinberg, and S. Silverstein. 1992. Restriction fragment length polymorphism of polymerase chain reaction products from vaccine and wild-type varicella-zoster virus isolates. J. Virol. 66:1016-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee, B. R., S. L. Feaver, C. A. Miller, C. W. Hedberg, and K. R. Ehresmann. 1996. An elementary school outbreak of varicella attributed to vaccine failure: policy implications. J. Infect. Dis. 190:477-483. [DOI] [PubMed] [Google Scholar]
- 12.Loparev, V. N., T. Agra, P. R. Krause, M. Takeaway, and D. S. Schmidt. 2000. Improved identification and differentiation of varicella-zoster virus (VZV) wild-type strains and an attenuated varicella vaccine strain using a VZV open reading frame 62-based PCR. J. Clin. Microbiol. 38:3156-3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Loparev, V. N., A. Gonzalez, M. Deleon-Carnes, G. Tipples, H. Fickenscher, E. G. Torfason, and D. S. Schmid. 2004. Global identification of three major genotypes of varicella-zoster virus: longitudinal clustering and strategies for genotyping. J. Virol. 78:8349-8358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quinlivan, M. L., A. A. Gershon, S. P. Steinberg, and J. Breuer. 2005. An evaluation of single nucleotide polymorphisms used to differentiate vaccine and wild type strains of varicella-zoster virus. J. Med. Virol. 75:174-180. [DOI] [PubMed] [Google Scholar]
- 15.Sharrar, R. G., P. LaRussa, S. A. Galea, S. P. Steinberg, A. R. Sweet, R. M. Keatley, M. E. Wells, W. P. Stephenson, and A. A. Gershon. 2000. The postmarketing safety profile of varicella vaccine. Vaccine 19:916-923. [DOI] [PubMed] [Google Scholar]
- 16.Takada, M., T. Suzutani, I. Yoshida, M. Matoba, and M. Azuma. 1995. Identification of varicella-zoster virus strains by PCR analysis of three repeat elements and a PstI-site-less region. J. Clin. Microbiol. 33:658-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tipples, G. A., D. Safronetz, and M. Gray. 2003. Real-time PCR assay for the detection of varicella-zoster virus DNA and differentiation of vaccine, wild-type and control strains. J. Virol. Methods. 113:113-116. [DOI] [PubMed] [Google Scholar]
- 18.Wagenaar, T., V. T. K. Chow, C. Buranathai, P. Thawatsupha, and C. Grose. 2003. The out of Africa model of varicella-zoster virus evolution: single nucleotide polymorphisms and private alleles distinguish Asian clades from European/North American clades. Vaccine 21:1072-1081. [DOI] [PubMed] [Google Scholar]
- 19.Weibel, R. E., B. J. Neff, B. J. Kuter, H. A. Guess, C. A. Rothenberger, A. J. Fitzgerald, K. A. Connor, A. A. McLean, M. R. Hilleman, and E. B. Buynak. 1984. Live attenuated varicella virus vaccine: efficacy trial in healthy children. N. Engl. J. Med. 310:1409-1411. [DOI] [PubMed] [Google Scholar]