Abstract
Sixty-two isolates of Enterobacteriaceae (35 Escherichia coli and 27 Klebsiella pneumoniae isolates) producing CTX-M-type β-lactamases were collected between March 2000 and June 2003 in different wards of Charles Nicolle Hospital in Tunis (Tunisia). Sequencing identified the blaCTX-M-15 determinant in 55 isolates and blaCTX-M-16 in 7 isolates. The CTX-M-15-producing strains were isolated in several wards and consisted mainly of two successive clonal groups of E. coli and a major clonal group of K. pneumoniae. The second clonal group of E. coli belonged to phylogenetic group B2 and harbored more virulence factors than the first clonal group. Among the 22 transconjugants or electroporants obtained with selected E. coli and K. pneumoniae CTX-M-15-producing strains, a predominant plasmid restriction pattern was obtained with 17 isolates. The four CTX-M-16-producing strains of E. coli yielded the same pulsed-field gel electrophoresis (PFGE) pattern, while the three CTX-M-16-producing strains of K. pneumoniae yielded two different PFGE patterns. All of the CTX-M-16-producing isolates were recovered in the pediatric ward and had the same plasmid restriction pattern.
First reported in Argentina and France (3, 7), the CTX-M-type enzymes were subsequently found in several European countries as well as in Asia and North America (3). Recent studies have shown the presence of these enzymes in African countries (2, 13, 14, 24, 31, 37, 39). In Tunisia, the first identified CTX-M-producing strain (CTX-M-3), Salmonella enterica serovar Wien, was recovered in Tunis in 2001 (1). Later, a strain (Salmonella enterica serovar Livingstone) producing CTX-M-27 caused a nosocomial outbreak in a neonatal ward in Sousse in 2002 (4).
One of these enzymes, CTX-M-15, is now found worldwide, mainly in Escherichia coli isolates recovered in hospitals and in the community and responsible for outbreaks in France, the United Kingdom, Sweden, and Canada (11, 12, 25, 28, 29, 30, 33, 40). Molecular characterization of plasmids encoding CTX-M-15 from E. coli strains involved in outbreaks in different countries showed that they additionally carried other antibiotic resistance genes, such as blaOXA-1, blaTEM-1, tetA, aac(6′)-Ib, and aac(3)-II, and sometimes a class 1 integron (6, 23, 27).
Phylogenetic studies of E. coli isolates producing CTX-M enzymes indicate that most belong to phylogenetic group D, except that CTX-M-15 producers often belong to group B2 (8, 25, 28, 32).
Since the first isolation of an extended-spectrum β-lactamase (ESBL)-producing Klebsiella pneumoniae strain at Charles Nicolle Hospital, Tunis, in 1984, a growing variety of Enterobacteriaceae and ESBL enzymes have been detected; 60% of the isolates were Klebsiella spp. and 12.5% were E. coli (5). In March 2000, an ESBL-producing clinical isolate of E. coli exhibiting an unusual resistance phenotype (a higher level of resistance to cefotaxime than to ceftazidime) was recovered. The aims of this retrospective study of all ESBL-producing E. coli and K. pneumoniae isolates recovered in Charles Nicolle Hospital from March 2000 to June 2003 were (i) to detect and identify CTX-M enzymes and associated resistance genes, (ii) to conduct an epidemiological investigation using chromosome and plasmid fingerprint analyses, and (iii) to determine the phylogenetic group and virulence factors of E. coli isolates.
MATERIALS AND METHODS
Bacterial strains.
All ESBL-producing strains of E. coli and K. pneumoniae recovered at Charles Nicolle Hospital between March 2000 and June 2003 were collected and identified with API 20E systems (bioMérieux, Marcy l'Etoile, France). E. coli J53-2 (pro met Rifr) and E. coli DH10B (Invitrogen SARL, Cergy-Pontoise, France) were used for conjugation and electroporation, respectively.
The nosocomial character of the infections was defined when the first CTX-M isolate was recovered from clinical samples obtained 3 days or more after admission.
Study of antibiotic consumption.
Broad-spectrum cephalosporin (cefotaxime, ceftazidime, and ceftriaxone) use data were collected from the pharmaceutical department of the Charles Nicolle Hospital between 2000 and 2003. Data were expressed in grams of active substance and then in the number of defined daily doses (DDD) according to the Anatomic Therapeutic Chemical classification from WHO Index 2006 (http://www.whocc.no/atcddd/). The number of hospitalization days was used to calculate the penetration index (ratio number of DDD to 1,000 hospitalization days).
Antibiotic susceptibility testing.
Antibiotic susceptibility was tested with the agar disk diffusion method according to CLSI (formerly NCCLS) guidelines (10). ESBLs were detected using a standard double-disk synergy test (17). The CTX-M phenotype of ESBL producers screened in this study was based on a similar or smaller inhibition zone with cefotaxime than with ceftazidime. The MICs of the following antibiotics were determined by a dilution method in Mueller-Hinton agar (Bio-Rad, Marnes-la-Coquette, France): ticarcillin, cefotaxime, and ceftazidime alone and combined with clavulanic acid (2 mg/liter), cefoxitin, and cefepime. E. coli ATCC 25922 and Pseudomonas aeruginosa ATCC 27853 were used as control strains.
Characterization of β-lactamases and associated resistance genes.
All ESBL-producing strains with a CTX-M phenotype were subjected to CTX-M consensus PCR (groups M-1, M-2, and M-9) using primers MA1 and MA2 as previously described (Table 1) (36). All PCR-positive strains were subjected to genomic DNA extraction with a QIAGEN mini kit (QIAGEN). Genes encoding TEM and CTX-M-1-type, CTX-M-2-type, and CTX-M-9 β-lactamases were amplified by PCR and sequenced as previously described (11). The nucleotide sequences and deduced protein sequences were analyzed with the BLAST and Clustal W programs (multiple-sequence alignment, pair-wise comparisons of sequences, and dendrograms).
TABLE 1.
Primers for phylogenetic studies, virulence factors, and resistance genes used for PCR assays
| Target | Sequence (5′-3′) | Primer name | Reference |
|---|---|---|---|
| chuA | GACGAACCAACGGTCAGGAT | chuA.1 | 9 |
| TGCCGCCAGTACCAAAGACA | chuA.2 | ||
| yjaA | TGAAGTGTCAGGAGACGCTG | yjaA.1 | 9 |
| ATGGAGAATGCGTTCCTCAAC | yjaA.2 | ||
| TspE4.C2 | GAGTAATGTCGGGGCATTCA | TspE4C2.1 | 9 |
| CGCGCCAACAAAGTATTACG | TspE4C2.2 | ||
| papG allele II | GGGATGAGCGGGCCTTTGAT | AlleleII-f | 18 |
| CGGGCCCCCAAGTAACTCG | AlleleII-r | ||
| papG allele III | GGCCTGCAATGGATTTACCTGG | AlleleIII-f | 18 |
| CCACCAAATGACCATGCCAGAC | AlleleIII-r | ||
| sfa/foc | CTCCGGAGAACTGGGTGCATCTTAC | sfa1 | 18 |
| CGGAGGAGTAATTACAAACCTGGCA | sfa2 | ||
| afa/draBC | GGCAGAGGGCCGGCAACAGGC | Afa f | 18 |
| CCCGTAACGCGCCAGCATCTC | Afa r | ||
| fimH | TGCAGAACGGATAAGCCGTGG | FimH f | 18 |
| GCAGTCACCTGCCCTCCGGTA | FimH r | ||
| hlyA | AACAAGGATAAGCACTGTTCTGGCT | hly f | 18 |
| ACCATATAAGCGGTCATTCCCGTCA | hly r | ||
| cnf1 | AAGATGGAGTTTCCTATGCAGGAG | cnf1 | 18 |
| CATTCAGAGTCCTGCCCTCATTATT | cnf2 | ||
| fyuA | TGATTAACCCCGCGACGGGAA | FyuA f′ | 18 |
| CGCAGTAGGCACGATGTTGTA | FyuA r | ||
| iutA | GGCTGGACATCATGGGAACTGG | AerJ f | 18 |
| CGTCGGGAACGGGTAGAATCG | AerJ r | ||
| kpsMT II | GCGCATTTGCTGATACTGTTG | kpsII f | 18 |
| CATCCAGACGATAAGCATGAGCA | kpsII r | ||
| traT | GGTGTGGTGCGATGAGCACAG | TraT f | 18 |
| CACGGTTCAGCCATCCCTGAG | TraT r | ||
| sat | ACTGGCGGACTCATGCTGT | Sat 1 | 35 |
| AACCCTGTAAGAAGACTGAGC | Sat 2 | ||
| iha | CTGGCGGAGGCTCTGAGATCA | IHA f | 19 |
| TCCTTAAGCTCCCGCGGCTGA | IHA r | ||
| iroN | AAGTCAAAGCAGGGGTTGCCCG | IRONEC-F | 19 |
| GACGCCGACATTAAGACGCAG | IRONEC-R | ||
| tetA | GTTTCGGGTTCGGGATGGTC | tetA up | This study |
| GCAGGCAGAGCAAGTAGAGG | tetA low | ||
| blaOXA-1 | TATCAACTTCGCTATTTTTTTA | OXA-1 up | This study |
| TTTAGTGTGTTTAGAATGGTGA | OXA-1 low | ||
| aac(6′)-Ib | ATGACTGAGCATGACCTT | AAC6′-Ib up | 28 |
| GAAGGGTTAGGCATCACT | AAC6′-Ib low | ||
| aac(3)-II | CAATAACGGAGGCAATTCG | AAC3-II up | 28 |
| GATTATCATTGTCGACGG | AAC3-II low | ||
| sul1 | CGGCGTGGGCTACCTGAACG | Sul 1-F | 26 |
| GCCGATCGCGTGAAGTTCCG | Sul 1-B | ||
| sul2 | GCGCTCAAGGCAGATGGCATT | Sul 2-F | 26 |
| GCGTTTGATACCGGCACCCGT | Sul 2-B |
Other antibiotic resistance genes, often found associated with blaCTX-M-15, blaOXA-1, aac(3)-II, aac(6′)-Ib, and tetA as well as sul1 and sul2 genes, were screened by PCR using the primers listed in Table 1.
Fingerprinting analysis.
Repetitive extragenic palindromic sequence PCR was performed with primers rep-1R and rep-2T for all of the E. coli isolates as previously described (11). Enterobacterial repetitive intergenic consensus sequence PCR was performed with primer ERIC-2 for all of the K. pneumoniae isolates as previously described (11). If isolates had similar patterns, they were subjected to pulsed-field gel electrophoresis (PFGE). PFGE was performed using a GenePath system (Bio-Rad, Marnes-la-Coquette, France) with genomic DNA digested with XbaI (Ozyme, Saint Quentin en Yvelines, France) at 14°C and 6 V/cm for 20 h, with pulse times of 5.3 to 49.9 s. Clonal relationships based on PFGE patterns were interpreted using the criteria established by Tenover et al. (38).
β-Lactam resistance transfer assays and plasmid fingerprint analysis.
Conjugation was carried out in Trypticase soy broth (Bio-Rad), with E. coli J53-2 as the recipient. Mating broths were incubated at 37°C for 18 h. Transconjugants were selected on Mueller-Hinton agar plates containing rifampin (250 mg/liter) and cefotaxime (2.5 mg/liter).
For transformation, plasmid DNA extracted from donors with a QIAGEN plasmid midi kit (QIAGEN, Courtaboeuf, France) were used to transform E. coli DH10B cells by electroporation following the manufacturer's instructions (Bio-Rad). Transformants were incubated for 1.5 h at 37°C and then mated on Drigalski agar (Bio-Rad) supplemented with 2.5 mg/liter cefotaxime.
For plasmid fingerprinting, plasmid DNA was extracted from the transconjugants and transformants with the QIAGEN plasmid midi kit and digested with EcoRI or HpaI. The resulting fragments were subjected to electrophoresis on a 0.8% agarose gel.
Phylotyping and virulence genotyping of E. coli.
The phylogenetic group of the E. coli isolates was determined by the PCR method developed by Clermont et al. (9), using a combination of three DNA markers (chuA, yjaA, and TspE4.C2). All isolates were screened for 14 virulence factors often found in extraintestinal pathogenic E. coli (ExPEC), namely, fimH, sfa/foc, papG allele II and allele III, afa, hlyA, cnf1, fyuA, iutA, kpsM II, traT, sat, iroN, and iha, using single or multiplex PCR assays (18, 19, 35) and the primers listed in Table 1.
Three archetypal ExPEC strains, CFT073, ECOR66, and EC7372, producing various virulence factors were used as positive controls (16, 20, 22).
Statistical analysis.
Factorial analysis of correspondence (FAC) was used to describe associations among clinical and bacterial data (15). FAC uses a covariance matrix based on χ2 distances. The computation determines a plane defined by two principal axes of the analysis; the first axis, F1, accounts for most of the variance, and the second axis, F2, orthogonal to F1, accounts for the largest part of the variance not accounted for by F1. The F1/F2 plane allowed the positioning of the variables according to their coordinates on each of these factors. This positioning describes the relation between the variables. When two variables are closely related on the plane, they are strongly associated. On the contrary, when they are distantly related, they are not associated. FAC was conducted with SPAD.N software (Cisia, Saint Mandé, France) from two two-way tables. A first table was constructed for the E. coli strains and had 35 rows (one for each E. coli strain) and 31 columns corresponding to the following 31 variables: the 4 years of isolation (2000 to 2003), the four wards (surgery, urology, general medicine, and pediatrics), the three types of infection (urine, blood, and others), the three phylogenetic groups (A, B2, and D), the seven molecular profiles (E1 to E7), the CTX-M type (15 or 16), and the nine discriminating virulence factors (Table 1). A second table was constructed for the K. pneumoniae strains. It had 27 rows (one for each K. pneumoniae strain) and 18 columns corresponding to the following 18 variables: the five wards (surgery, general medicine, urology, pediatrics, and intensive care unit [ICU]), the three types of infection (urine, blood, and others), the nine molecular profiles (K1 to K9), and the CTX-M type (15 or 16). In each column, each strain was coded as a binary variable (present = 1, absent = 0).
RESULTS
Clinical isolates.
The first isolate found to produce a CTX-M-type β-lactamase was an E. coli strain recovered from the urine of a surgical patient (8 March 2000). Two months later, other isolates harboring the CTX-M PCR consensus sequence were found in the general medicine ward and subsequently in other wards. By June 2003, 35 E. coli and 27 K. pneumoniae strains recovered from different patients and positive for CTX-M consensus PCR were detected. CTX-M-producing E. coli and K. pneumoniae strains represented 8% and 2.7% of ESBL producers, respectively, belonging to the same species in 2000, compared to 22% and 1.25% in 2001, 28.5% and 8% in 2002, and 27% and 30% in 2003 (January to June).
Their ward distribution was as follows: 28% general medicine, 28% surgery, 21% urology, 15% pediatrics, and 8% ICU. They were associated with urinary tract infections (52%), bacteremia (22%), wound infections (14%), lower respiratory tract infections (7%), and catheter colonization (5%) (Table 2). All of these infections were nosocomial.
TABLE 2.
Epidemiological data, resistance determinants, and plasmid backbones of the different clones of E. coliand K. pneumoniae producing CTX-M-15 or CTX-M-16 β-lactamase at the Charles Nicolle Hospital in Tunis
| Clone (n) | CTX-M allele | Period of isolation (mo/yr) | Ward(s) | No. of clones with indicated type of infection:
|
No. of clones witha:
|
Plasmid backbone(s)b | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Urine | Blood | Pus | Respiratory | Catheter | blaTEM-1 | blaOXA-1 | aac(3)-II | aac(6′)-Ib | tetA | sul1 | sul2 | Cipr | |||||
| E1 (10) | 15 | 03/00-01/02 | Surgery, medicine 1, urology | 9 | 0 | 1 | 0 | 0 | 10 | 8 | 9 | 10 | 10 | 0 | 10 | 10 | P1 (2/2) |
| E2 (3) | 16 | 07/01-12/01 | Pediatric | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | P3 (3/3) |
| E3 (3) | 15 | 09/01 | Medicine 1 | 3 | 0 | 0 | 0 | 0 | 3 | 0 | 3 | 3 | 0 | 0 | 2 | 3 | P1 (3/3) |
| E4 (1) | 16 | 03/02 | Pediatric | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | P3 (1/1) |
| E5 (1) | 15 | 10/02 | Surgery | 0 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | P1 (1/1) |
| E6 (16) | 15 | 12/02-06/03 | Surgery, medicine 1 and 2, urology | 4 | 5 | 3 | 2 | 2 | 15 | 15 | 15 | 15 | 0 | 14 | 0 | 16 | P1 (2/2), P5 (1/1) |
| E7 (1) | 15 | 12/02 | Medicine 1 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | P1 (1/1) |
| K1 (2) | 16 | 05/00-06/00 | Pediatric | 2 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 2 | 2 | 2 | 0 | 0 | P3 (2/2) |
| K2 (1) | 16 | 08/01 | Pediatric | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 0 | 0 | P3 (1/1) |
| K3 (1) | 15 | 05/02 | Medicine 3 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | P6 (1/1) |
| K4 (17) | 15 | 10/02-06/03 | Surgery, medicine 1, urology, ICU | 4 | 8 | 4 | 2 | 1 | 14 | 17 | 17 | 17 | 0 | 1 | 17 | 17 | P1 (5/5) |
| K5 (1) | 15 | 12/02 | Urology | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | P1 (1/1) |
| K6 (2) | 15 | 12/02-03/03 | Surgery, medicine 1 | 0 | 2 | 0 | 1 | 1 | 0 | 2 | 2 | 2 | 2 | 2 | 0 | 2 | P1 (2/2) |
| K7 (1) | 15 | 03/03 | Pedriatric | 0 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | P7 (1/1) |
| K8 (1) | 15 | 03/03 | Surgery | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | P2 (1/1) |
| K9 (1) | 15 | 05/03 | ICU | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | P4 (1/1) |
These resistances were detected by PCR as described in Materials and Methods, except for the resistance to ciprofloxacin (Cipr), which was detected only by the disk diffusion method.
The plasmid backbone has been analyzed only in transconjugants or electroporants. Values in parentheses are numbers of plasmid analyses/numbers of transconjugants or electroporants.
During the same period, the total number of ESBL-producing E. coli and K. pneumoniae strains were, respectively, 50 and 73 in 2000, 54 and 81 in 2001, 35 and 113 in 2002, and 37 and 54 from January to June 2003.
Broad-spectrum cephalosporin consumption.
The evolution of the consumption of broad-spectrum cephalosporins, evaluated by the penetration index (number of DDD per 1,000 hospitalization days), showed a global increase of 27% between 2000 and 2003 (from 34.5 to 43.9). The penetration index of cefotaxime increased from 29.8 to 34.9, and that of ceftazidime increased from 4.7 to 8.9. The consumption of ceftriaxone was very small (less than 1%).
β-Lactam susceptibility.
All of the strains were highly resistant to ticarcillin (MIC > 1,024 mg/liter). The cefotaxime MICs ranged from 256 to >2,048 mg/liter (MIC90, 1,024 mg/liter), and those of ceftazidime ranged from 64 to >2,048 mg/liter (MIC90, 128 mg/liter). Thirty E. coli (83%) and seven K. pneumoniae (22%) isolates had a higher level of resistance to cefotaxime than to ceftazidime. However, 5 E. coli and 20 K. pneumoniae isolates showed similar levels of resistance to cefotaxime and ceftazidime (256 to >2,048 mg/liter). Clavulanic acid partially or completely restored the activities of cefotaxime (0.5 to 64 mg/liter) and ceftazidime (1 to 128 mg/liter). All of the isolates were resistant to cefepime and aztreonam (16 to >128 mg/liter) but remained susceptible to imipenem.
Characterization of β-lactamase-encoding genes and other resistance genes.
The results of PCR and sequence analysis are summarized in Table 2. CTX-M-encoding genes were detected in all of the isolates and in their transconjugants/electroporants. The deduced amino acid sequences corresponded to CTX-M-15 in 55 isolates (31 E. coli and 24 K. pneumoniae isolates) and CTX-M-16 in 7 isolates (4 E. coli and 3 K. pneumoniae isolates). The blaTEM-1 gene was identified in 30 E. coli and 19 K. pneumoniae isolates; all but one (an E. coli isolate, Ec7) of the seven isolates carrying blaCTX-M-16 were negative for blaTEM. The blaOXA-1 gene was detected in 52 isolates (25 E. coli and 27 K. pneumoniae isolates). The aminoglycoside resistance genes aac(3)-II and aac(6′)-Ib were found in 55 and 58 isolates, respectively. tetA was found in 11 E. coli isolates and 7 K. pneumoniae isolates. sul2 was detected in 13 E. coli isolates and 18 K. pneumoniae isolates, whereas sul1 was detected in 15 E. coli and 6 K. pneumoniae isolates. Only two K. pneumoniae isolates produced both sul1 and sul2. Thirty-one E. coli isolates and 21 K. pneumoniae isolates were resistant to ciprofloxacin. None of the pediatric isolates was resistant to ciprofloxacin.
Epidemiological results.
The 35 E. coli isolates yielded seven distinct repetitive extragenic palindromic sequence PCR patterns, and the 27 K. pneumoniae isolates yielded nine different enterobacterial repetitive intergenic consensus sequence PCR patterns (Table 2 and data not shown). Isolates with similar patterns were subjected to PFGE and were classified as clonally related (Fig. 1) (38). Two major clones producing CTX-M-15 were observed among E. coli isolates and were designated clone E1 (10 isolates, 28.6%) and clone E6 (16 isolates, 45.7%). Two minor clones were observed (E2 and E3), each comprising three isolates; one of them, E3, which produced CTX-M-16, was recovered only in the pediatric ward. Clone E1 predominated in 2000 to 2001, and clone E6 predominated in 2002 to 2003 (Fig. 1). Seventeen K. pneumoniae strains producing CTX-M-15 had the same profile, designated K4 (63%); all were isolated in 2002 to 2003 (Table 2 and Fig. 2). The three K. pneumoniae strains producing CTX-M-16 (clones K1 and K2) were recovered, all in the pediatric ward (Table 2). tetA was associated with the E. coli clonal strain E1. sul2 was also detected in clonal strain E1 and in all K. pneumoniae isolates belonging to clone K4, whereas sul1 was detected only in E. coli clonal group E6 (Table 2).
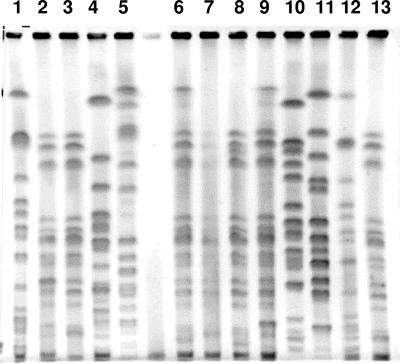
FIG. 1.
PFGE patterns of K. pneumoniae strains carrying blaCTX-M genes. Lanes 1 to 13, Kp19, Kp20, Kp65, Kp18, Kp15, Kp61, Kp67, Kp62, Kp69, Kp13, Kp14, Kp56, and Kp30 corresponding to molecular types K1, K4, K4, K9, K6, K4, K4, K4, K4, K3, K7, K1, and K4, respectively.
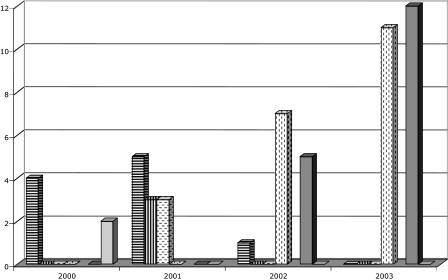
FIG. 2.
Number of patients infected by clonal strains of E. coli and K. pneumoniae producing CTX-M-type β-lactamases and isolated at Charles Nicolle Hospital between March 2000 and June 2003. Bar with horizontal hatching, E. coli clone E1 (CTX-M-15); bar with vertical hatching, E. coli clone E2 (CTX-M-16); bar with horizontal dashes, E. coli clone E3 (CTX-M-15); bar with vertical dashes, E. coli clone E6 (CTX-M-15); light-gray bar, K. pneumoniae clone K1 (CTX-M-16); dark-gray bar, K. pneumoniae clone K4 (CTX-M-15).
Transferability of CTX-M determinants and plasmid fingerprint analysis.
Fourteen E. coli and 15 K. pneumoniae isolates were selected according to their blaCTX-M gene, their fingerprint, and their antimicrobial resistance pattern. Four and eight E. coli isolates with the E1 and E6 fingerprints, respectively, were selected. All 29 strains were tested for conjugal transfer of cefotaxime resistance, and 19 (9 E. coli and 10 K. pneumoniae isolates) were positive. Electroporation of plasmid DNA from the other 10 strains into E. coli DH10B successfully transferred cefotaxime resistance. Large plasmids were found in K. pneumoniae and E. coli transconjugants or electroporants. EcoRI restriction of plasmids from transconjugants and electroporants of the 22 strains producing CTX-M-15-type enzymes yielded six different patterns, with a major plasmid restriction pattern (P1) in 17 strains (data not shown). This P1 plasmid was found in all E. coli clones and in three K. pneumoniae clones, including epidemic clone K4. Plasmids isolated from the six electroporants and one transconjugant of the CTX-M-16-producing strains yielded similar restriction patterns, named P3, after digestion with HpaI (Fig. 3).
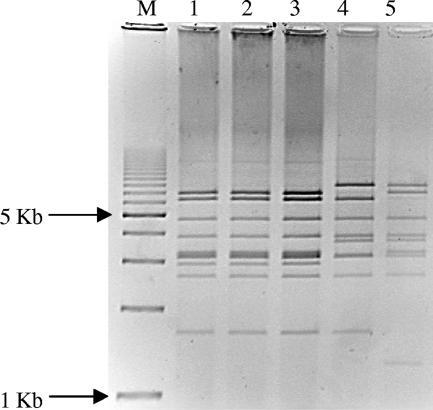
FIG. 3.
HpaI-digested plasmid profiles of transconjugants or electroporants producing CTX-M-16. Lanes 1 to 4, electroporants of Ec3, Ec38, Kp12, and Kp19, respectively; lane 5, transconjugant Ec7; lane M, molecular weight marker, 1-kb DNA ladder (Bio-Rad).
Phylogenetic analysis and virulence genotyping of E. coli isolates.
The results of phylogenetic studies and virulence factor (Vf) determination are reported in Table 3. The two largest clones, E1 and E6, belong to phylogenetic groups A and B2, respectively. The two minor clones, E2 and E3, belong to groups B2 and D, respectively. Clone E2 (group B2) expressed virulence factors often encountered in ExPEC strains (fyuA, papG allele III, hlyA, cnf1, kpsMT II, iha, and sat), whereas clone E6, which belongs to the same phylogenetic group, expressed fewer Vfs and did not produce cytotoxin or hemolysin. Curiously, major clone E6 exhibited the same Vf profile as minor clone E3 belonging to group D.
TABLE 3.
Phylogenetic groups and virulence factors of E. coli producing CTX-M β-lactamase at the Charles Nicolle Hospital in Tunisa
| Clone (n) | CTX-M allele | Phylogenetic group | Detection of indicated virulence factor:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| fimH | fyuA | iutA | papG allele | hlyA | cnf1 | KpsMTII | iha | sat | |||
| E1 (10) | 15 | A | − | − | − | − | − | − | − | − | − |
| E2 (3) | 16 | B2 | + | + | − | III | + | + | + | − | − |
| E3 (3) | 15 | D | + | + | + | − | − | − | + | + | + |
| E4 (1) | 16 | D | + | + | − | − | − | − | + | − | − |
| E5 (1) | 15 | A | − | + | − | − | − | − | − | − | − |
| E6 (16) | 15 | B2 | + | + | + | − | − | − | + | + | + |
| E7 (1) | 15 | A | + | − | − | − | − | − | − | − | − |
All strains were negative for papG allele II, sfa/foc, iroN, and afa genes, and all strains were positive for the tra gene.
Statistical analysis.
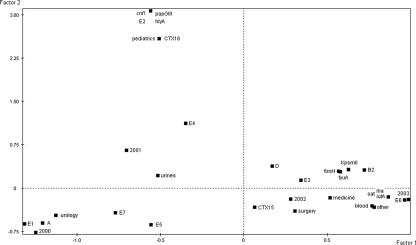
To explore the associations among bacterial characteristics and epidemiological characteristics, FACs were done. A first FAC was conducted on the E. coli data. The F1/F2 plane accounted for 55% of the total variance (Fig. 4). The planes obtained from the other factors of the FAC accounted for lower percentages of the variance than F1 and F2 and did not significantly improve the data interpretation. The projections of the variables on the F1/F2 plane distinguished three groups of variables: (i) CTX-M-16 type, pediatrics, Vfs cnf1, hlyA, and papG allele III, and clone E2, which were projected on the positive values of the second factor, F2, are closely related on the plane, and thus are strongly associated; (ii) phylogenetic group A, clones E1, E5, and E7, urology, and urinary tract infection, which were distinguished by the negative values of F1 and are associated; and (iii) phylogenetic group B2, the Vfs fyuA, fimH, KpsMT II, iha, iutA, and sat, the type of infection (blood and other), the surgery and medicine wards, and clones E3 and E6, which were distinguished by the positive values of F1 and are associated. Moreover, the projections of the variable “year of isolation” followed the increasing values of F1 from 2000 and 2001, projected on its negative values, to 2002 and 2003, projected on its positive values.
FIG. 4.
Projections of the bacterial and clinical variables of the 35 E. coli strains on F1/F2 planes computed by factorial analysis of correspondence. A, B2, and D, phylogenetic groups A, B2, and D; E1 to E7, E. coli molecular types 1 to 7; CTX15 and CTX16, β-lactamase types CTX-M-15 and CTX-M-16; urine, blood, and other, urinary tract infection, bacteremia, and other infections; 2000 to 2003, years of isolation. The nine VFs are named as in Table 1.
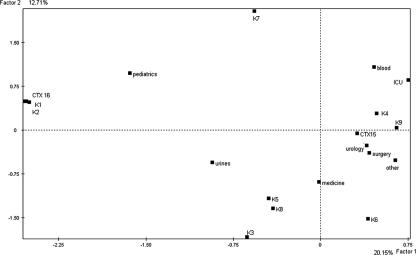
A second FAC was conducted on the K. pneumoniae data. The projections of the variables on plane F1/F2, which accounted for 32.86% of the total variance, distinguished three groups of variables: (i) CTX-M-16 type, clones K1 and K2, and pediatrics, which were projected on the negative values of the first factor, F1; (ii) clones K3, K5, and K8 and the origin of infection (urine), which were projected on the negative values of F1 and F2; and (iii) clone K4, the origin of infection (blood), and the ward (ICU), which were distinguished by the positive values of the two factors (Fig. 5).
FIG. 5.
Projections of the bacterial and clinical variables of the 27 K. pneumoniae strains on F1/F2 planes computed by factorial analysis of correspondence. K1 to K9, K. pneumoniae molecular types 1 to 9; CTX15 and CTX16, β-lactamase types CTX-M-15 and CTX-M-16; urine, blood, and other, urinary tract infection, bacteremia, and miscellaneous infections; 2000 to 2003, years of isolation.
DISCUSSION
At the Charles Nicolle Hospital in Tunis, 62 enterobacterial strains producing CTX-M β-lactamase were collected between March 2000 and June 2003. The isolation rate increased during this period, from 6 isolates in 2000 to 13 in 2001, 18 in 2002, and 26 between January and June 2003. All of our isolates produce CTX-M-15 or CTX-M-16, both of which harbored the substitution of Asp-240→Gly which increases the activity against ceftazidime (5, 6, 7, 14, 34). During this period, the increase of consumption of cefotaxime and ceftazidime could have contributed to the emergence of ESBLs and particularly to these CTX-M-type enzymes. This is the first report of CTX-M-15-type β-lactamases in Tunisia and the first report of CTX-M-16-producing Enterobacteriaceae in an African country. All of these strains were multiresistant, producing other β-lactamases (e.g., TEM-1 and OXA-1) and aminoglycoside-modifying enzymes. They were resistant to ciprofloxacin, except for the strains recovered from the pediatric ward. Multiresistance has often been described for ESBL (and particularly CTX-M)-producing clinical isolates (3, 6, 7, 11, 23, 27, 28).
K. pneumoniae and E. coli isolates producing CTX-M-16 were isolated only in the pediatric ward. Three of the four E. coli (clone E2) and two of the three K. pneumoniae (clone K1) isolates producing CTX-M-16 were epidemiologically related, suggesting probable clonal spread. This clonal grouping was illustrated on the two FACs (Fig. 4 and 5). For E. coli isolates, clone E2 was closely related to three particular Vfs (cnf1, hlyA, and papG allele III) and to the pediatric ward by their projections on the positive values of the second factor, F2 (Fig. 4). For K. pneumoniae isolates, clones K1 and K2 and pediatric origin were grouped according to the negative values of the first factor, F1 (Fig. 5). Furthermore, all of the CTX-M-16-producing strains were found to carry very similar HpaI restriction P3 plasmid patterns, pointing to the existence of one common backbone for the CTX-M-16-encoding plasmids. Epidemiological studies showed that the dissemination of CTX-M-16 could be the consequence of both strain spreading and plasmid diffusion.
CTX-M-15-producing E. coli and K. pneumonaie isolates were recovered in all of the other wards. Twenty-six of the 31 CTX-M-15-producing E. coli isolates belong to two major clones. The first clone, E1 (phylogenetic group A), was predominant in the years 2000 to 2001 and was essentially recovered from urine (9/10). The second clone, E6 (phylogenetic group B2), appeared in December 2002, and 7 of the 16 isolates were recovered from blood and the lower respiratory tract (Table 2). In the FAC performed on E. coli data, the first axis opposed the variables (phylogenetic group A, clone E1, urology ward, and year of isolation [2000]) projected on its negative values with the variables (phylogenetic group B2, clone E6, blood and other sites of infection, several Vfs [iha, iutA, fuyA, kpsM II, and fimH], and year of isolation [2003]) projected on its positive values (Fig. 4). This well-established opposition between two levels of intrinsic virulence among ExPEC strains (8) illustrated the fact that CTX-M-15 resistance had been transferred from a less virulent E. coli clone (E1) to a more virulent E. coli clone (E6). In the same period (October 2002), we observed the emergence of the predominant and multiresistant clone K4 of K. pneumoniae. Similarly to that of E. coli, the FAC indicated the close relatedness between K. pneumoniae clone K4 and the variables blood and ICU (Fig. 5), illustrating the association between clinical virulence and antibiotic resistance.
As suggested by plasmid fingerprinting, the same plasmid (same backbone) encoding CTX-M-15 could have been transferred first from clone E1 to the K. pneumoniae clone K4 and then from clone K4 to clone E6 (which emerged 2 months later). This shift (from clone E1 to clone E6) could be explained by the virulence genotype. Clone E1 is a typical commensal strain with fewer than two virulence factors, unlike clone E6 (18-20). Interestingly, clone E6 showed lower intrinsic virulence than archetypal ExPEC strains and particularly the absence of toxins (18-20). But the virulence traits of this clone could be involved in colonization, infection, and persistence in humans (nonspecific adhesin, siderophore, and resistance to the serum and to phagocytosis) (18, 19). These factors could simultaneously explain the spread and the persistence of this “successful” E. coli clone. This recalls the results of two French studies that investigated nosocomial outbreaks in long-term-care facilities due to E. coli isolates producing CTX-M-15 β-lactamase and similar virulence traits (25, 28). Previous studies on the relation between antibiotic resistance and virulence in human isolates of E. coli suggested that antibiotic-resistant clones (except those resistant to fluoroquinolones) were less virulent than susceptible strains (21). In our study, we observed the success of a multiresistant and virulent clone of E. coli.
In summary, clonal spread of strains, multiresistance, several virulence factors, plasmid transfer, and broad-spectrum cephalosporin consumption have contributed to the nosocomial dissemination of the CTX-M-encoding genes among E. coli and K. pneumoniae strains in our hospital.
Acknowledgments
This study received financial support from the Ministry of Science and Technology, Tunisia, and grants from Faculté de Médecine Pierre et Marie Curie (site Saint-Antoine), Université Paris VI, and the European Community, contract LSHM-CT 2003-503335.
The control strains for virulence factors were kindly provided by Erick Denamur and Olivier Clermont. We thank Dominique Decré for critical reading of the manuscript.
Footnotes
Published ahead of print on 6 September 2006.
REFERENCES
- 1.Armand-Lefevre, L., V. Leflon-Guibout, J. Bredin, F. Barguellil, A. Amor, J. M. Pagès, and M.-H. Nicolas-Chanoine. 2003. Imipenem resistance in Salmonella enterica serovar Wien related to porin loss and CMY-4 β-lactamase production. Antimicrob. Agents Chemother. 47:1165-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blomberg, B., R. Jureen, K. P. Manji, B. S. Tamim, D. S. Mwakagile, W. K. Urassa, M. Fataki, V. Msangi, M. G. Tellevik, S. Y. Maselle, and N. Langeland. 2005. High rate of fatal cases of pediatric septicemia caused by gram-negative bacteria with extended-spectrum beta-lactamases in Dar es Salaam, Tanzania. J. Clin. Microbiol. 43:745-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonnet, R. 2004. Growing group of extended-spectrum β-lactamases: the CTX-M enzymes. Antimicrob. Agents Chemother. 48:1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bouallègue-Godet, O., Y. Ben Salem, L. Fabre, M. Demartin, P. A. Grimont, R. Mzoughi, and F.-X. Weill. 2005. Nosocomial outbreak caused by Salmonella enterica serotype Livingstone producing CTX-M-27 extended-spectrum β-lactamase in a neonatal unit in Sousse, Tunisia. J. Clin. Microbiol. 43:1037-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boutiba-Ben Boubaker, I., R. Ghozzi, H. Ben Abdallah, K. Mamlouk, A. Kamoun, and S. Ben Redjeb. 2004. Evolution of acquired-resistance to third-generation cephalosporins in Enterobacteriaceae in a Tunisian hospital 1993-2001. Clin. Microbiol. Infect. 10:665-667. [DOI] [PubMed] [Google Scholar]
- 6.Boyd, D. A., S. Tyler, S. Christianson, A. McGeer, M. P. Muller, B. M. Willey, E. Bryce, M. Gardam, P. Nordmann, and M. R. Mulvey. 2004. Complete nucleotide sequence of a 92-kilobase plasmid harboring the CTX-M-15 extended-spectrum β-lactamase involved in an outbreak in long-term-care facilities in Toronto, Canada. Antimicrob. Agents Chemother. 48:3758-3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bradford, P. A. 2001. Extended-spectrum β-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin. Microbiol. Rev. 14:933-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Branger, C., O. Zamfir, S. Geoffroy, G. Laurans, G. Arlet, H. V. Thien, S. Gouriou, B. Picard, and E. Denamur. 2005. Genetic background of Escherichia coli and extended-spectrum β-lactamase type. Emerg. Infect. Dis. 11:54-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clermont, O., S. Bonacorsi, and E. Bingen. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66:4555-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clinical and Laboratory Standards Institute. 2005. Performance standards for antimicrobial susceptibility testing; 15th informational supplement. Approved standard M2-A8 and M7-A6. Clinical and Laboratory Standards Institute, Wayne, Pa.
- 11.Eckert, C., V. Gautier, M. Saladin-Allard, N. Hidri, C. Verdet, Z. Ould-Hocine, G. Barnaud, F. Delisle, A. Rossier, T. Lambert, A. Philippon, and G. Arlet. 2004. Dissemination of CTX-M-type β-lactamases among clinical isolates of Enterobacteriaceae in Paris, France. Antimicrob. Agents Chemother. 48:1249-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fang, H., C. Lundberg, B. Olsson-Liljequist, G. Hedin, E. Lindback, A. Rosenberg, and J. Struwe. 2004. Molecular epidemiological analysis of Escherichia coli isolates producing extended-spectrum β-lactamases for identification of nosocomial outbreaks in Stockholm, Sweden. J. Clin. Microbiol. 42:5917-5920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frank, T., G. Arlet, V. Gautier, A. Talarmin, and R. Bercion. 2006. Extended-spectrum β-lactamase-producing Enterobacteriaceae, Central African Republic. Emerg. Infect. Dis. 12:863-865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gangoue-Pieboji, J., V. Miriagou, S. Vourli, E. Tzelepi, P. Ngassam, and L. S. Tzouvelekis. 2005. Emergence of CTX-M-15-producing enterobacteria in Cameroon and characterization of a blaCTX-M-15-carrying element. Antimicrob. Agents Chemother. 49:441-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greenacre, M. 1992. Correspondence analysis in medical research. Stat. Methods Med. Res. 1:97-117. [DOI] [PubMed] [Google Scholar]
- 16.Guignot, J., J. Breard, M.-F. Bernet-Camard, I. Peiffer, B. J. Nowicki, A. L. Servin, and A.-B. Blanc-Potard. 2000. Pyelonephritogenic diffusely adhering Escherichia coli EC7372 harboring Dr-II adhesin carries classical uropathogenic virulence genes and promotes cell lysis and apoptosis in polarized epithelial caco-2/TC7 cells. Infect. Immun. 68:7018-7027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jarlier, V., M. H. Nicolas, G. Fournier, and A. Philippon. 1988. Extended broad-spectrum beta-lactamases conferring transferable resistance to newer beta-lactam agents in Enterobacteriaceae: hospital prevalence and susceptibility patterns. Rev. Infect. Dis. 10:867-878. [DOI] [PubMed] [Google Scholar]
- 18.Johnson, J. R., and A. L. Stell. 2000. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J. Infect. Dis. 181:261-272. [DOI] [PubMed] [Google Scholar]
- 19.Johnson, J. R., T. A. Russo, P. I. Tarr, U. Carlino, S. S. Bilge, J. C. Vary, Jr., and A. L. Stell. 2000. Molecular epidemiological and phylogenetic associations of two novel putative virulence genes, iha and iroNE. coli, among Escherichia coli isolates from patients with urosepsis. Infect. Immun. 68:3040-3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson, J. R., P. Delavari, M. Kuskowski, and A. L. Stell. 2001. Phylogenetic distribution of extraintestinal virulence-associated traits in Escherichia coli. J. Infect. Dis. 183:78-88. [DOI] [PubMed] [Google Scholar]
- 21.Johnson, J. R., M. A. Kuskowski, K. Owens, A. Gajewski, and P. L. Winokur. 2003. Phylogenetic origin and virulence genotype in relation to resistance to fluoroquinolones and/or extended-spectrum cephalosporins and cephamycins among Escherichia coli isolates from animals and humans. J. Infect. Dis. 188:759-768. [DOI] [PubMed] [Google Scholar]
- 22.Kao, J. S., D. M. Stucker, J. W. Warren, and H. L. Mobley. 1997. Pathogenicity island sequences of pyelonephritogenic Escherichia coli CFT073 are associated with virulent uropathogenic strains. Infect. Immun. 65:2812-2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karisik, E., M. J. Ellington, R. Pike, R. E. Warren, D. M. Livermore, and N. Woodford. 2006. Molecular characterization of plasmids encoding CTX-M-15 β-lactamase from Escherichia coli strains in the United Kingdom. J. Antimicrob. Chemother. 58:665-668. [DOI] [PubMed] [Google Scholar]
- 24.Kariuki, S., J. E. Corkill, G. Revathi, R. Musoke, and C. A. Hart. 2001. Molecular characterization of a novel plasmid-encoded cefotaximase (CTX-M-12) found in clinical Klebsiella pneumoniae isolates from Kenya. Antimicrob. Agents Chemother. 45:2141-2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kassis-Chikhani, N., S. Vimont, K. Asselat, C. Trivalle, B. Minassian, C. Sengelin, V. Gautier, D. Mathieu, E. Dussaix, and G. Arlet. 2004. CTX-M beta-lactamase-producing Escherichia coli in long-term care facilities, France. Emerg. Infect. Dis. 10:1697-1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kerrn, M. B., T. Klemmensen, N. Frimodt-Moller, and F. Espersen. 2002. Susceptibility of Danish Escherichia coli strains isolated from urinary tract infections and bacteraemia, and distribution of sul genes conferring sulphonamide resistance. J. Antimicrob. Chemother. 50:513-516. [DOI] [PubMed] [Google Scholar]
- 27.Lavollay, M., K. Mamlouk, T. Frank, A. Akpabie, B. Burghoffer, S. Ben Redjeb, R. Bercion, V. Gautier, and G. Arlet. 2006. Clonal dissemination of a CTX-M-15 β-lactamase-producing Escherichia coli strain in the Paris area, Tunis, and Bangui. Antimicrob. Agents Chemother. 50:2433-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leflon-Guibout, V., C. Jurand, S. Bonacorsi, F. Espinasse, M. C. Guelfi, F. Duportail, B. Heym, E. Bingen, and M. H. Nicolas-Chanoine. 2004. Emergence and spread of three clonally related virulent isolates of CTX-M-15-producing Escherichia coli with variable resistance to aminoglycosides and tetracycline in a French geriatric hospital. Antimicrob. Agents Chemother. 48:3736-3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muller, M., A. McGeer, B. M. Willey, D. Reynolds, R. Malanczyj, M. Silverman, M. A. Green, and M. Culf. 2002. Outbreaks of multi-drug resistant Escherichia coli in long-term care facilities in the Durham, York, and Toronto regions of Ontario, 2000-2002. Can. Commun. Dis. Rep. 28:113-118. [PubMed] [Google Scholar]
- 30.Mulvey, M. R., E. Bryce, D. Boyd, M. Ofner-Agostini, S. Christianson, A. E. Simor, S. Paton, and The Canadian Hospital Epidemiology Committee of The Canadian Nosocomial Infection Surveillance Program, Health Canada. 2004. Ambler class A extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella spp. in Canadian hospitals. Antimicrob. Agents Chemother. 48:1204-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Naas, T., A. Lezzar, C. Bentchouala, F. Smati, J. M. Scheftel, H. Monteil, and P. Nordmann. 2005. Multidrug-resistant Salmonella enterica serotype Senftenberg isolates producing CTX-M β-lactamases from Constantine, Algeria. J. Antimicrob. Chemother. 56:439-440. [DOI] [PubMed] [Google Scholar]
- 32.Pitout, J. D. D., K. B. Laupland, D. L. Church, M. L. Menard, and J. R. Johnson. 2005. Virulence factors of Escherichia coli that produce CTX-M-type extended-spectrum β-lactamases. Antimicrob. Agents Chemother. 49:4667-4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pitout, J. D. D., D. B. Gregson, D. L. Church, S. Elsayed, and K. B. Laupland. 2005. Community-wide outbreaks of clonally related CTX-M-14 β-lactamase-producing Escherichia coli strains in the Calgary Health Region. J. Clin. Microbiol. 43:2844-2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Poirel, L., M. Gniadkowski, and P. Nordmann. 2002. Biochemical analysis of the ceftazidime-hydrolysing extended-spectrum beta-lactamase CTX-M-15 and of its structurally related beta-lactamase CTX-M-3. J. Antimicrob. Chemother. 50:1031-1034. [DOI] [PubMed] [Google Scholar]
- 35.Ruiz, J., K. Simon, J. P. Horcajada, M. Velasco, M. Barranco, G. Roig, A. Moreno-Martinez, J. A. Martinez, T. Jimenez de Anta, J. Mensa, and J. Vila. 2002. Differences in virulence factors among clinical isolates of Escherichia coli causing cystitis and pyelonephritis in women and prostatitis in men. J. Clin. Microbiol. 40:4445-4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saladin, M., V. T. Cao, T. Lambert, J. L. Donay, J. L. Herrmann, Z. Ould-Hocine, C. Verdet, F. Delisle, A. Philippon, and G. Arlet. 2002. Diversity of CTX-M beta-lactamases and their promoter regions from Enterobacteriaceae isolated in three Parisian hospitals. FEMS Microbiol. Lett. 209:161-168. [DOI] [PubMed] [Google Scholar]
- 37.Soge, O. O., A. M. Queenan, K. K Ojo, B. A. Adeniyi, and M. C. Roberts. 2006. CTX-M-15 extended-spectrum β-lactamase from Nigerian Klebsiella pneumoniae. J. Antimicrob. Chemother. 57:24-30. [DOI] [PubMed] [Google Scholar]
- 38.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weill, F. X., J. D. Perrier-Gros-Claude, M. Demartin, S. Coignard, and P. Grimont. 2004. Characterization of extended-spectrum-β-lactamase (CTX-M-15)-producing strains of Salmonella enterica isolated in France and Senegal. FEMS Microbiol. Lett. 238:353-358. [DOI] [PubMed] [Google Scholar]
- 40.Woodford, N., M. E. Ward, M. E. Kaufmann, J. Turton, E. J. Fagan, D. James, A. P. Johnson, R. Pike, M. Warner, T. Cheasty, A. Pearson, S. Harry, J. B. Leach, A. Loughrey, J. A. Lowes, R. E. Warren, and D. M. Livermore. 2004. Community and hospital spread of Escherichia coli producing CTX-M extended-spectrum β-lactamases in the UK. J. Antimicrob. Chemother. 54:735-743. [DOI] [PubMed] [Google Scholar]