Abstract
The gastrointestinal microbiota of preterm infants in a neonatal intensive care unit differs from that of term infants. In particular, the colonization of preterm infants by bifidobacteria is delayed. A double-blind, placebo-controlled, randomized clinical study was performed on 69 preterm infants to investigate the role of Bifidobacterium lactis Bb12 supplementation in modifying the gut microbiota. Both culture-dependent and culture-independent approaches were used to study the gut microbiota. Bifidobacterial numbers, determined by fluorescence in situ hybridization, were significantly higher in the probiotic than in the placebo group (log10 values per g of fecal wet weight: probiotic, 8.18 + 0.54 [standard error of the mean]; placebo, 4.82 + 0.51; P < 0.001). A similar trend for bifidobacterial numbers was also obtained with the culture-dependent method. The infants supplemented with Bb12 also had lower viable counts of Enterobacteriaceae (log10 values of CFU per g of fecal wet weight: probiotic, 7.80 + 0.34; placebo, 9.03 + 0.35; P = 0.015) and Clostridium spp. (probiotic, 4.89 + 0.30; placebo, 5.99 + 0.32; P = 0.014) than the infants in the placebo group. Supplementation of B. lactis Bb12 did not reduce the colonization by antibiotic-resistant organisms in the study population. However, the probiotic supplementation increased the cell counts of bifidobacteria and reduced the cell counts of enterobacteria and clostridia.
The microbiota of the gastrointestinal tract of preterm infants in a neonatal intensive care unit differs considerably from that of normal-term infants (31). Use of antibiotics for extended periods, lack of full breastfeeding, need for artificial respiration, and repeated invasive interventions may contribute to this situation (30). Increased permeability of the intestinal epithelial lining may lead to bacterial translocation and, subsequently, sepsis. The neonatal intensive care unit environment predisposes these infants to nosocomial infections. Some clinical trials on preterm infants have demonstrated the safe use of probiotics as a preventive measure to create a commensal microbiota that suppresses the growth of pathogens (1, 6, 20). In these trials, different strains of probiotics and different regimens of administration have been used. In breastfed infants, bifidobacteria are a major component of the intestinal microbiota (35). Therefore, various strains of bifidobacteria have been used in intervention trials, including B. lactis Bb12. However, those trials have only investigated the possible effects of Bb12 on full-term infants and toddlers, and the possible effects on preterm infants have not been studied.
This study aimed to investigate with both culture-dependent and culture-independent methods whether the supplementation of preterm infants with Bifidobacterium lactis Bb12 results in the modification of their gut microbiota in such a way that the growth of potentially harmful bacteria is suppressed.
MATERIALS AND METHODS
Study design.
A double-blind, placebo-controlled, randomized clinical trial was performed on 69 preterm infants born with a gestational age of <37 weeks in the Ernst von Bergmann hospital, Potsdam, Germany, between August 2003 and June 2005. The infants were randomized into the placebo or the probiotic group with the help of the Randoma software version 4.3 (HaSoTec, Rostock, Germany) on the basis of birth weight, gestational age at birth, gender, arterial umbilical cord pH, and Apgar score at 5 min. The exclusion criteria were chromosomal aberration, human immunodeficiency virus infection in the mother, hydrops fetalis, and inborn malformation of the gastrointestinal tract. The formula-based placebo (Nestlé FM 2000B) and verum (Nestlé FM 2000A) preparations were supplied by Nestlé, Konolfingen, Switzerland. The verum contained 2 × 109 cells of Bifidobacterium lactis Bb12 per gram of powder. The concentration of Bb12 in 1 ml solution of verum in water was 4 × 108 with an osmolarity of 428 mosmol liter−1. The verum group received 1.6 × 109 cells on day 1 to 3 and 4.8 × 109 cells from day 4 onward. The administration of the study preparation started on the first day after birth and continued for 21 days. The study ended at the 35th day after birth or when the infant was discharged from the hospital, if earlier. Anamnestic and the routine clinical data were collected for all infants and their mothers.
Fecal samples were collected as fresh as possible during the study period. The poststudy sample was available only from a few infants because most of the infants had been discharged from the hospital by that time, and it was difficult to obtain a fresh sample for analysis. The samples from each infant were analyzed weekly for bacterial cell counts by both fluorescence in situ hybridization (FISH) and plating on the culture media. In many instances, the samples were not available on the sampling day, and therefore, the number of samples included for statistics differ for the various parameters investigated as indicated.
The study was approved by the local ethical committee, and informed consent was obtained from the parents before infants were enrolled in the study. For ethical reasons, only noninvasive methods were used.
Culturing of different bacterial groups on selective and nonselective media.
Stool samples were collected weekly for 3 weeks. The numbers of infants included in the analysis were as follows: probiotic group, week 1 = 18, week 2 = 24, week 3 = 18; placebo group, week 1 = 21, week 2 = 27, week 3 = 19. Samples were processed within 3 h of collection as follows: 100 mg of the sample was homogenized and 10-fold serial dilutions were made in Soerensen buffer (25 mM KH2PO4, 33 mM Na2HPO4 × 12H2O, 0.04% thioglycolic acid, pH 6.8). The dilutions (in duplicates, 100 μl each) were plated on different nonselective and selective media (Table 1) in an anoxic workstation (MK3; DW Scientific, Shipley, United Kingdom) which contained a gas atmosphere of N2-CO2-H2 (80:10:10, by vol) for anaerobes and was under the laminar flow hood for aerobes. Plates for anaerobic groups of bacteria were incubated in the anaerobic workstation, which was maintained at 37°C. Plates for aerobic and facultative aerobes were incubated aerobically at 37°C. The colonies were counted after 48 h, and the results are given as CFU per gram of fecal wet weight.
TABLE 1.
Microbiological media used for cultivation of different bacterial groups
| Culture medium (source) | Bacterial group(s) or use |
|---|---|
| Eosin methylene blue agar (Fluka, Germany) | Enterobacteriaceae |
| KF Enterococcus agar (Oxoid, Germany) | Enterococcus and Streptococcus spp. |
| Mannitol salt agar (Oxoid, Germany) | Staphylococcus spp. |
| Columbia agar with 5% sheep blood (Bio Mérieux, Germany) | Anaerobic bacteria |
| Albicans ID2 (Bio Mérieux, Germany) | Candida albicans |
| Clostridium difficile agar (Bio Mérieux, Germany) | Clostridia |
| Bacteroides bile esculin agar (BD Diagnostics Systems, Germany) | Bacteroides |
| Bifidus selective agar (Fluka, Germany) | Bifidobacterium spp. |
| Standard 1 nutrient agar (Merck, Germany) | Aerobic bacteria |
| Mueller-Hinton agar (Roth, Germany) | Antibiotic resistance tests |
Antibiotic susceptibility testing by the disk diffusion method.
In the neonatal intensive care unit of the Ernst von Bergmann hospital, the antibiotics commonly used for the treatment of infections in preterm infants are combinations of vancomycin (15 mg kg of body weight−1 day−1) and amikacin (15 mg kg−1 day−1) as well as of piperacillin (150 mg kg−1 day−1) and cefotaxime (200 mg kg−1 day−1). Imipenem (60 mg kg−1 day−1) was used occasionally. One antibiotic of each combination, namely vancomycin, piperacillin, or imipenem was used for the antibiotic resistance tests. Penicillin was also included in the analysis, as a large number of strains resistant to this antibiotic are commonly found in the nosocomial environment (36). The probiotic and placebo groups contained 24 infants each. Samples were collected in the second and third weeks. Samples from infants with and without antibiotic treatment were included in the analysis.
To check for antibiotic resistance, morphologically different colonies from the aerobic plates were picked and restreaked on Columbia blood agar until pure cultures were obtained. The purity was checked by gram staining. The strain to be tested for antibiotic resistance was grown overnight on Columbia blood agar. The optical density was adjusted to a 0.5 MacFarland's standard by diluting the colonies in 0.8% NaCl. The inoculum was spread on Mueller-Hinton agar (each plate contained 25 ml of media) with a cotton swab. The plates were dried, and the antibiotic disks (BioMérieux, Nürtingen, Germany) were placed on the agar surface with sterile forceps. They were then incubated overnight at 37°C, and the zone of inhibition around each disk was measured. The zone diameters were interpreted for resistance with the help of guidelines given by DIN58940-3 Bbl 1 (8). Imipenem (10 μg), piperacillin (75 μg), vancomycin (30 μg), and penicillin (10 U/E) disks were used for the test. Resistant strains were identified to the species level with the Vitek system (BioMérieux, Nürtingen, Germany) according to the manufacturer's instructions.
Enumeration of bacterial cells by FISH.
Fecal samples were collected weekly and processed within 3 h. The numbers of infants included in the analysis were as follows: probiotic, week 1 = 26, week 2 = 28, week 3 = 29; placebo, week 1 = 26, week 2 = 28, week 3 = 27. Fresh feces were diluted 10-fold with in phosphate-buffered saline (150 mM NaCl, 10 mM Na2HPO4, 20 mM NaH2PO4, pH 7.4) and fixed as described by Thiel and Blaut (33), except that the centrifugal force used for washing during fixation was 16,000 × g instead of 8,000 × g.
Microscopic slides were prepared according to the method described by Thiel and Blaut (33). The probes used in this study are listed in Table 2. An equimolar mixture of the probes EUB338, EUB785, EUB927, EUB1055, and EUB1088, referred to as Eubmix, was used to count the total bacterial cells. All probes were commercially synthesized (Thermo Hybaid, Ulm, Germany) and 5′ labeled with Cy3. They were used at a concentration of 10 pmol/μl, except for Bac303, which was used at a fourfold-higher concentration. The enumeration of the bacterial cells labeled with Clostridium lituseburense (Clit135) was done on cells both with and without lysozyme treatment, as different cell morphologies were observed in both cases. Overnight hybridization was done for all probes, except for Bac303, which was hybridized for 1 h.
TABLE 2.
Oligonucleotide used in the study
| Probe | Sequence (5′-3′) | OPD codea | Reference | Temp (°C) | Bacterial group(s) targeted |
|---|---|---|---|---|---|
| EUB338 | GCTGCCTCCCGTAGGAGT | S-D-Bact-0338-a-A-18 | 3 | 46 | Total bacteria |
| EUB785 | CTACCAGGGTATCTAATCC | S-D-Bact-0785-a-A-19 | 19 | 46 | Total bacteria |
| EUB927 | ACCGCTTGTGCGGGCCC | S-D-Bact-0927-a-A-17 | 10 | 46 | Total bacteria |
| EUB1055 | CACGAGCTGACGACAGCCAT | S-D-Bact-1055-a-A-20 | 19 | 46 | Total bacteria |
| EUB1088 | GCTCGTTGCGGGACTTAACC | S-D-Bact-1088-a-A-20 | 19 | 46 | Total bacteria |
| Erec482 | GCTTCTTAGTCARGTACCG | S-*-Erec-0482-a-A-19 | 9 | 50 | Eubacterium rectale cluster |
| Bac303 | CCAATGTGGGGGACCTT | S-*-Bacto-0303-a-A-17 | 22 | 46 | Bacteroides and Prevotella |
| Bif164 | CATCCGGCATTACCACCC | S-G-Bif-0164-a-A-18 | 17 | 46 | Bifidobacterium spp. |
| Lab158 | GGTATTAGCAYCTGTTTCCA | S-G-Lab-0158-a-A-20 | 12 | 46 | Lactobacillus and Enterococcus spp. |
| Veil223 | AGACGCAATCCCCTCCTT | S-*-Veil-0223-a-A-18 | 11 | 49 | Veillonellae |
| Str493 | GTTAGCCGTCCCTTTCTGG | S-*-Strc-0493-a-A-19 | 9 | 50 | Streptococcus and Lactococcus spp. |
| Ec1531 | CACCGTAGTGCCTCGTCATCA | L-S-Eco-1531-a-A-21 | 27 | 46 | Enterobacteriaceae |
| Sta697 | TCCTCCATATCTCTGCGC | S-*-Sta-0697-a-A-18 | 34 | 58 | Staphylococcus spp. |
| Chis150 | TTATGCGGTATTAATCTYCCTTT | S-*-Chis-0150-a-A-23 | 9 | 49 | Clostridium histolyticum group |
| Clit135 | GTTATCCGTGTGTACAGGG | S-*-Clit-0135-a-A-19 | 9 | 51 | Clostridium lituseburense group |
OPD code, Oligonucleotide Probe Database code (2).
Samples were analyzed with an Axioplan2 imaging microscope (Carl Zeiss, Oberkochen, Germany).
Statistics.
Statistical analyses were performed with the statistical software package SPSS 11.5 (SPSS, Inc., Chicago, Illinois). Fisher's exact test, linear-by-linear association, Levene's test, and the Mann-Whitney test were used to check the efficiency of the randomization program used. Colony counts and numbers of bacteria estimated with FISH were expressed as log10 counts and are given as geometric means in the graphs. A general linear model was used for analysis with subject as random factor; antibiotic therapy (yes or no), treatment (probiotic or placebo), and week of sample collection as fixed factors; and week × treatment and antibiotic therapy × treatment as interaction terms. The differences were considered significant at a P value of <0.05 for all analyses. The probiotic and placebo groups were further split to check the influence of antibiotic therapy.
RESULTS
Study group characteristics.
The probiotic and placebo groups contained 37 and 32 preterm infants, respectively. The characteristics of the study population are shown in Table 3. The probiotic and placebo groups were similar with regard to all parameters chosen for randomization (data not shown). Forty-six infants (26 probiotic, 20 placebo) received antibiotic therapy during the study period of 3 weeks. All of these infants were subjected to standard antibiotic therapy, which included cefotaxime and piperacillin in the first 3 days. After the third day, vancomycin and amikacin were given until the condition improved. Imipenem was given only to three infants. Feeding of the probiotic or placebo was started on the first or second day after birth. No adverse effect was observed in any of the infants supplemented with Bifidobacterium lactis Bb12. FISH analysis and enumeration of viable counts were done on 65 (33 probiotic, 32 placebo) and 58 (26 probiotic, 32 placebo) infants, respectively, due to unavailability of fresh samples from all infants at the time of collection.
TABLE 3.
Study group characteristicsa
| Parameter | Result (%) for group:
|
|
|---|---|---|
| Probiotic | Placebo | |
| Gender (female) | 62.2 | 65.6 |
| Mode of delivery | ||
| Spontaneous | 13.5 | 9.4 |
| Cesarean | 86.5 | 90.6 |
| Birth type | ||
| Single | 56.8 | 53.1 |
| Twins | 37.8 | 34.4 |
| Triplets | 5.4 | 12.5 |
The probiotic and placebo groups contained 37 and 32 infants, respectively.
Culturing on different selective and nonselective media.
The infants in the placebo group had higher numbers of total bacteria than those in the probiotic group (aerobic, P = 0.02; anaerobic, P = 0.008) (Fig. 1). Conversely, the counts of bifidobacteria were significantly higher in the probiotic group than in the placebo group (P = 0.002).
FIG. 1.
Effect of probiotic and placebo supplementation on CFU of bacterial groups tested (probiotic, n = 26; placebo, n = 32). The empty and filled bars represent the placebo and the probiotic groups, respectively. The error bars represent the standard errors of the means. P values are as follows: 0.005, total bacteria; 0.02, total aerobic bacteria; 0.008, total anaerobic bacteria; 0.002, bifidobacteria; 0.015, Enterobacteriaceae; 0.014, clostridia. *, P < 0.05; **, P < 0.01.
The placebo group had higher numbers of enterobacteria and clostridia than the probiotic group (P = 0.015 and 0.014, respectively). There were no significant differences between the probiotic and placebo groups in the numbers of Staphylococcus spp., Streptococcus spp., Bacteroides spp., and Candida spp. (Fig. 1). Candida spp. were found in only 4 infants (2 isolates from each probiotic and placebo group) and in very low numbers. Colonies with a morphology characteristic of Clostridium difficile (nonhemolytic, gray, slightly raised, filamentous edge on blood agar) were found in almost 70% of the samples analyzed. This preliminary identification was further supported by automatic identification by the Vitek system. Probiotic supplementation had no influence on the occurrence of C. difficile, and it was equally present in both study groups. It was isolated from first week onwards, although the occurrence rate increased with time (week 1, 50%; week 2, 61%; week 3, 73%).
At different time points, significant differences in cell counts were found between the placebo and probiotic groups for total aerobic bacteria, total anaerobic bacteria, bifidobacteria, streptococci, and clostridia in the first week, bifidobacteria in the second week, and Enterobacteriaceae in the third week (data not shown).
Antibiotic resistance testing of the strains isolated from the plates.
Antibiotic resistance tests were performed on samples from 24 infants of each group. Bacteria resistant to either one of the four tested antibiotics were isolated from all but three infants. The most commonly isolated antibiotic-resistant strains included Escherichia coli (48% of the infants), Staphylococcus epidermidis (46%), Staphylococcus aureus (19%), Enterococcus faecium (29%), Klebsiella pneumoniae (17%), Staphylococcus scuiri (15%), and Enterococcus faecalis (15%). Other antibiotic-resistant isolates were identified as Enterobacter cloacae (8%), Staphylococcus simulans (4%), Staphylococcus capitis (2%), Klebsiella ornithinolytica (2%), Staphylococcus xylosus (2%), Streptococcus agalactiae (2%), Streptococcus equines (2%), Pasteurella multocida (2%), Pasteurella haemolytica (2%), Streptococcus bovis (2%) and Staphylococcus warneri (2%). The probiotic supplementation did not influence the presence of these antibiotic-resistant strains. Most of the antibiotic-resistant strains identified as S. sciuri, S. simulans, and K. pneumoniae were isolated from infants born close to each other in time and present in the intensive care unit at the same time.
Resistance to penicillin was most widespread, with 42% of the infants harboring one or more completely resistant strains. Of these, 23% and 19% of infants in the probiotic and placebo groups, respectively, had penicillin-resistant organisms. Organisms resistant to imipenem, piperacillin, and vancomycin were isolated from 32%, 28%, and 35% of the infants, respectively. There were no significant differences between the probiotic and placebo groups with regard to the number of infants colonized with antibiotic-resistant strains.
Fluorescence in situ hybridization.
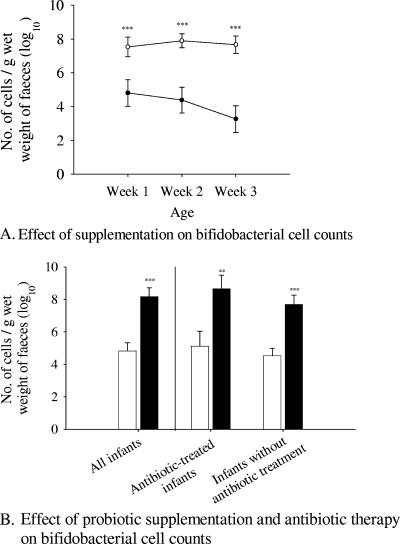
There were no significant differences between the probiotic and placebo groups in the total number of bacteria, as detected by hybridization with Eubmix. Numbers of bifidobacteria (Bif164) were higher in the probiotic group over all three weeks (P = 0.003, <0.001, and <0.001, respectively) (Fig. 2A). Considering all time points, the infants in the probiotic group had higher numbers of bifidobacteria (P < 0.001) than those in the placebo group. This effect was independent of the antibiotic treatment (P = 0.006) (Fig. 2B). There were no differences between the antibiotic-treated and nontreated infants when they were compared independent of the probiotic effect (P = 0.282).
FIG. 2.
(A) Weekly changes in the numbers of bifidobacteria targeted with Bif164 during probiotic and placebo supplementation: week 1, probiotic n = 18, placebo n = 21; week 2, probiotic n = 24, placebo n = 27; week 3, probiotic n = 18, placebo n = 19. Open and closed circles refer to the probiotic and placebo groups, respectively. The error bars represent the standard errors of the means. The P value is <0.001 in all cases. (B) Effect of probiotic supplementation and antibiotic therapy on bifidobacteria targeted by Bif164. The effect is shown for all infants (probiotic n = 32, placebo n = 33), antibiotic-treated infants (probiotic n = 23, placebo n = 20), and infants without antibiotic treatment (probiotic n = 10, placebo n = 12). The empty and filled bars represent the placebo and the probiotic groups, respectively. The error bars represent the standard errors of the means. P values are as follows: <0.001, all infants; 0.006, infants with antibiotic treatment; <0.001, infants without antibiotic treatment. **, P < 0.01; ***, P < 0.001.
Considering the overall effect of probiotic supplementation, there were no significant differences between the probiotic and placebo groups in terms of bacteria targeted with Ec1531, Lab158, and Str493. Interestingly, the bacteria belonging to the C. lituseburense group showed different cell morphologies when the hybridization was done with or without lysozyme treatment. Short and long rods were observed when the sample was not treated with lysozyme prior to hybridization, while only long rods were seen with lysozyme treatment (Fig. 3). This effect was not found in the four adult samples which we tested for comparison. In the probiotic group, higher numbers of bacteria were targeted by Clit135 upon preparation of the sample with lysozyme treatment (P = 0.010), while there were no differences between the groups when no lysozyme treatment was used (Table 4).
FIG. 3.
FISH signals obtained upon hybridization of the fecal sample of an infant (subject number 35) with the Clit135 probe with (A) and without (B) lysozyme treatment.
TABLE 4.
FISH counts in infant feces after supplementation with placebo or Bb12
| Probe | FISH count for groupa:
|
P value | |
|---|---|---|---|
| Probiotic (n = 32) | Placebo (n = 31) | ||
| Eub mix | 9.95 + 0.06 | 9.83 + 0.08 | NS |
| Bif164 | 7.73 + 0.27 | 4.25 + 0.43 | <0.001 |
| Ec1531 | 4.38 + 0.48 | 4.94 + 0.47 | NS |
| Lab158 | 4.05 + 0.46 | 5.07 + 0.43 | NS |
| Str493 | 4.20 + 0.44 | 4.62 + 0.45 | NS |
| Sta697 | 2.41 + 0.36 | 1.89 + 0.33 | NS |
| Veil223 | 2.53 + 0.43 | 2.24 + 0.39 | NS |
| Chis150 | 2.26 + 0.38 | 2.99 + 0.42 | NS |
| Clit135 (without lysozyme treatment) | 2.47 + 0.39 | 2.87 + 0.41 | NS |
| Clit135 (with lysozyme treatment) | 4.03 + 0.40 | 2.60 + 0.36 | 0.01 |
| Bac303 | 0.43 + 0.18 | 0.59 + 0.21 | NS |
| Erec482 | 0.09 + 0.08 | 0.00b | <0.001 |
Cell counts are given as log10 values per g (wet weight) feces. Each number is the geometric mean from all time points+standard error. NS, nonsignificant.
Below the limit of detection.
The bacteria targeted by the probes Chis150, Bac303, Sta697, Veil223, and Erec482 were present in very low numbers and large interindividual differences were observed (Table 4). The probiotic supplementation did not have any significant effect on the occurrence and numbers of bacteria in these groups. The bacteria targeted by Erec482 were detected in only one infant from the probiotic group.
Antibiotic therapy significantly lowered the total bacterial cell numbers (antibiotic-treated infants, 9.80 + 0.08; infants without antibiotic treatment, 10.14 + 0.14; P = 0.03) when all three weeks were considered. Bifidobacteria were detected in 92.30%, 96.42%, and 89.65% of the infants in the probiotic group in week 1, week 2, and week 3, respectively, while the proportion of bifidobacterium-positive infants in the placebo group was lower (69.23%, 53.57%, and 48.14%).
DISCUSSION
Bifidobacterium lactis Bb12 was chosen for the trial because it has been shown to have the highest adhesion to human mucus of all bifidobacteria tested, and hence, a high colonization capacity has been found for this strain (14-16). The major findings of this intervention trial were the increase in the cell numbers of Bifidobacterium spp. and the reduction in the cell numbers of Enterobacteriaceae and Clostridium spp. in the gut of preterm infants in response to B. lactis Bb12 supplementation.
Effect of probiotic supplementation on intestinal microbiota assessed with both culture-independent and culture-dependent methods.
Microorganisms can be isolated as pure cultures and identified to the species level by the use of culture-dependent methods. Although this provides useful information, it has to be considered that a considerable proportion of the dominant fecal microbiota of adults cannot be cultured because their growth requirements are unknown (32). Furthermore, enumeration based on culture-dependent methods is prone to error because the media used for cultivation are never truly selective. Therefore, both approaches were used to monitor the development of intestinal microbiota in the present study. Only a few studies assessed the microbial colonic microbiota of premature infants by molecular methods, preferentially denaturing gradient gel electrophoresis (7, 25, 31). This is the first time that FISH has been applied to the study of the microbiota of premature infants.
In the present study, we observed for all bacterial groups significantly higher cell counts with plating than with FISH when all samples from both the probiotic and placebo groups were considered (data not shown). Considering the effect of probiotic supplementation, the cell counts of total bacteria for all three weeks obtained by FISH and plate counts were significantly different but nevertheless in the same range (FISH placebo, 9.89 + 0.11; probiotic, 10.05 + 0.12; CFU placebo, 10.23 + 0.05; probiotic, 10.03 + 0.04).
The probiotic group displayed higher numbers of bifidobacteria throughout the trial independent of the enumeration method used, although the numbers estimated by FISH were much lower. Discrepancies between plate counts and FISH for bifidobacteria have been shown in two other studies (13, 24). At the hybridization conditions used, Bif164 is highly specific for Bifidobacterium spp. On the other hand, the media used to cultivate the bifidobacteria are not really specific, and some species of lactobacilli may have been counted along with the bifidobacteria.
The numbers of enterobacteria enumerated with Ec1531 increased with age, and there were no differences between the probiotic and placebo groups. Colony counts of Enterobacteriaceae as determined by plating on EMB agar were higher for the placebo group throughout all three study weeks. The counts of enterobacteria determined by plating (EMB agar) were significantly higher than those determined by FISH (Ec1531) in both study groups. This indicates that higher numbers of one or several bacterial groups that grow on EMB agar but are not targeted by Ec1531 were present in the infants belonging to the placebo group. Reduction in the numbers of enterobacteria upon administration of Bifidobacterium breve has been shown previously (20).
Veillonella spp. were counted with Veil223, which targets Veillonella dispar, Veillonella parvula, and Veillonella atypica. Cell counts and occurrence of this group were in agreement with the plate counts reported by Sakata et al. (30). This bacterial group was not enumerated by plating in the present study.
The FISH counts for Staphylococcus spp. (Sta967), Streptococcus spp. (Str493) and Bacteroides spp. (Bac303) were significantly lower than the corresponding colony counts. This can be attributed to the low specificity of the culture media used. Independent of the enumeration method used, there were no differences in the numbers of these bacterial groups between the probiotic and placebo groups. However, the Bacteroides counts were much lower in both study groups than reported earlier (30).
In most previous studies, clostridia were observed with low frequency during the first two weeks after birth (5, 30). In this study, they were detected in most of the infants in low numbers already in the first week. In general, plating resulted in higher clostridial counts than FISH (Chis150 and Clit135 together). This difference can be explained by the fact that the clostridial probes used in this study (Clit135 and Chis150) target only a fraction of the bacteria growing on the Clostridium-specific agar. Selective media are rarely really selective. We therefore assume that organisms not detected by the two probes, such as members of the Eubacterium rectale/Clostridium coccoides cluster, also grow on this medium. Bacterial groups defined by growth requirements usually differ from those defined by phylogeny. The placebo group had higher cell counts of cultivable Clostridium spp. (plate counts) than the probiotic group.
Higher numbers of bacteria targeted by Clit135 were found in the probiotic group than in the placebo group only when lysozyme treatment was used in sample preparation. The observation that the cell counts of bacteria targeted by Clit135 were higher upon treatment with lysozyme is not surprising, as lysozyme treatment is important for the oligonucleotide probe to cross the cell wall of gram-positive bacteria and to hybridize with the target 16S rRNA. From our results, it appears that the preterm infants are colonized by bacteria that are targeted by Clit135 but that are sensitive to lysozyme treatment.
C. difficile was found in almost 70% of the infants enrolled in this study. The carriage rates of C. difficile in normal neonates under 1 year was found to be 84.4% with an ensuing decrease to 30.3% by 2 years of age (23). The presence of C. difficile in healthy newborn infants in contrast to adults has not been linked to any disease (21). This shows that the pathogenicity of a given organism may differ significantly depending on habitat.
The acquisition of Candida spp. in neonates occurs by two possible routes: via mother-neonate transmission and via the environmental route (cross-contamination) (28-30). In the present study, Candida spp. was isolated from only 5.7% of the infants compared to the 26.7% colonization rate found in the study by Baley et al. (4).
Effect of probiotic supplementation on occurrence of antibiotic-resistant strains.
The determination of the antimicrobial susceptibility of a microorganism is an important prerequisite for its approval as a probiotic. B. lactis Bb12 is resistant to cloxacillin, vancomycin, gentamicin, kanamycin, neomycin, streptomycin, fusidic acid, nalidixic acid, and polymyxin B (37). The transfer of vancomycin resistance from lactobacilli and bifidobacteria to other bacteria has not been observed yet (18). With regard to general concerns on the safety of probiotics, i.e., potential transferability of antibiotic resistance determinants, bifidobacteria appear safe for use in the general healthy population with their low natural and acquired resistance to antibiotics (26).
With regard to the antibiotics used in this study, Bifidobacterium lactis Bb12 was found to be sensitive to penicillin, piperacillin, and imipenem but resistant to vancomycin. Resistance to vancomycin partly explains the observation that the bifidobacterial numbers were not different between the antibiotic-treated and -nontreated infants (as observed by enumeration with FISH). The supplementation of Bb12 did not reduce the occurrence of antibiotic-resistant organisms in this study. This indicated that Bb12 does not suppress the growth of such organisms.
Conclusion.
The use of probiotics represents a noninvasive method which aims to create an intestinal microbial community whose composition is closer to that of breastfed term infants. In this study, supplementation of preterm infants with B. lactis Bb12 had a beneficial effect on gut microbiota composition. The number of bifidobacteria (by both methods) was increased, while the numbers of enterobacteria and clostridia (cultivable counts), which include many potential pathogens, were reduced. However, the supplementation did not reduce the fecal reservoir of antibiotic-resistant bacteria.
Footnotes
Published ahead of print on 13 September 2006.
REFERENCES
- 1.Agarwal, R., N. Sharma, R. Chaudhry, A. Deorari, V. K. Paul, I. H. Gewolb, and P. Panigrahi. 2003. Effects of oral Lactobacillus GG on enteric microflora in low-birth-weight neonates. J. Pediatr. Gastroenterol. Nutr. 36:397-402. [DOI] [PubMed] [Google Scholar]
- 2.Alm, E. W., D. B. Oerther, N. Larsen, D. A. Stahl, and L. Raskin. 1996. The oligonucleotide probe database. Appl. Environ. Microbiol. 62:3557-3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amann, R. I., L. Krumholz, and D. A. Stahl. 1990. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J. Bacteriol. 172:762-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baley, J. E., R. M. Kliegman, B. Boxerbaum, and A. A. Fanaroff. 1986. Fungal colonization in the very low birth weight infant. Pediatrics 78:225-232. [PubMed] [Google Scholar]
- 5.Blakey, J. L., L. Lubitz, G. L. Barnes, R. F. Bishop, N. T. Campbell, and G. L. Gillam. 1982. Development of gut colonisation in pre-term neonates. J. Med. Microbiol. 15:519-529. [DOI] [PubMed] [Google Scholar]
- 6.Costalos, C., V. Skouteri, A. Gounaris, S. Sevastiadou, A. Triandafilidou, C. Ekonomidou, F. Kontaxaki, and V. Petrochilou. 2003. Enteral feeding of premature infants with Saccharomyces boulardii. Early Hum. Dev. 74:89-96. [DOI] [PubMed] [Google Scholar]
- 7.de la Cochetiere, M. F., H. Piloquet, C. des Robert, D. Darmaun, J. P. Galmiche, and J. C. Roze. 2004. Early intestinal bacterial colonization and necrotizing enterocolitis in premature infants: the putative role of Clostridium. Pediatr. Res. 56:366-370. [DOI] [PubMed] [Google Scholar]
- 8.Deutsches Institut Fur Normung. 2000. DIN 58940-3 Bbl 1. Medical microbiology. Susceptibility testing of pathogens to antimicrobial agents. Part 3: agar diffusion test. Data for the interpretation of inhibition zone diameters, p. 175-184. Deutsches Institut Fur Normung, Berlin, Germany.
- 9.Franks, A. H., H. J. Harmsen, G. C. Raangs, G. J. Jansen, F. Schut, and G. W. Welling. 1998. Variations of bacterial populations in human feces measured by fluorescent in situ hybridization with group-specific 16S rRNA-targeted oligonucleotide probes. Appl. Environ. Microbiol. 64:3336-3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giovannoni, S. J., E. F. DeLong, G. J. Olsen, and N. R. Pace. 1988. Phylogenetic group-specific oligodeoxynucleotide probes for identification of single microbial cells. J. Bacteriol. 170:720-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harmsen, H. J., G. C. Raangs, T. He, J. E. Degener, and G. W. Welling. 2002. Extensive set of 16S rRNA-based probes for detection of bacteria in human feces. Appl. Environ. Microbiol. 68:2982-2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harmsen, H. J., P. Elfferich, F. Schut, and G. W. Welling. 1999. A 16S rRNA-targeted probe for detection of lactobacilli and enterococci in fecal samples by fluorescent in situ hybridization. Microb. Ecol. Health Dis. 11:3-12. [Google Scholar]
- 13.Harmsen, H. J., A. C. Wildeboer-Veloo, G. C. Raangs, A. A. Wagendorp, N. Klijn, J. G. Bindels, and G. W. Welling. 2000. Analysis of intestinal flora development in breast-fed and formula-fed infants by using molecular identification and detection method. J. Pediatr. Gastroenterol. Nutr. 30:61-67. [DOI] [PubMed] [Google Scholar]
- 14.He, F., A. C. Ouwehand, E. Isolauri, H. Hashimoto, Y. Benno, and S. Salminen. 2001. Comparison of mucosal adhesion and species identification of bifidobacteria isolated from healthy and allergic infants. FEMS Immunol. Med. Microbiol. 30:43-47. [DOI] [PubMed] [Google Scholar]
- 15.He, F., A. C. Ouwehand, E. Isolauri, M. Hosoda, Y. Benno, and S. Salminen. 2001. Differences in composition and mucosal adhesion of bifidobacteria isolated from healthy adults and healthy seniors. Curr. Microbiol. 43:351-354. [DOI] [PubMed] [Google Scholar]
- 16.Juntunen, M., P. V. Kirjavainen, A. C. Ouwehand, S. J. Salminen, and E. Isolauri. 2001. Adherence of probiotic bacteria to human intestinal mucus in healthy infants and during rotavirus infection. Clin. Diagn. Lab. Immunol. 8:293-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Langendijk, P. S., F. Schut, G. J. Jansen, G. C. Raangs, G. R. Kamphuis, M. H. Wilkinson, and G. W. Welling. 1995. Quantitative fluorescence in situ hybridization of Bifidobacterium spp. with genus-specific 16S rRNA-targeted probes and its application in fecal samples. Appl. Environ. Microbiol. 61:3069-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leclercq, R., and E. Derlot. 1988. Plasmid-mediated resistance to vancomycin and teicoplanin in Enterococcus faecium. N. Engl. J. Med. 21:157-161. [DOI] [PubMed] [Google Scholar]
- 19.Lee, S. H., and C. Malone. 1993. Use of multiple 16S rRNA-targeted fluorescent probes to increase signal strength and measure cellular RNA from natural planktonic bacteria. Mar. Ecol. Prog. Ser. 101:193-201. [Google Scholar]
- 20.Li, Y., T. Shimizu, A. Hosaka, N. Kaneko, Y. Ohtsuka, and Y. Yamashiro. 2004. Effects of Bifidobacterium breve supplementation on intestinal flora of low birth weight infants. Pediatr. Int. 46:509-515. [DOI] [PubMed] [Google Scholar]
- 21.Mackie, R. I., A. Sghir, and H. R. Gaskins. 1999. Developmental microbial ecology of the neonatal gastrointestinal tract. Am. J. Clin. Nutr. 69(Suppl):1035S-1045S. [DOI] [PubMed] [Google Scholar]
- 22.Manz, W., R. Amann, W. Ludwig, M. Vancanneyt, and K. H. Schleifer. 1996. Application of a suite of 16S rRNA-specific oligonucleotide probes designed to investigate bacteria of the phylum cytophaga-flavobacter-bacteroides in the natural environment. Microbiology 142(Pt 5):1097-1106. [DOI] [PubMed] [Google Scholar]
- 23.Matsuki, S., E. Ozaki, M. Shozu, M. Inoue, S. Shimizu, N. Yamaguchi, T. Karasawa, T. Yamagishi, and S. Nakamura. 2005. Colonization by Clostridium difficile of neonates in a hospital, and infants and children in three day-care facilities of Kanazawa, Japan. Int. Microbiol. 8:43-48. [PubMed] [Google Scholar]
- 24.Mikkelsen, L. L., C. Bendixen, M. Jakobsen, and B. B. Jensen. 2003. Enumeration of bifidobacteria in gastrointestinal samples from piglets. Appl. Environ Microbiol. 69:654-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Millar, M. R., C. J. Linton, A. Cade, D. Glancy, M. Hall, and H. Jalal. 1996. Application of 16S rRNA gene PCR to study bowel flora of preterm infants with and without necrotizing enterocolitis. J. Clin. Microbiol. 34:2506-2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moubareck, C., F. Gavini, L. Vaugien, M. J. Butel, and F. Doucet-Populaire. 2005. Antimicrobial susceptibility of bifidobacteria. J. Antimicrob. Chemother. 55:38-44. [DOI] [PubMed] [Google Scholar]
- 27.Poulsen, L. K., F. Lan, C. S. Kristensen, P. Hobolth, S. Molin, and K. A. Krogfelt. 1994. Spatial distribution of Escherichia coli in the mouse large intestine inferred from rRNA in situ hybridization. Infect. Immun. 62:5191-5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reef, S. E., B. A. Lasker, D. S. Butcher, M. M. McNeil, R. Pruitt, H. Keyserling, and W. R. Jarvis. 1998. Nonperinatal nosocomial transmission of Candida albicans in a neonatal intensive care unit: prospective study. J. Clin. Microbiol. 36:1255-1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rotimi, V. O., S. A. Olowe, and I. Ahmed. 1985. The development of bacterial flora of premature neonates. J. Hyg. (London) 94:309-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sakata, H., H. Yoshioka, and J. Fujita. 1985. Development of the intestinal flora in very low birth weight infants compared to normal full term newborns. Eur. J. Pediatr. 144:186-190. [DOI] [PubMed] [Google Scholar]
- 31.Schwiertz, A., B. Gruhl, M. Löbnitz, P. Michel, M. Radke, and M. Blaut. 2003. Development of the intestinal bacterial composition in hospitalized preterm infants in comparison with breast-fed, full term infants. Pediatr. Res. 54:393-399. [DOI] [PubMed] [Google Scholar]
- 32.Suau, A., R. Bonnet, M. Sutren, J. J. Godon, G. R. Gibson, M. D. Collins, and J. Dore. 1999. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl. Environ. Microbiol. 65:4799-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thiel, R., and M. Blaut. 2005. An improved method for the automated enumeration of fluorescently labelled bacteria in human feces. J. Microbiol. Methods 61:369-379. [DOI] [PubMed] [Google Scholar]
- 34.Trebesius, K., L. Leitritz, K. Adler, S. Schubert, I. B. Autenrieth, and J. Heesemann. 2000. Culture independent and rapid identification of bacterial pathogens in necrotising fasciitis and streptococcal toxic shock syndrome by fluorescence in situ hybridisation. Med. Microbiol. Immunol. (Berlin) 188:169-175. [DOI] [PubMed] [Google Scholar]
- 35.Yoshioka, H., K. Iseki, and K. Fujita. 1983. Development and differences of intestinal flora in the neonatal period in breast-fed and bottle-fed infants. Pediatrics 72:317-321. [PubMed] [Google Scholar]
- 36.Yu, J. L., S. X. Wu, and H. Q. Jia. 2001. Study on antimicrobial susceptibility of bacteria causing neonatal infections: a 12 year study (1987-1998). Singapore Med. J. 42:107-110. [PubMed] [Google Scholar]
- 37.Zhou, J. S., C. J. Pillidge, P. K. Gopal, and H. S. Gill. 2005. Antibiotic susceptibility profiles of new probiotic Lactobacillus and Bifidobacterium strains. Int. J. Food Microbiol. 98:211-217. [DOI] [PubMed] [Google Scholar]