Abstract
Crimean-Congo hemorrhagic fever virus (CCHFV) is a tick-borne virus in the family Bunyaviridae, genus Nairovirus. The virus is transmitted to humans through infected tick bites or from direct contact with viremic animals or humans. In the present study, a total of 1,015 adult ticks were collected from cattle (603 specimens), sheep (17 specimens), and goats (395 specimens) in the Kelkit Valley in Turkey. Four tick species were recognized on the animals in the surveyed region. The most abundant species were Rhipicephalus bursa and Hyalomma marginatum marginatum, at 47.68% (484/1,015) and 46.40% (471/1,015), respectively. Reverse transcriptase PCR was used to recover partial sequences of the CCHFV small (S) genome segment. The presence of CCHFV was determined in 3 of 33 (9.09%) R. bursa pools and in 1 of 31 (3.22%) H. m. marginatum pools. Virus sequences from R. bursa were extremely different from those of the Greek CCHFV strain (U04958) isolated from an R. bursa tick. Phylogenetic analysis indicated that the CCHFV isolates obtained in this study clustered in group 5, whose range encompasses southwestern Russian and Kosovo. This is the first evidence of CCHFV in ticks from Turkey. Even though Hyalomma is the main vector for CCHFV, R. bursa may play a role in CCHFV transmission.
Crimean-Congo hemorrhagic fever virus (CCHFV) is a tick-borne virus that causes severe hemorrhagic disease in humans (5). The CCHFV genome consists of three molecules of negative-sense single-stranded RNA, each encapsulated separately. The virion particle contains viral RNA polymerase (the L segment), surface glycoproteins G1 and G2 (the M segment), and a nucleocapsid (N) protein (the S segment) (5, 21, 22).
CCHF is a zoonotic disease typified by severe hemorrhagic fever in humans. After an incubation period of 2 to 7 days, the patient has flu-like symptoms that rapidly turn into a hemorrhagic state with bleeding from the mucous membranes and petechiae, often asssociated with thrombocytopenia and leukopenia. High mortality rates from the disease, varying from 10% to 60%, make CCHF a public health concern (5, 9, 22).
The virus is transmitted to humans through infected tick bites or from direct contact with viremic animals or humans (9). Although Hyalomma ticks are one of the major vectors of CCHFV, at least 31 tick species harbor the virus (9). The disease is distributed in Asia, the Middle East, Africa, and southeastern Europe (9, 23, 26).
Although serological evidence of CCHFV was detected years ago, clinical CCHFV infection was first recognized in 2002, and outbreaks have occurred in Turkey in subsequent years (12, 19). Genetic analysis of the virus isolated from patients shows that it is closely related to Kosovo and Russian strains (19). Ticks are very important to the ecology of CCHFV. The biological role of the ticks is noteworthy, not only as virus vectors, but also as reservoirs of the virus (5, 9, 21, 22). However, there has been no report of tick fauna or of what tick species carry the virus in the region of endemicity where the outbreaks occurred in Turkey. The aims of this study were (i) to evaluate the tick fauna of the region of endemicity, (ii) to establish what tick species harbor the virus, and (iii) to determine the genetic lineage of the virus in Turkey.
MATERIALS AND METHODS
Study area.
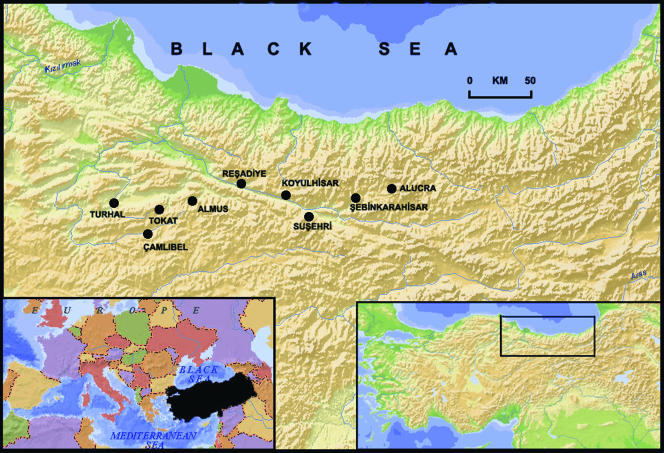
The study was conducted in nine locations (Tokat, Camlibel, Almus, Turhal, Resadiye, Koyulhisar, Susehri, Sebinkarahisar, and Alucra) in the region of the Kelkit Valley, an area of about 12,100 km2 in the middle Black Sea region of Turkey (Fig. 1). The valley is a transitional zone between central Anatolia and the middle and eastern Black Sea regions geographically. The valley is the northernmost point and the longest valley of the Yesilirmak River basin (246 km). Kelkit Valley starts from the Giresun Mountains and lies in an east-west direction along the Yesilirmak and Canik Mountains. The mean altitudes of these mountains are between 1,400 and 1,500 m. At the bottom of the valley, there is a clear decrease in height in an east-west direction. The altitude is about 650 m in Koyulhisar and 450 m in Resadiye. Mediterranean climate is found in the study area along the valley, but in the upper part of the valley, the effect of Mediterranean climate begins to decrease and an oceanic climate becomes dominant. A semiarid, cold Mediterranean climate is found in Koyulhisar and Resadiye. Annual rainfall is low in the central basin of the region (400 to 500 mm/year). There are four vegetation types in the region, namely, forest, macchie, degraded forest, and hygrophilous (dominated by Pinus brutia, Phillyrea lotifolia, Quercus coccifera, Pinus nigra, Pinus sylvestris, and Vitex agnus-castus) (18).
FIG. 1.
Map of Turkey and of the Kelkit Valley showing the locations of the study sites. (Copyright Selcuk Hayli; reproduced with permission.)
Tick collection.
Ticks were collected from domestic ruminants (cattle, sheep, and goats) in nine previously selected collecting points where the families of the identified patients were living (Fig. 1). The local department of the Ministry of Agriculture treated the domestic animals with acaricidal treatments soon after the outbreak was noted. As a result, we were not able to collect a sufficient number of ticks in some identified patients' homes, and we extended our tick collection to include the animals of the patients' neighbors.
The sampling was done in spring (between May and June 2005). The whole body of each sampled animal was inspected for the presence of tick infestations by palpation, mainly on the ears and along the nape of the neck, perineum, scrotum or udder, and tail base. Ticks collected from each animal were kept alive in separate vials and labeled, with collection points noted, and were then taken to the laboratory for species identification. The ticks were morphologically examined under a stereomicroscope and identified using a guide to the identification of species (11). Although immature stages were present in the samples from some locations, only adult stages were classified to species level to avoid misidentification of the ticks. The identified ticks were pooled into groups of 8 to 20 according to species, host, and geographic origin and were stored at −70°C for a week.
Extraction of RNA from tick species.
Ticks from each pool were washed twice with phosphate-buffered saline and crushed with a mortar and pestle in 1.5 ml of phosphate-buffered saline containing 10% fetal calf serum, 500 IU/ml penicillin, and 500 μg/ml streptomycin. After homogenization, the suspension was incubated at 4°C for 1 h and centrifuged at 2,500 × g for 5 min. The samples were kept at −70°C until they were used. Total RNA was extracted from the samples by using the RNAeasy kit (QIAGEN, GmbH, Hilden, Germany) according to the recommendations of the supplier. The RNA was dissolved in 50 ml of RNase-free water and stored at −70°C until it was needed.
RT-PCR.
Reverse transcription of RNA was conducted as described previously (7). Briefly, a 30-μl reaction mixture contained 5 μl of extracted RNA preparation, 2.5 μmol/liter of random hexamers, and 100 U of Moloney murine leukemia virus reverse transcriptase (RT) (Superscript II; Invitrogen, Life Technologies). The reaction mixtures were incubated for 60 min at 42°C, and the reaction was stopped by heating the mixture at 95°C for 5 min and chilling it on ice. The PCR was conducted in a 50-μl reaction mixture containing 5 μl of cDNA template, 10 mM Tris-HCl, 50 mM KCl, 1.5 mM MgCl2, 20 pmol R3 primer (5′-GACAAATTCCCTGCACCA-3′), 20 pmol F2 primer (5′-TGGACACCTTCACAAACTC-3′), 1 U of Taq polymerase (Promega Corporation, Madison, WI), and 0.2 mmol/liter of each deoxynucleoside triphosphate. The cycling conditions were 94°C for 3 min; 33 cycles of 94°C for 45 s, 55°C for 45 s, and 72°C for 1 min; and a final extension at 72°C for 10 min. Nested PCR was performed with F3 primer (5′-GAATGTGCATGGGTTAGCTC-3′) and R2 primer (5′-GACATCACAATTTCACCAGG-3′), and second-round PCR products were visualized by ethidium bromide staining after 1.5% agarose gel electrophoresis. The purified PCR products were cloned into the Topo XL cloning vector (Invitrogen Corp., Carlsbad, CA). All plasmid stocks were prepared by using the Plasmid Maxi kit (QIAGEN), and concentrations were determined by averaging the A260s (1 = 50 μl ml−1) of multiple dilutions. The copy number of each plasmid was calculated from the average molecular mass of 660 Da for 1-bp double-stranded DNA. Plasmid standards were made by preparing serial 10-fold dilutions in a final volume of 1 ml water, and 10 μl of each diluted plasmid was added to a 50-μl PCR mixture.
DNA sequence comparisons and phylogenetic analysis.
Cycle sequencing was conducted by the chain termination method using fluorescent dye termination (PRISMTM; ABI, Foster City, CA), with R2 or F3 used as a sequencing primer. The products were separated in an automated DNA genetic analyzer (ABI 310 Prism; Perkin-Elmer Corporation, Foster City, CA). Each construct was sequenced at least three times. The phylogenetic tree generated by a neighbor-joining method with Kimura two-parameter distances by using MEGA software (version 3.1) showed a geographic clustering of the sequences of CCHF viruses. Bootstrap confidence limits were based on 500 replicates (29, 30).
Nucleotide sequence accession numbers.
The nucleotide sequences were deposited in the EMBL/GenBank databases under the accession numbers DQ397823, DQ397824, DQ397825, and DQ397826.
RESULTS
Tick species and distribution.
A total of 1,015 adult ticks were collected from cattle (603 specimens), sheep (17 specimens), and goats (395 specimens). The numbers and distributions of adult tick species according to the collecting points on the farms visited are documented in Table 1. Four tick species were recognized on the animals in the surveyed region. The most abundant species were Rhipicephalus bursa and Hyalomma marginatum marginatum with 47.68% (484/1,015) and 46.40% (471/1,015), respectively. Boophylus annulatus ticks represented 4.53% (46/1,015) of the total number of ticks. Hemaphysalis sulcata was the minor species and represented 1.37% (14/1,015) of the tick population. R. bursa and H. m. marginatum were found in all locations in the surveyed region. B. annulatus was encountered in three localities (Almus, Turhal, and Resadiye), while H. sulcata was found in only one location (Resadiye). R. bursa and H. m. marginatum were found widely distributed in the surveyed region, whereas B. annulatus and H. sulcata showed narrow distributions.
TABLE 1.
Numbers and distributions of adult tick species according to the collecting points of farms visited
| Location | No. of ticks
|
||||
|---|---|---|---|---|---|
| H. m. marginatum | R. bursa | B. annulatus | H. sulcata | Total | |
| Tokat | 43 | 36 | 79 | ||
| Camlibel | 39 | 9 | 48 | ||
| Almus | 73 | 1 | 40 | 114 | |
| Turhal | 87 | 75 | 2 | 164 | |
| Resadiye | 149 | 4 | 4 | 14 | 171 |
| Koyulhisar | 37 | 205 | 242 | ||
| Susehri | 21 | 4 | 25 | ||
| Sebinkarahisar | 8 | 150 | 158 | ||
| Alucra | 14 | 14 | |||
| Total | 471 | 484 | 46 | 14 | 1,015 |
Detection of the S segment of CCHFV RNA and sequence analysis.
A total of 69 tick pools were included in this study. Most of the tick pools were R. bursa (33 pools; 47.83%) and H. m. marginatum (31 pools; 44.92%). B. annulatus and H. sulcata represented 4 (5.8%) and 1 (1.44%) pools, respectively (Table 2). All of the tick pools were examined by nested RT-PCR. The presence of CCHFV was detected in 3 of 33 (9.09%) R. bursa pools and in 1 of 31 (3.22%) H. m. marginatum pools. The positive samples produced bands of the expected size (260 nucleotides [nt]) with the F2-R3/F3-R2 primer sets. Neither B. annulatus nor H. sulcata gave any positive nested-RT-PCR result against CCHFV (Table 2). The detection sensitivity of the assay was determined for each of the CCHFV isolates cloned into the plasmid vectors. It varied from 35 nanograms to 52 nanograms. Nucleotide alignments between the isolates (DQ3978223 and DQ3978225) from R. bursa pools revealed that they were almost identical, except for one nucleotide substitution that did not result in an amino acid change in the isolate DQ3978225. However, the CCHFV sequence obtained from H. m. marginatum (DQ3978226) differed from those of the other strains by 2.3% (DQ3978223 and DQ3978224) and 2.8% (DQ3978225), which resulted in an amino acid change.
TABLE 2.
Detection of CCHFV by nested RT-PCR in adult ticks collected from domestic ruminants in the Kelkit Valley in the middle Black Sea region of Turkey
| Tick species and host | Total no. of ticks | No. of pools | No. of positive pools |
|---|---|---|---|
| H. m. marginatum | |||
| Cattle | 424 | 28 | 1 |
| Sheep | 9 | 1 | 0 |
| Goats | 39 | 2 | 0 |
| R. bursa | |||
| Cattle | 119 | 8 | 1 |
| Sheep | 8 | 1 | 0 |
| Goats | 357 | 24 | 2 |
| B. annulatus | |||
| Cattle | 46 | 4 | 0 |
| H. sulcata | |||
| Cattle | 14 | 1 | 0 |
| Total | 1,015 | 69 | 4 |
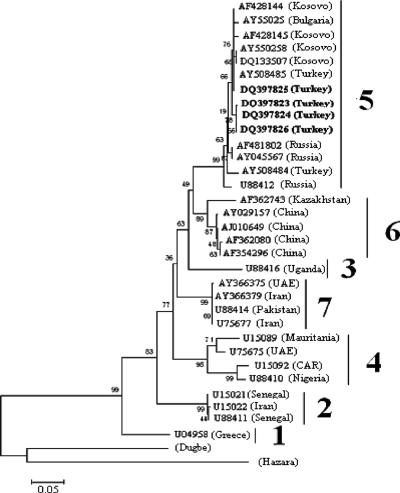
To investigate the genetic relationships of the CCHFV strains circulating in Turkey, we performed phylogenetic analysis based on a 212-nt fragment from the S segment sequences of the CCHFV strains obtained in this study, which showed that they clustered with the Kosovo (1335079), Bulgarian (AY550258), and Russian (AF481802) strains in clade 5 (Fig. 2). Thus, our analysis provides strong evidence for the circulation of closely related genetic variants of CCHFV in the Balkan region from Kosovo and Bulgaria to the southern regions of Russia. Nucleotide and amino acid divergences of the respective S segment sequences of CCHFV and two other nairoviruses, Dugbe and Hazara, are shown in Table 3.
FIG. 2.
Phylogenetic tree based on 216 nt of the small (S) segment of CCHFV. The tree was generated by the neighbor-joining method with Kimura two-parameter distances using MEGA software (version 3.1). Bootstrap confidence limits were based on 500 replicates. The numerical values in the tree represent bootstrap results. The GenBank accession number and the geographic origin are given for each isolate. Newly sequenced strains from Turkey described in this paper are shown in boldface. Dugbe and Hazara viruses were used as the outgroups. The genetic lineages of CCHFV are numbered 1 to 7. UAE, United Arab Emirates; CAR, Central African Republic.
TABLE 3.
Nucleotide and amino acid differences between CCHF, Dugbe, and Hazara virus strains
| Strain | % Nucleotide or amino acid difference compared to indicated straina
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AF362080 | AF362743 | DQ133507 | DQ397823 | M25150 | M86624 | U04958 | U15021 | U15022 | U75675 | U75677 | U88410 | U88414 | U88416 | |
| AF362080 | 2.8 | 8.9 | 8.9 | 13.7 | 8.9 | 10.4 | 5.8 | 4.3 | 4.3 | 8.9 | 4.3 | 9.0 | 14.4 | |
| AF362743 | 6.4 | 8.9 | 8.9 | 12.1 | 8.9 | 10.4 | 5.8 | 7.3 | 7.3 | 8.9 | 4.3 | 13.5 | 16.9 | |
| DQ13350 | 11.8 | 13.5 | 0.0 | 13.7 | 7.3 | 8.9 | 5.8 | 7.3 | 7.3 | 7.3 | 5.8 | 14.7 | 15.6 | |
| DQ39782 | 11.7 | 13.4 | 1.9 | 13.7 | 7.3 | 8.9 | 5.8 | 7.3 | 7.3 | 7.3 | 5.8 | 13.5 | 16.8 | |
| M25150 | 26.4 | 26.3 | 24.2 | 24.8 | 12.1 | 13.7 | 12.1 | 10.4 | 10.4 | 12.1 | 12.1 | 65.2 | 64.2 | |
| M86624 | 18.2 | 20.1 | 18.4 | 19.6 | 25.5 | 1.4 | 2.8 | 7.3 | 7.3 | 0.0 | 5.8 | 142.5 | 226.2 | |
| U04958 | 18.9 | 20.8 | 19.0 | 20.3 | 26.2 | 0.5 | 4.3 | 8.9 | 8.9 | 1.4 | 7.3 | 22.2 | 27.5 | |
| U15021 | 17.1 | 20.4 | 19.8 | 19.8 | 29.5 | 20.2 | 20.9 | 4.3 | 7.3 | 5.8 | 2.8 | 16.9 | 19.4 | |
| U15022 | 17.0 | 18.3 | 21.8 | 21.7 | 28.9 | 24.2 | 25.0 | 10.2 | 0.0 | 7.3 | 4.3 | 17.5 | 20.0 | |
| U75675 | 18.3 | 19.1 | 20.5 | 19.8 | 27.2 | 27.3 | 28.1 | 11.3 | 3.3 | 7.3 | 4.3 | 12.2 | 18.7 | |
| U75677 | 18.2 | 20.1 | 18.4 | 19.6 | 25.5 | 0.0 | 0.5 | 20.2 | 24.2 | 27.3 | 5.8 | 0.0 | 16.2 | |
| U88410 | 14.4 | 16.9 | 15.6 | 16.8 | 27.5 | 19.4 | 20.0 | 22.0 | 22.1 | 22.2 | 19.4 | 16.4 | 22.2 | |
| U88414 | 12.1 | 13.7 | 17.1 | 17.1 | 87.8 | 73.3 | 20.7 | 15.4 | 17.1 | 13.7 | 8.9 | 13.7 | 15.7 | |
| U88416 | 4.3 | 4.3 | 5.8 | 5.8 | 65.8 | 69.2 | 12.1 | 5.8 | 7.3 | 2.8 | 2.8 | 4.3 | 12.1 | |
The percentages of nucleotide (above the diagonal) and amino acid (below the diagonal) differences are presented.
DISCUSSION
Crimean-Congo hemorrhagic fever was first documented in the Crimean region of Russia in 1944 (9). Currently, the distribution of CCHFV comprises over 30 countries in Asia, the Middle East, southeastern Europe, and Africa (9). In spite of serological evidence of the infection, it was first recognized in Turkey in 2002, and an increasing number of cases were reported between 2002 and 2005 (as reported by the Turkish Ministry of Health). Most of the cases have been reported in the middle Black Sea and northern inner Anatolia regions (12, 19) (Fig. 1), indicating that CCHF is endemic in that region of Turkey.
The virus is transmitted to livestock and humans by the bite of infected ticks or by exposure to the tissues or blood of infected animals. CCHFV, like other tick-borne zoonotic agents, circulates in nature in tick-vertebrate-tick (5, 6, 9, 21, 22). Since 2002, a total of 504 cases have occurred in Turkey, 26 of them fatal (as reported by the Turkish Ministry of Health). Most cases had been bitten by a tick or handled livestock, and a few were infected through nosocomial infections (as reported by the Turkish Ministry of Health) (12, 19). However, there has been no report of tick fauna, what tick species carry the virus, or why the CCHF outbreaks have occurred mainly in the Kelkit Valley region of Turkey. To address those questions, a total of 1,015 adult ticks were collected from domestic ruminants in the region of endemicity. The most abundant species were R. bursa and H. m. marginatum (Table 1). R. bursa is a typical representative of the tick fauna commonly found on domestic ruminants in different regions of Turkey (4, 15, 17, 27, 28). However, the high prevalence of H. m. marginatum in our samples is surprising, because it is not a common tick on domestic ruminants in Turkey (1, 2, 3, 4, 16, 33, 34). In this sense, H. m. marginatum seems to be geographically restricted to parts of the middle Black Sea region of Turkey. There are suitable habitats in the region for the ticks to survive, as immature stages of H. m. marginatum are common parasites of wild and domestic birds, e.g., rooks, hooded crows, and partridges, as well as European brown hares, eared hedgehogs, and wild ungulates (13, 20). Also, hunting has been banned by the Turkish government for many years, which may have caused the populations of tick species in the region to increase.
From the phylogenetic tree (Fig. 2), it was noted that the CCHFV isolates obtained in this study clustered in group 5, which encompasses Russian and Eastern European strains and is more closely related to Kosovo (DQ133507), southwestern Russian (AF481802 and AY045567), Bulgarian (AY550258), and Turkish (AY508484) strains. In this study, the presence of CCHFV was detected in three R. bursa pools and in one H. m. marginatum pool. Nucleotide alignments between the isolates (DQ3978223 to DQ3978225) from R. bursa pools indicated that they were almost identical. The CCHFV sequence obtained from H. m. marginatum (DQ3978226) differed from those of other strains by only 2.3% (DQ3978223 and DQ3978224) and 2.8% (DQ3978225). The data on the genetic diversity of CCHFV in Turkey are very limited, and only two CCHFV strains (AY508484 and AY508485) were obtained from humans in 2003 in Gumushane, which is located in the Kelkit Valley (19). The strains isolated during the 2003 outbreak were very closely related to all four strains isolated in the present study (Fig. 2), with nucleotide identity of up to 99.1% among them. Our results are consistent with those of Karti et al. (19), showing that the CCHFV circulating in Turkey was associated with the Kosovo-Black Sea region rather than having been introduced from Iran or the Middle East by infected ticks or livestock movement (8, 10).
Several CCHFV S segment sequences from different regions of the world indicate that there are considerable genetic differences between CCHFV isolates (9, 14, 23, 31). Yashina et al. (32) hypothesized that an explanation of the genetic diversity between CCHFV strains depends not only on different geographical locations, but also on different tick species. In the present study, CCHFV isolates in geographical locations very close to each other were found in two different tick species, and nucleotide identity among them was more than 97%. To our knowledge, this is the second report of CCHFV obtained from R. bursa ticks. The Greek CCHFV strain (U04958) isolated from an R. bursa tick is extremely different from our isolates (23.1% nucleotide difference) (24, 25). Taken together, our results are consistent with different geographical locations being important in the genetic variability of CCHFV, but they do not support the hypothesis of different tick species.
In conclusion, the results of the present study demonstrate the presence of CCHFV in R. bursa and H. m. marginatum ticks. This is the first evidence of CCHFV in ticks from Turkey. Even though Hyalomma is the main vector for CCHFV, R. bursa may play a role in CCHFV transmission. Phylogenetic analysis provides strong evidence for the circulation in Turkey of CCHFV closely related to southwestern Russian and Kosovo strains. An explanation of the source of these outbreaks in Turkey might be the migration of virus-infected or tick-infested birds from Russia to Turkey, but further studies are needed to support this hypothesis.
Acknowledgments
This research was financially supported and approved by the Scientific and Technological Research Council of Turkey (TUBITAK), project number V 104 O 392.
We are grateful to Ali Mirazimi and Anna Papa for providing RT-PCR controls. We are also grateful to Suleyman Felek and Mehmet Ziya Doymaz for valuable discussions. We thank Senel Cavusoglu, Sinan Aybek, Ahmet Kocak, and Muhsin Tastan for helping to collect tick species. We also thank Selcuk Hayli for generation of Fig. 1.
REFERENCES
- 1.Aktas, M., K. Altay, and N. Dumanli. 2006. A molecular survey of bovine Theileria parasites among apparently healthy cattle with a note on the distribution of ticks in eastern Turkey. Vet. Parasitol. 138:179-185. [DOI] [PubMed] [Google Scholar]
- 2.Aktas, M., N. Dumanli, and M. Angın. 2004. Cattle infestation by Hyalomma ticks and prevalence of Theileria in Hyalomma species in the east of Turkey. Vet. Parasitol. 119:1-8. [DOI] [PubMed] [Google Scholar]
- 3.Arslan, M. O., S. Umur, and L. Aydın. 1999. Prevalence of Ixodidae on cattle in Kars province. J. Turk. Parazitol. 23:331-335. (In Turkish.) [Google Scholar]
- 4.Aydin, L. 2000. Distribution and species of ticks on ruminants in the southern Marmara region. J. Turk. Parazitol. 24:194-200. (In Turkish.) [Google Scholar]
- 5.Bishop, D. H. 1996. Biology and molecular biology of bunyaviruses, p. 19-61. In R. M. Elliott (ed.), The Bunyaviridae. Plenum Press, New York, N.Y.
- 6.Burney, M. I., A. Ghafoor, M. Saleen, P. A. Webb, and J. Casals. 1980. Nosocomial outbreak of viral hemorrhagic fever caused by Crimean hemorrhagic fever-Congo virus in Pakistan, January 1976. Am. J. Trop. Med. Hyg. 29:941-947. [DOI] [PubMed] [Google Scholar]
- 7.Burt, F. J., P. A. Leman, J. F. Smith, and R. Swanepoel. 1998. The use of a reverse transcription-polymerase chain reaction for the detection of viral nucleic acid in the diagnosis of Crimean-Congo haemorrhagic fever. J. Virol. Methods 70:129-137. [DOI] [PubMed] [Google Scholar]
- 8.Chinikar, S., S. M. Persson, M. Johansson, B. Linda, M. Goya, B. Houshmand, A. Mirazimi, A. Plyusnin, A. Lundkvist, and M. Nilsson. 2004. Genetic analysis of Crimean-Congo hemorrhagic fever virus in Iran. J. Med. Virol. 73:404-411. [DOI] [PubMed] [Google Scholar]
- 9.Chris, A. 2004. Whitehouse Crimean-Congo hemorrhagic fever. Rev. Antivir. Res. 64:145-160. [DOI] [PubMed] [Google Scholar]
- 10.Drosten, C., D. Minnak, P. Emmerich, H. Schmitz, and T. Reinicke. 2002. Crimean-Congo hemorrhagic fever in Kosovo. J. Clin. Microbiol. 40:1122-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Estrada-Peña, A., A. Bouattour, J. L. Camicas, and A. R. Walker. 2004. Ticks of domestic animals in the Mediterranean region, 1st ed., p. 131. University of Zaragoza, Zaragoza, Spain.
- 12.Gozalan, A., L. Akin, J. M. Rolain, F. S. Tapar, O. Oncul, H. Yoshikura, H. Zeller, D. Raoult, and B. Esen. 2004. Epidemiological evaluation of a possible outbreak in and nearby Tokat province. Microbiol. Bull. 38:33-34. (In Turkish.) [PubMed] [Google Scholar]
- 13.Grigor'ev, M. P., I. M. Evchenko, L. I. Shaposhnikova, K. V. Shchenetts, A. Emel'ianov, N. V. Ermolova, N. I. Tikhenko, and B. I. Levchenko. 2001. Role of some wild birds and mammals in the natural foci of Crimean haemorrhagic fever in Stavropol' region. Zh. Mikrobiol. Epidemiol. Immunobiol. 6:92-95. [PubMed] [Google Scholar]
- 14.Hewson, R., J. Chamberlain, V. Mioulet, G. Lloyd, B. Jamil, R. Hasan, A. Gmyl, L. Gmyl, S. E. Smirnova, A. Lukashev, G. Karganova, and C. Clegg. 2004. Crimean-Congo haemorrhagic fever virus: sequence analysis of the small RNA segments from a collection of viruses world wide. Virus Res. 102:185-189. [DOI] [PubMed] [Google Scholar]
- 15.Inci, A., S. Nalbantoglu, Y. Cam, A. Atasever, Z. Karaer, A. Cakmak, F. Sayin, B. A. Yukari, A. Ica, and A. Deniz. 2003. Theileriosis and tick infestations in sheep and goats around kayseri. Turk. J. Vet. Anim. Sci. 27:57-60. (In Turkish.) [Google Scholar]
- 16.Inci, A., A. Cakmak, Z. Karaer, S. Dincer, F. Sayin, and A. Ica. 2002. Seroprevalence of bovine babesiosis around Kayseri. Turk. J. Vet. Anim. Sci. 26:1345-1350. (In Turkish.) [Google Scholar]
- 17.Inci, A., Z. Karaer, and A. Ica. 2002. Babesiosis in sheep and goats around Kayseri. Erciyes Univ. J. Health Sci. (Veterinary) 16:1, 79-83. (In Turkish.) [Google Scholar]
- 18.Karaer, F., M. Kilinc, and H. Kutbay. 1999. The woody vegetation of Kelkit Valley. Turk. J. Bot. 23:319-344. [Google Scholar]
- 19.Karti, S., S. Odabasi. Z. Korten, V. Yilmaz, M. Sonmez, M. Caylan, R. Akdogan, E. Eren, N. Koksal, I. Ovali, E. B. Erickson, M. J. Vincent, S. T. Nichol, J. A. Comer, P. E. Rollin, and T. G. Ksiazek. 2004. Crimean-Congo hemorrhagic fever in Turkey. Emerg. Infect. Dis. 10:1379-1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kotti, B. K., L. I. Shaposhnikova, I. M. Evchenko, B. I. Levchenko, D. B. Surkhaev, P. N. Korzhov, and I. M. Tokhov. 2001. Hyalomma marginatum Koch in Stavropol' region. Zh. Mikrobiol. Epidemiol. Immunobiol. 6:105-108. [PubMed] [Google Scholar]
- 21.Marriott, A. C., and P. A. Nuttall. 1996a. Molecular biology of nairoviruses, p. 91-104. In R. M. Elliott (ed.), The Bunyaviridae. Plenum Press, New York, N.Y.
- 22.Nichol, S. T. 2001. Bunyaviruses, p. 1603-1633. In D. M. Knipe and P. M. Howley (ed.), Fields virology, vol. 1, 4th ed. Lippincott, Williams & Wilkins, Philadelphia, Pa. [Google Scholar]
- 23.Papa, A., E. Papadimitriou, B. Boz'ovic, and A. Antoniadis. 2005. Genetic characterization of the M RNA segment of a Balkan Crimean-Congo hemorrhagic fever virus strain. J. Med. Virol. 75:466-469. [DOI] [PubMed] [Google Scholar]
- 24.Papa, A., B. Bozovi, V. Pavlidou, E. Papadimitriou, M. Pelemis, and A. Antoniadis. 2002. Genetic detection and isolation of Crimean-Congo hemorrhagic fever virus, Kosovo, Yugoslavia. Emerg. Infect. Dis. 8:852-854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Papa, A., S. Bino, A. Llagami, B. Brahimaj, E. Papadimitriou, V. Pavlidou, E. Velo, G. Cahani, M. Hajdini, A. Pilaca, A. Harxhi, and A. Antoniadis. 2001. Crimean-Congo hemorrhagic fever in Albania. Eur. J. Clin. Microbiol. Infect. Dis. 21:603-606. [DOI] [PubMed] [Google Scholar]
- 26.Rodriguez, L. L., G. O. Maupin, T. G. Ksiazek, P. E. Rollin, A. S. Khan, T. F. Schwarz, R. S. Lofts, J. F. Smith, A. M. Noor, C. J. Peters, and S. T. Nichol. 1997. Molecular investigation of a multisource outbreak of Crimean-Congo hemorrhagic fever in the United Arab Emirates. Am. J. Trop. Med. Hyg. 57:512-518. [DOI] [PubMed] [Google Scholar]
- 27.Sayin, F., and N. Dumanli. 1982. Ticks (Ixodidae) of domestick animals in the province of Elazig, Turkey. Ankara Univ. Vet. Fak. Derg. 29:344-362. (In Turkish.) [Google Scholar]
- 28.Sayin, F., S. Dincer, Z. Karaer, N. Dumanli, A. Cakmak, A. Inci, B. A. Yukari, and Z. Vatansever. 1997. Status of the tick infestation of sheep and goats in Turkey. Parasitologia 39:145-152. [PubMed] [Google Scholar]
- 29.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 30.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yashina, L., O. Vyshemirskii, S. Seregin, I. Petrova, E. Samokhvalov, D. Lvov, V. Gutorov, I. Kuzina, G. Tyunnikov, Y. W. Tang, S. Netesov, and V. Petrov. 2003. Genetic analysis of Crimean-Congo hemorrhagic fever virus in Russia. J. Clin. Microbiol. 41:860-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yashina, L., I. Petrova, S. Seregin, O. Vyshemirskii, D. Lvov, V. Aristova, J. Kuhn, S. Morzunov, V. Gutorov, I. Kuzina, G. Tyunnikov, S. Netesov, and V. Petrov. 2003. Genetic variability of Crimean-Congo haemorrhagic fever virus in Russia and Central Asia. J. Gen. Virol. 84:1199-1206. [DOI] [PubMed] [Google Scholar]
- 33.Yay, M., S. Yazar, L. Aydin, and I. Sahin. 2004. Investigation of tick species on sheep and cattle around of Kayseri. Erciyes Univ. J. Health Sci. 13:25-29. [Google Scholar]
- 34.Yukari, B. A., and S. Umur. 2002. The prevalence of tick species (Ixodoidea) in cattle, sheep and goats in the Burdur region, Turkey. Turk. J. Vet. Anim. Sci. 26:1260-1270. [Google Scholar]