Abstract
We undertook this study to assess the accuracy of the clindamycin-erythromycin disk approximation test (D-test) for detection of inducible clindamycin resistance in Staphylococcus spp. One hundred sixty-three Staphylococcus aureus and 68 coagulase-negative Staphylococcus (CoNS) spp. which were erythromycin nonsusceptible but clindamycin susceptible were tested using the D-test performed at both 15-mm and 22-mm disk separations and compared with genotyping as the “gold standard.” The rate of inducible clindamycin resistance was 96.3% for S. aureus and 33.8% for CoNS spp. The sensitivities of the D-tests performed at 15 mm and 22 mm were 100% and 87.7%, respectively, and specificities were 100% for both. The use of 22-mm disk separation for the D-test to detect inducible clindamycin resistance results in an unacceptably high very major error rate (12.3%). All isolates with false-negative results harbored the ermA gene, and the majority were methicillin-resistant Staphylococcus aureus. False-negative results were associated with smaller clindamycin zone sizes and double-edged zones. We recommend using a disk separation distance of ≤15 mm. There is wide geographic variation in the rates of inducible clindamycin resistance, and each laboratory should determine the local rate before deciding whether to either perform the D-test routinely or else report that all erythromycin-resistant S. aureus isolates are also clindamycin resistant.
Acquired staphylococcal resistance to lincosamides such as clindamycin is largely mediated by ribosomal methylases encoded by one of several erm genes. These enzymes methylate the bacterial ribosome at the binding site for macrolide, lincosamide, and streptogramin B (MLSB) antibiotics, thus inhibiting antibiotic activity. Such resistance may be inducible (iMLSB) or constitutive. Induction occurs in the presence of erythromycin but not clindamycin (16). However, if clindamycin is used for treatment of an isolate with iMLSB resistance, selection for a mutation in the macrolide-responsive promoter region upstream of the erm gene may occur, leading to constitutive clindamycin resistance and treatment failure (5, 16-18, 23, 32, 35). It is thus recommended by most experts that clindamycin therapy be avoided for Staphylococcus sp. isolates that display iMLSB resistance, despite a low clindamycin MIC (5, 16, 27, 32).
Macrolide resistance due to active efflux encoded by the msrA gene is also found in Staphylococcus spp. It results in resistance to macrolides and streptogramin B antibiotics, but not lincosamides (MS phenotype), and clindamycin is active against such isolates (16). The prevalence of this type of resistance shows great geographical variation but is generally less common than iMLSB resistance (9, 20, 29).
It is important for laboratories to distinguish between MS and iMLSB resistance before reporting an erythromycin-nonsusceptible Staphylococcus sp. isolate as clindamycin susceptible. However, the two are indistinguishable when routine broth microdilution (BMD) testing is used. They can be distinguished by the erythromycin-clindamycin disk approximation test or D-test. When an organism expressing iMLSB resistance is tested according to Clinical and Laboratory Standards Institute (CLSI) methods with a 15-μg erythromycin disk placed close to a 2-μg clindamycin disk, the zone of inhibition around the clindamycin disk is flattened to form a “D” shape (positive D-test), whereas in the MS phenotype, the clindamycin zone remains circular (7). It is not clear from the literature what distance between antibiotics disks is ideal for the D-test. A false-negative D-test will result in reporting an isolate as clindamycin susceptible when it should be reported as resistant (very major error), while a false-positive test will result in reporting an isolate as resistant when it should be reported as susceptible (major error).
The need to test each Staphylococcus sp. isolate for inducible clindamycin resistance depends on the local prevalence of the two phenotypes. If MS resistance is uncommon, many laboratories will not perform the D-test but simply report all erythromycin-resistant isolates as clindamycin resistant (31). There are few published data on the prevalence of the two phenotypes in Australia, but what information is available suggests the iMLSB phenotype predominates (25; S. Aurangabadkar, V. Sintchenko, and D. Rankin, Abstr. Natl. Conf. Aust. Soc. Microbiol., abstr. PP02.2, 2004).
The aims of this study were (i) to determine the accuracy of the D-test when disks were placed 15 mm apart manually or 22 mm apart using an automated dispenser and (ii) to establish the prevalence of the two mechanisms of resistance in isolates obtained in our laboratory.
MATERIALS AND METHODS
Collection of isolates.
The Centre for Infectious Diseases and Microbiology provides microbiology laboratory services to hospitals, general practitioners, and other health facilities in metropolitan Sydney, Australia, as well as regional and rural New South Wales. Between 24 July and 5 November 2005, all significant isolates (i.e., those for which antibiotic susceptibility testing was performed) of Staphylococcus spp. which were erythromycin resistant (MIC, ≥8 mg/liter) or intermediate (MIC, 1 to 4 mg/liter) and clindamycin susceptible (MIC ≤ 0.5 mg/liter) by BMD were collected. Duplicate isolates from the same patient were excluded. Identification and BMD were performed using the Phoenix Automated Microbiology System (Becton Dickinson, North Ryde, New South Wales, Australia). Tube coagulase testing was also performed on each isolate using an established method (37). All phenotypic and genotypic testing was performed on-site at the Centre for Infectious Diseases and Microbiology.
Phenotypic testing.
Disk approximation testing was performed twice for each isolate according to the CLSI method using 2-μg clindamycin disks and 15-μg erythromycin disks placed on inoculated Mueller-Hinton agar (2). For the first test, disks were placed 15 mm apart edge to edge manually and, for the second, 22 mm apart using a six-disk disk dispenser (Bio-Rad, Hercules, California). A single 0.5 McFarland suspension in normal saline was used to inoculate both plates. Part of this suspension was also inoculated onto blood agar to ensure purity. Plates were incubated at 35°C in atmospheric conditions for 18 h. To minimize bias, plates containing the 15-mm D-test were identified by the accession number of the specimen only and placed in numerical order prior to reading while plates containing the 22-mm D-test were identified by an alphanumeric code and placed in alphabetical order. The D-tests were read independently by three observers using reflected light. Tests showing flattening of the clindamycin zone adjacent to the erythromycin disk were classified as D-test positive, while those with a circular zone were classified as D-test negative. The result recorded by two or more was taken as correct. Clindamycin zone sizes were recorded for all D-test-negative isolates as well as the last 84 consecutive isolates collected. Testing was performed in weekly batches, and Staphylococcus aureus ATCC BAA-976 (negative) and ATCC BAA-977 (positive) were included with each batch for quality control.
Genetic testing. (i) DNA extraction.
Single colonies from the susceptibility purity plate were used for extraction of bacterial DNA. Colonies were placed in 100 μl of digestion buffer (10 mM Tris-HCl [pH 8.0], 0.45% Triton X-100, 0.45% Tween 20), heated to 100°C for 10 min, frozen at −20°C, and then thawed and centrifuged. The supernatant containing DNA was then stored at −20°C until required for PCR.
(ii) PCR.
Genes encoding MLSB resistance (ermA, ermB, ermC, and ermTR) and MS resistance (msrA) as well as mecA (for methicillin resistance) and nuc (specific for S. aureus) were amplified in multiplex reactions using the primers indicated in Table 1. Reaction mixtures consisted of 5 μl of template, 12.5 pmol of each primer, 2.5 mM of each deoxynucleoside triphosphate, 2.5 μl 10× PCR buffer, 3 mM additional MgCl2 (final concentration, 4.5 mM), 0.5 U QIAGEN HotStart Taq polymerase, and water to make a final volume of 25 μl. Cycling conditions were 95°C for 15 min and then 35 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 60 s, followed by a final extension at 72°C for 10 min. A reverse line blot assay was performed as described elsewhere (38). Two oligonucleotide probes for each amplified gene were used (Table 1). A test was regarded as positive only if both probes for the gene were positive. If no erythromycin resistance gene was detected, repeat PCR using single oligonucleotide primer pairs was carried out and amplification products were detected using agarose gel electrophoresis.
TABLE 1.
Primers used in multiplex PCRs, and corresponding oligonucleotide probes used in the reverse line blot assay
| Gene | Primer: sequence | Probe: sequence |
|---|---|---|
| ermA | ermA/TRSb: 35690 TCA GGA AAA GGA CAT TTT ACC 35671 | ermAAp: 35647 TCG ACT CAT TTT GAC TAG CTC TT 35669 |
| ermAAb: 35320 ATA TAG TGG TGG TAC TTT TTT GAG C35344 | ermASp: 35387 GAG CTT TGG GTT TAC TAT TAA TGG 35364 | |
| ermB | ermBSb: 739 GGT AAA GGG CAT TTA ACG AC 758 | ermBAp: 787 TTA CCT GTT TAC TTA TTT TAG CCA G 763 |
| ermBAb: 1232 CGA TAT TCT CGA TTG ACC C 1214 | ermBSP: 1147 CTT ACC CGC CAT ACC ACA 1164 | |
| ermC | ermCsb: 655 CTTGTTGATCACGATAATTTCC 676 | ermCAp: 700 GCA ATA TAT CCT TGT TTA AAA CTT GG 675 |
| ermCab: 838 TAGCAAACCCGTATTCCACG 819 | ermCSp: 756 CATAAGTACGGATATAATACGCA 778 | |
| ermTR | ermA/TRSb: 35690 TCA GGA AAA GGA CAT TTT ACC 35671 | ermTRAp: 398 CCT TCA TCA ATC TCT ATA GCA TTC 375 |
| ermTRAb: 699 AAA ATA TGC TCG TGG CAC 682 | ermTRSp: 641 TGC TGT TAA TGG TGG AAA TG 660 | |
| msrA | msrASb: 2895 TCCAATCATTGCACAAAATC 2914 | msrAAp: 2938 TTT GAC TTC CTT TAA CCA ATG TTA G 2914 |
| msrAAb: 3058 CAATTCCCTCTATTTGGTGGT 3038 | msrASp: 3006 TGT AGG TAA GAC AAC TTT ACT TGA AGC 3032 | |
| nuc | nucSb: 511 GCGATTGATGGTGATACGGTT 531 | nucAp: 558 CATTGGTTGACCTTTGTACATTAA 535 |
| nucAb: 789 AGCCAAGCCTTGACGAACTAAAGC 766 | nucSp: 745 GATGGAAAAATGGTAAACGAAG 766 | |
| mecA | mecASb: 1190 TCC AGA TTA CAA CTT CAC CAG G 1211 | mecAAp: 1236 TGCTGTTAATATTTTTTGAGTTGAAC 1211 |
| mecAAb: 1357 CAT TTA CCA CTT CAT ATC TTG TAA CG 1332 | mecASp: 1309 GAT AAA TCT TGG GGT GGT TAC AAC 1332 |
Data analysis and statistics.
Data were analyzed using Microsoft Access 2000 and statistical calculations performed using Microsoft Excel 2000, Statistical Program for the Social Sciences (SPSS) for Windows, Confidence Interval Analysis for Windows (available at http://www.medschool.soton.ac.uk/cia), and Program for Reliability Assessment with Multiple Coders (PRAM). The Student t test, Mann-Whitney test, chi-squared test, and Cohen's kappa test were used where appropriate. The very major error rate was defined as the percentage of isolates carrying one or more erm genes that were incorrectly identified as having the MS phenotype by D-test (12).
RESULTS
Isolate collection.
During the study period, 1,012 S. aureus (632 methicillin susceptible [MSSA] and 380 methicillin resistant [MRSA]) and 139 coagulase-negative staphylococcus (CoNS) isolates were identified and tested for antibiotic susceptibility, excluding duplicate isolates from the same patient. Of these, 60 MSSA (9.5%), 103 MRSA (27.0%), and 68 CoNS (48.9%) isolates were erythromycin nonsusceptible but clindamycin susceptible by BMD and so were included in the study. All study isolates were erythromycin resistant (MIC ≥ 8 mg/liter) except one MSSA isolate which had intermediate susceptibility.
D-test.
Using 15-mm edge-to-edge disk separation, 180 of the 231 isolates were recorded as D-test positive, with 100% consensus between the three observers (kappa value of 1; Table 2). Using 22-mm disk separation, there was discordance between the three observers for 33 of 231 tests (14.3%; kappa value of 0.77; Table 2). All 51 of the isolates that were D-test negative at 15 mm were also D-test negative at 22 mm. However, of the 180 isolates that were D-test positive at 15 mm, 22 were said to be D-test negative at 22 mm (20 S. aureus isolates and 2 CoNS isolates). Control strains were read correctly on each occasion by all three observers at both distances.
TABLE 2.
Interobserver agreement for the D-testa
| Resultb | 15-mm disk separation
|
22-mm disk separation
|
||||
|---|---|---|---|---|---|---|
| No. (%) | No. of:
|
No. (%) | No of:
|
|||
| False negatives | False positives | False negatives | False positives | |||
| +++ | 180 (71.7) | NAc | 0 | 150 (64.9) | NA | 0 |
| ++− | 0 | NA | 0 | 8 (3.4) | NA | 0 |
| −−+ | 0 | 0 | NA | 25 (10.8) | 10 | NA |
| −−− | 51 (22.1) | 0 | NA | 48 (20.8) | 12 | NA |
| Total | 231 | 0 | 0 | 231 | 22 | 0 |
Genotyping was considered the gold standard.
+++, all observers read test as positive; ++−, positive reading for two observers, negative reading for one observer (considered a positive result); −−+, negative reading for two observers, positive reading for one observer (considered a negative result); −−−, all observers read test as negative.
NA, not applicable.
Genotyping.
The frequency of the various genotypes among the study isolates is shown in Table 3. No macrolide resistance gene was detectable in 1 of the 231 study isolates (0.4%), a MSSA isolate that showed iMLSB resistance by D-test. This is similar to the rate described in other studies (6, 19, 30). Possible explanations include mutations in the primer binding site and the presence of rare resistance mechanisms (21, 26, 30, 36).
TABLE 3.
Genotype frequency among MSSA, MRSA, and CoNS isolates, and D-test results for each genotype
| Genotype | No. (%) of isolates with genotype
|
No. of isolates with indicated D-test result
|
|||||
|---|---|---|---|---|---|---|---|
| MSSA | MRSA | CoNS | 15 mm
|
22 mm
|
|||
| Negative | Positive | Negative | Positive | ||||
| ermA | 42 (68.9) | 46 (45.1) | 2 (2.9) | 0 | 90 | 22 | 68 |
| ermA msrA | 1 (1.6) | 0 | 1 (1.5) | 0 | 2 | 0 | 2 |
| ermC | 14 (23.0) | 53 (52.0) | 20 (29.4) | 0 | 87 | 0 | 87 |
| msrA | 3 (4.9) | 3 (2.9) | 45 (66.2) | 51 | 0 | 51 | 0 |
| No gene amplified | 1 (1.6) | 0 | 0 | 0 | 1 | 0 | 1 |
| Total | 61 | 102 | 68 | 51 | 180 | 73 | 158 |
Using the presence of at least one erm gene as the “gold standard” for detection of inducible resistance and the presence of msrA in the absence of an erm gene as the negative gold standard, the 15-mm D-test had a 100% sensitivity (95% confidence interval [CI], 98.0% to 100%) and specificity (95% CI, 93.0% to 100%) while the 22-mm D-test had a sensitivity of 87.7% (95% CI, 82.9% to 92.5%) and specificity of 100% (95% CI, 93% to 100%). The very major error rate using 22 mm was 12.3% and was higher for MRSA isolates (18.2%) than for MSSA (3.5%) or CoNS (8.7%) isolates.
The median zone diameter for false-negative tests was 21 mm (range, 20 to 26 mm) versus 27 mm (range, 20 to 31 mm) for true negatives (P < 0.0001), indicating that false-negative results occur when the edge of the clindamycin zone is too far from the erythromycin disk for induction to occur. Likewise, ermA-containing isolates which produced false-negative results were more likely to have a smaller zone diameter than ermA-containing isolates which produced true-positive results (median diameter, 21 mm [range, 20 to 26 mm] versus 27 mm [range 20 to 37 mm]; P < 0.0001). For the sample of consecutive isolates, there were no significant differences in clindamycin zone sizes between the iMLSB and MS phenotypes or between the ermA and ermC genotypes.
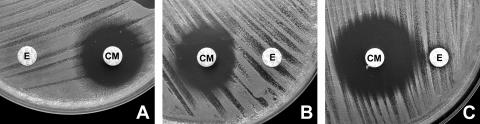
A “double zone” around the clindamycin disk was present in 9 of 22 (40.9%) false-negative D-tests at 22 mm compared with 3 of 51 (5.9%) true-negative tests (P = 0.0002). An example of the double zone and small zone size associated with false-negative tests is given in Fig. 1.
FIG. 1.
A. False-negative D-tests were more likely at 22 mm when the clindamycin zone diameter was smaller and showed a double zone of inhibition. B. The same isolate tested at 15 mm. C. D-test-negative isolate tested at 15 mm. E, erythromycin disk; CM, clindamycin disk.
Only ermA-containing isolates were read as false negative at 22 mm (22 of 92, 23.9%), while all 87 ermC-containing isolates were read correctly (P < 0.0001). Eighteen of the 22 (81.8%) false-negative isolates were MRSA isolates, while 2 were MSSA and 2 were CoNS.
DISCUSSION
This study confirmed a high frequency (96.3%) of iMLSB resistance among S. aureus isolates with an erythromycin-nonsusceptible, clindamycin-susceptible phenotype in an Australian laboratory. This high rate was observed in both MSSA and MRSA isolates, from community- and hospital-acquired infections, and from metropolitan and rural areas (data not shown). The frequency of iMLSB resistance in such isolates shows marked geographic variability, ranging from 7 to 100% in published studies (1, 4, 7-11, 14, 15, 18, 20, 22, 24, 25, 28, 29, 31-33). There are fewer published data for CoNS, but generally iMLSB resistance in erythromycin-nonsusceptible but clindamycin-susceptible CoNS isolates is less common than in S. aureus. Rates in published studies range from 28 to 84% (4, 7, 8, 11, 14, 28, 31).
It is important for laboratories to be aware of the local prevalence of iMLSB isolates so they can choose whether to perform the D-test routinely or whether to report all erythromycin-resistant Staphylococcus sp. isolates as also being clindamycin resistant. This prevalence may change over time with the emergence of strains with different sensitivity patterns, so periodic surveys should be performed if testing is not routine (1).
Currently in our laboratory, approximately 28 isolates of S. aureus would need to be tested to detect one for which clindamycin susceptibility could be reported. As a result, we perform the D-test only in special circumstances. Such circumstances include instances where clindamycin therapy is most likely to be useful, such as bone and joint infections and infections due to nonmultiresistant MRSA, particularly where the patient is known to have hypersensitivity to beta-lactams.
A number of authors suggest that a range of distances up to 28 mm may be used for performance of the D-test, and the CLSI states that edge-to-edge distances of 15 to 26 mm may be used (3). The greater distance is more convenient since automated disk dispensers generally place disks 20 to 26 mm apart. We chose to compare the minimum distance recommended by the CLSI (15 mm) to the distance produced by automated dispensers used in our laboratory (22 mm) since there is conflicting opinion about the accuracy of the D-test when distances above 20 mm are used (7, 13, 34). One study which validated a distance of 26 mm found that all but one of 59 Staphylococcus sp. isolates with iMLSB resistance were positive by D-test using 26 mm and that all were positive using 20 mm and 15 mm (7). The low rate of very major errors in that study may relate to the nature of resistance determinants in local strains: only 8 of 28 iMLSB S. aureus isolates genotyped in the study harbored ermA, and the remainder harbored ermC, which we found to be less associated with false-negative D-test results. Furthermore, since all three distances were compared on a single plate, the possibility of observer bias cannot be excluded.
The present study has found that the D-test using a 22-mm distance is inaccurate, with a sensitivity of only 87.7% when compared with genotyping, and a very major error rate of 12.3% (18.2% for MRSA isolates). Moreover, the results are more difficult to read, with a significant level of disagreement between observers, which was not apparent when the shorter separation distance was used. We recommend that a distance of ≤15 mm should be used for disk approximation testing.
Acknowledgments
Many thanks to Heather Gidding for statistical analysis; Stella Pendle, Elaine Cheong, Elaine Finch, Julie Sheedy, Maureen Lynch, Marion Yuen, and all staff of the bacteriology laboratory at CIDM for assistance in collection of isolates and reading of D-tests; and Zhongsheng Tong for assistance with genotyping.
Footnotes
Published ahead of print on 27 September 2006.
REFERENCES
- 1.Chavez-Bueno, S., B. Bozdogan, K. Katz, K. L. Bowlware, N. Cushion, D. Cavuoti, N. Ahmad, G. H. McCracken, Jr., and P. C. Appelbaum. 2005. Inducible clindamycin resistance and molecular epidemiologic trends of pediatric community-acquired methicillin-resistant Staphylococcus aureus in Dallas, Texas. Antimicrob. Agents Chemother. 49:2283-2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clinical and Laboratory Standards Institute. 2003. Performance standards for antimicrobial disk susceptibility tests. Approved standard M2-A8 (ISBN 1-56238-485-6), 8th ed. Clinical and Laboratory Standards Institute, Wayne, Pa.
- 3.Clinical and Laboratory Standards Institute. 2005. Performance standards for antimicrobial susceptibility testing. Fifteenth international supplement M100-S15, p. 44-51. Clinical and Laboratory Standards Institute, Wayne, Pa.
- 4.Delialioglu, N., G. Aslan, C. Ozturk, V. Baki, S. Sen, and G. Emekdas. 2005. Inducible clindamycin resistance in staphylococci isolated from clinical samples. Jpn. J. Infect. Dis. 58:104-106. [PubMed] [Google Scholar]
- 5.Drinkovic, D., E. R. Fuller, K. P. Shore, D. J. Holland, and R. Ellis-Pegler. 2001. Clindamycin treatment of Staphylococcus aureus expressing inducible clindamycin resistance. J. Antimicrob. Chemother. 48:315-316. [DOI] [PubMed] [Google Scholar]
- 6.Eady, E. A., J. I. Ross, J. L. Tipper, C. E. Walters, J. H. Cove, and W. C. Noble. 1993. Distribution of genes encoding erythromycin ribosomal methylases and an erythromycin efflux pump in epidemiologically distinct groups of staphylococci. J. Antimicrob. Chemother. 31:211-217. [DOI] [PubMed] [Google Scholar]
- 7.Fiebelkorn, K. R., S. A. Crawford, M. L. McElmeel, and J. H. Jorgensen. 2003. Practical disk diffusion method for detection of inducible clindamycin resistance in Staphylococcus aureus and coagulase-negative staphylococci. J. Clin. Microbiol. 41:4740-4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fokas, S., M. Tsironi, M. Kalkani, and M. Dionysopouloy. 2005. Prevalence of inducible clindamycin resistance in macrolide-resistant Staphylococcus spp. Clin. Microbiol. Infect. 11:337-340. [DOI] [PubMed] [Google Scholar]
- 9.Frank, A. L., J. F. Marcinak, P. D. Mangat, J. T. Tjhio, S. Kelkar, P. C. Schreckenberger, and J. P. Quinn. 2002. Clindamycin treatment of methicillin-resistant Staphylococcus aureus infections in children. Pediatr. Infect. Dis. J. 21:530-534. [DOI] [PubMed] [Google Scholar]
- 10.Gadepalli, R., B. Dhawan, S. Mohanty, A. Kapil, B. K. Das, and R. Chaudhry. 2006. Inducible clindamycin resistance in clinical isolates of Staphylococcus aureus. Indian J. Med. Res. 123:571-573. [PubMed] [Google Scholar]
- 11.Hamilton-Miller, J. M., and S. Shah. 2000. Patterns of phenotypic resistance to the macrolide-lincosamide-ketolide-streptogramin group of antibiotics in staphylococci. J. Antimicrob. Chemother. 46:941-949. [DOI] [PubMed] [Google Scholar]
- 12.Hindler, J. F., and S. Munro. 2004. Evaluating antimicrobial susceptibility test systems, p. 5.17.1-5.17.11. In H. D. Isenberg (ed.), Clinical microbiology procedures handbook, vol. 2. ASM Press, Washington, D.C. [Google Scholar]
- 13.Jenssen, W. D., S. Thakker-Varia, D. T. Dubin, and M. P. Weinstein. 1987. Prevalence of macrolides-lincosamides-streptogramin B (sic) resistance and erm gene classes among clinical strains of staphylococci and streptococci. Antimicrob. Agents Chemother. 31:883-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kader, A. A., A. Kumar, and A. Krishna. 2005. Induction of clindamycin resistance in erythromycin-resistant, clindamycin susceptible and methicillin-resistant clinical staphylococcal isolates. Saudi Med. J. 26:1914-1917. [PubMed] [Google Scholar]
- 15.Kim, H. B., B. Lee, H. C. Jang, S. H. Kim, C. I. Kang, Y. J. Choi, S. W. Park, B. S. Kim, E. C. Kim, M. D. Oh, and K. W. Choe. 2004. A high frequency of macrolide-lincosamide-streptogramin resistance determinants in Staphylococcus aureus isolated in South Korea. Microb. Drug Resist. 10:248-254. [DOI] [PubMed] [Google Scholar]
- 16.Leclercq, R. 2002. Mechanisms of resistance to macrolides and lincosamides: nature of the resistance elements and their clinical implications. Clin. Infect. Dis. 34:482-492. [DOI] [PubMed] [Google Scholar]
- 17.Leclercq, R., and P. Courvalin. 1991. Bacterial resistance to macrolide, lincosamide, and streptogramin antibiotics by target modification. Antimicrob. Agents Chemother. 35:1267-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levin, T. P., B. Suh, P. Axelrod, A. L. Truant, and T. Fekete. 2005. Potential clindamycin resistance in clindamycin-susceptible, erythromycin-resistant Staphylococcus aureus: report of a clinical failure. Antimicrob. Agents Chemother. 49:1222-1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lim, J. A., A. R. Kwon, S. K. Kim, Y. Chong, K. Lee, and E. C. Choi. 2002. Prevalence of resistance to macrolide, lincosamide and streptogramin antibiotics in gram-positive cocci isolated in a Korean hospital. J. Antimicrob. Chemother. 49:489-495. [DOI] [PubMed] [Google Scholar]
- 20.Marr, J. K., A. T. Lim, and L. G. Yamamoto. 2005. Erythromycin-induced resistance to clindamycin in Staphylococcus aureus. Hawaii Med. J. 64:6-8. [PubMed] [Google Scholar]
- 21.Matsuoka, M., K. Endou, H. Kobayashi, M. Inoue, and Y. Nakajima. 1998. A plasmid that encodes three genes for resistance to macrolide antibiotics in Staphylococcus aureus. FEMS Microbiol. Lett. 167:221-227. [DOI] [PubMed] [Google Scholar]
- 22.Navaneeth, B. V. 2006. A preliminary in vitro study on inducible and constitutive clindamycin resistance in Staphylococcus aureus from a south Indian tertiary care hospital. Int. J. Infect. Dis. 10:184-185. [DOI] [PubMed] [Google Scholar]
- 23.Panagea, S., J. D. Perry, and F. K. Gould. 1999. Should clindamycin be used as treatment of patients with infections caused by erythromycin-resistant staphylococci? J. Antimicrob Chemother. 44:581-582. [DOI] [PubMed] [Google Scholar]
- 24.Patel, M., K. B. Waites, S. A. Moser, G. A. Cloud, and C. J. Hoesley. 2006. Prevalence of inducible clindamycin resistance among community- and hospital-associated Staphylococcus aureus isolates. J. Clin. Microbiol. 44:2481-2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pimentel, J. D., and G. D. Lum. 2006. Use of the D-test to determine the prevalence of inducible clindamycin-resistant Staphylococcus aureus in a Northern Territory hospital. Pathology 38:258-259. [DOI] [PubMed] [Google Scholar]
- 26.Prunier, A. L., B. Malbruny, D. Tande, B. Picard, and R. Leclercq. 2002. Clinical isolates of Staphylococcus aureus with ribosomal mutations conferring resistance to macrolides. Antimicrob. Agents Chemother. 46:3054-3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rayner, C., and W. J. Munckhof. 2005. Antibiotics currently used in the treatment of infections caused by Staphylococcus aureus. Intern. Med. J. 35(Suppl. 2):S3-S16. [DOI] [PubMed] [Google Scholar]
- 28.Sanchez, M. L., K. K. Flint, and R. N. Jones. 1993. Occurrence of macrolide-lincosamide-streptogramin resistances among staphylococcal clinical isolates at a university medical center. Is false susceptibility to new macrolides and clindamycin a contemporary clinical and in vitro testing problem? Diagn. Microbiol. Infect. Dis. 16:205-213. [DOI] [PubMed] [Google Scholar]
- 29.Sattler, C. A., E. O. Mason, Jr., and S. L. Kaplan. 2002. Prospective comparison of risk factors and demographic and clinical characteristics of community-acquired, methicillin-resistant versus methicillin-susceptible Staphylococcus aureus infection in children. Pediatr. Infect. Dis. J. 21:910-917. [DOI] [PubMed] [Google Scholar]
- 30.Schmitz, F. J., J. Petridou, A. C. Fluit, U. Hadding, G. Peters, and C. von Eiff. 2000. Distribution of macrolide-resistance genes in Staphylococcus aureus blood-culture isolates from fifteen German university hospitals. Multicentre Study on Antibiotic Resistance in Staphylococci. Eur. J. Clin. Microbiol. Infect. Dis. 19:385-387. [DOI] [PubMed] [Google Scholar]
- 31.Schreckenberger, P. C., E. Ilendo, and K. L. Ristow. 2004. Incidence of constitutive and inducible clindamycin resistance in Staphylococcus aureus and coagulase-negative staphylococci in a community and a tertiary care hospital. J. Clin. Microbiol. 42:2777-2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Siberry, G. K., T. Tekle, K. Carroll, and J. Dick. 2003. Failure of clindamycin treatment of methicillin-resistant Staphylococcus aureus expressing inducible clindamycin resistance in vitro. Clin. Infect. Dis. 37:1257-1260. [DOI] [PubMed] [Google Scholar]
- 33.Spiliopoulou, I., E. Petinaki, P. Papandreou, and G. Dimitracopoulos. 2004. erm(C) is the predominant genetic determinant for the expression of resistance to macrolides among methicillin-resistant Staphylococcus aureus clinical isolates in Greece. J. Antimicrob. Chemother. 53:814-817. [DOI] [PubMed] [Google Scholar]
- 34.Steward, C. D., P. M. Raney, A. K. Morrell, P. P. Williams, L. K. McDougal, L. Jevitt, J. E. McGowan, Jr., and F. C. Tenover. 2005. Testing for induction of clindamycin resistance in erythromycin-resistant isolates of Staphylococcus aureus. J. Clin. Microbiol. 43:1716-1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Werckenthin, C., S. Schwarz, and H. Westh. 1999. Structural alterations in the translational attenuator of constitutively expressed ermC genes. Antimicrob. Agents Chemother. 43:1681-1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wondrack, L., M. Massa, B. V. Yang, and J. Sutcliffe. 1996. Clinical strain of Staphylococcus aureus inactivates and causes efflux of macrolides. Antimicrob. Agents Chemother. 40:992-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.York, M. K., M. M. Traylor, J. Hardy, and M. Henry. 2004. Coagulase test-rapid plasma method, p. 3.17.14.1-3.17.14.3. In H. D. Isenberg (ed.), Clinical microbiology procedures handbook, vol. 1. ASM Press, Washington, D.C. [Google Scholar]
- 38.Zeng, X., F. Kong, H. Wang, A. Darbar, and G. L. Gilbert. 2006. Simultaneous detection of nine antibiotic resistance-related genes in Streptococcus agalactiae using multiplex PCR and reverse line blot hybridization assay. Antimicrob. Agents Chemother. 50:204-209. [DOI] [PMC free article] [PubMed] [Google Scholar]