Abstract
Since the announcement of the WHO program for the global eradication of poliomyelitis and the establishment of epidemiological and virological surveillance, the emergence and circulation of pathogenic vaccine-derived polioviruses (VDPV) presenting >1% nucleotide divergence from the sequence of the original vaccine strain have been demonstrated in certain regions. We developed and used a multiple restriction fragment length polymorphism (RFLP) method to investigate the frequency of these VDPV in a population with a high level of oral poliovirus vaccine coverage in northwestern Russia. Modified RFLP profiles were found to be strongly correlated with the presence of mutations and recombination events in vaccine strains. We found that a substantial proportion of vaccine strains had high percentages of nucleotide substitutions (>0.5%), including a type 3 VDPV with 1.4% nucleotide divergence. These findings indicate that VDPV or pre-VDPV strains are not rare in certain populations with high levels of vaccine coverage. The multiple RFLP method appears to be a simple and rapid tool for monitoring such strains, which could jeopardize the benefits of the eradication program.
The oral poliovirus vaccine (OPV) is widely used in routine immunization programs. The World Health Organization (WHO) decided to use OPV as the principal means of vaccination for the global eradication of poliomyelitis because this vaccine limits the circulation of wild polioviruses. This vaccine, prepared with attenuated Sabin strains of all three serotypes (Sabin 1, 2, and 3 strains), induces durable intestinal and humoral immune responses and is a safe means of protecting against infection with wild polioviruses. OPV has been used successfully in mass vaccination campaigns. Thanks to WHO strategies, including national immunization days, subnational immunization days, and “mopping-up” strategies, considerable progress has been made towards the goal of stopping wild poliovirus transmission. Three WHO regions—the Americas and the Western Pacific and European regions—have been certified free of wild poliovirus. Only a small number of countries, including India, Pakistan, Afghanistan, Egypt, Nigeria, and Niger, still have endemic poliovirus (28).
However, the genetic variability of live poliovirus vaccine strains may lead to the formation and excretion into the environment of new variants of polioviruses differing from the original Sabin strains. Two natural evolution mechanisms, i.e., nucleotide substitution (mutation) and molecular recombination, generate genetic variation in polioviruses. The poliovirus genome is a positive single-stranded RNA genome comprising about 7,500 nucleotides. Most of the poliovirus RNA, with the exception of two untranslated regions at the extremities (5′ untranslated region [5′UTR] and 3′UTR), is an open reading frame encoding a single large polyprotein that is cleaved into structural capsid proteins (from VP1 to VP4) and nonstructural proteins, including the 3D protein, the RNA-dependent RNA polymerase. The high frequency of errors of this RNA polymerase, which introduces nucleotide mutations with a mean frequency of about 10−4, together with a lack of proofreading mechanisms, may result in the accumulation of nucleotide substitutions during viral replication.
The simultaneous administration of three poliovirus serotypes in the OPV may lead to multiple infection of intestinal cells, facilitating intermolecular recombination between the genomes of the three serotypes of polioviruses used for vaccination and the emergence of intertypic recombinant strains. Such recombinants have frequently been reported for patients with vaccine-associated paralytic poliomyelitis (VAPP) (11, 12, 19, 23) as well as for healthy vaccinated subjects (5, 10, 25).
The genetic instability of OPV strains is one of the most serious disadvantages of this vaccine. The extensive use of OPV may, under certain conditions, lead to the development of VAPP in rare patients. VAPP has been observed in unvaccinated or incompletely immunized children living in close contact with recently vaccinated children and in vaccinated subjects. Paralysis may occur following primary vaccination of immunocompetent subjects at a frequency of 1 case per 750,000 subjects vaccinated (29). Immunodeficient patients are particularly at risk following vaccination with OPV, as they may go on to develop VAPP and may excrete vaccine-derived poliovirus strains over long periods of time. For such individuals, the WHO recommends vaccination with an inactivated poliovirus vaccine. In most cases, poliovirus vaccine strains are excreted by immunocompetent children over a period of 3 to 4 weeks following primary immunization with OPV and over a shorter period following subsequent OPV administration (1). The prolonged replication of vaccine-derived poliovirus strains (VDPV; OPV strains presenting >1% nucleotide divergence from the sequence of the original vaccine strain) for more than 1 year in patients with immunodeficiency has been reported in various countries (3, 8, 13, 21, 22, 26, 27).
In addition, VDPV have been shown to circulate for several years in the last phases of poliomyelitis eradication. Outbreaks of acute flaccid paralysis (AFP) caused by circulating VDPV strains were observed in various geographical areas in communities with low levels of or gaps in vaccine coverage. Three outbreaks of poliomyelitis associated with circulating VDPV of serotype 1 or 2 with 1.9% to 3.0% nucleotide substitutions were registered from 2000 to 2002, in the Dominican Republic and Haiti, in the Philippines, and in Madagascar (31). According to the reported molecular clock of poliovirus strains, with about 1% nucleotide substitutions per year (3, 13, 24), these VDPV had circulated or multiplied for about 22 to 36 months. The prolonged circulation of type 2 VDPV, responsible for 30 cases of AFP from 1983 to 1993, has been demonstrated in Egypt (39). The VDPV strains isolated in these outbreaks were found to have recombined with other enterovirus strains that probably belonged to the coxsackie A viruses of enterovirus species C.
The risk of VDPV emergence, environmental dissemination, and prolonged circulation in certain population groups also exists elsewhere. This risk, complicating the task of eradicating poliomyelitis worldwide, has encouraged the WHO Polio Laboratory Network, which is responsible for extensive virological surveillance, to look for ways of detecting VDPV early. The goal of the WHO program for the global eradication of poliomyelitis is to stop the transmission of wild polioviruses by vaccination with OPV and then, once wild polioviruses have been eliminated throughout the world, to stop immunization against poliomyelitis. Determining when to stop immunization is a difficult issue requiring further research, including studies of VDPV emergence and evaluation of the frequency of VDPV circulation in countries with different epidemiological situations and levels of vaccine coverage.
The objective of this study was to devise a simple method for rapidly detecting mutant and recombinant poliovirus strains and to use this method to determine the frequency of VDPV strain circulation in northwestern Russia. We used a multiple restriction fragment length polymorphism (RFLP) method because this method does not require nucleotide sequencing and is thus accessible to most laboratories involved in poliomyelitis and AFP surveillance. Modified RFLP profiles were found to be strongly correlated with the presence of a certain number of nucleotide substitutions. All strains with modified restriction profiles for one or two of the analyzed genomic regions encoding capsid proteins had numerous mutations, and 12.8% of the strains studied had high percentages of nucleotide substitutions (>0.5% nucleotide substitutions). This indicates the frequent circulation or multiplication of vaccine strains for more than 6 months in this population of individuals with a high level of vaccine coverage in northwestern Russia.
MATERIALS AND METHODS
Strains.
Since 1998, the Saint Petersburg Subnational Poliomyelitis Laboratory has conducted surveillance for poliomyelitis and acute flaccid paralysis in 14 administrative territories of Russia with a population of over 25 million. During the period from 1998 to 2002, the laboratory investigated 1,040 stool samples from 424 patients with AFP and VAPP and from 191 healthy children in contact with AFP and VAPP patients.
VAPP was diagnosed for patients with clinical symptoms of typical poliomyelitis with residual paralysis on day 60 after the onset of paralysis and from whom polioviruses genetically very similar to the Sabin vaccine strain were isolated. Other cases were classified as AFP because of the absence of residual paralysis.
From all these samples, 97 poliovirus strains of different serotypes were isolated, and 39 were chosen for this study based on predefined criteria (described in Results).
Virus isolation and antigen characterization.
Viruses were isolated in accordance with standard WHO procedures (37) from three cell cultures, including human rhabdomyosarcoma (RD) and epidermoid carcinoma (HEp-2c) cells and murine L20B cells. L20B cells, which are mouse L cells genetically engineered to express the human poliovirus receptor, are used for the specific isolation of polioviruses (30). The isolated polioviruses were identified by seroneutralization with type-specific neutralizing antisera supplied by the Specialized Poliomyelitis Laboratory, National Institute of Public Health and the Environment, Bilthoven, The Netherlands. This method, based on pools of different sera, can be used to identify strain serotypes and to separate mixtures of polioviruses of different serotypes.
Intratypic differentiation of polioviruses was carried out according to WHO recommendations by the Regional Reference Laboratory for Poliomyelitis (M. P. Chumakov Institute of Poliomyelitis and Viral Encephalitidis, Moscow, Russia), using two methods, namely, an enzyme-linked immunosorbent assay (ELISA) with cross-adsorbed polyclonal antisera and PCR with Sabin strain-specific primers (36, 38). Almost all strains were classified as vaccine-like strains. Only one strain, of serotype 1, was characterized as non-Sabin-like by ELISA; two strains of serotype 3 were not neutralized by either serum (nonreactive), and two strains were neutralized by both sera (double reactive) (37). Reverse transcription-PCR (RT-PCR) with Sabin-specific primers indicated that these four strains, like the other strains studied, were of vaccine origin.
Intratypic differentiation using MAbs.
A third method for the intratypic differentiation of polioviruses was also used. This method was based on the use of monoclonal antibodies (MAbs) specific for wild or vaccine types of poliovirus (9). A microneutralization assay with a panel of MAbs (1o and Io, 2o and IIa, and 3o and IIIo, specific for wild and vaccine types of polioviruses of serotypes 1, 2, and 3, respectively), obtained from the Specialized Poliomyelitis Laboratory at the Institut Pasteur, Paris, France, was carried out. Serial 10-fold dilutions of the tested poliovirus strains and of the reference strains were prepared. We dispensed 50-μl aliquots of diluted MAbs (the dilution was determined according to the neutralization results obtained with reference strains used as virus controls) into 96-well microtiter plates. Fifty microliters of diluted virus was added, and the plates were incubated at 37°C for 2 h. Following the addition of a suspension of HEp-2c cells, plates were incubated at 37°C until a cytopathic effect developed for virus controls. The strains studied were considered Sabin-like or non-Sabin-like according to their neutralization indexes (NIs), based on the difference between log virus titers in the presence and absence of MAbs (NIs of ≥3.00 were considered positive). Strains were classified as Sabin-like or non-Sabin-like according to which specific MAbs gave a positive NI. Strains were considered nonneutralized if they were not neutralized by either of the MAbs.
RT-PCR.
Viral RNA was reverse transcribed directly from the cell-free supernatant of infected cells. A mixture of 2 μl of viral suspension, 10 pmol of antisense primer, 0.5 μl of RNasin (40 U/μl; Promega), and distilled water was heated at 80°C for 5 min for denaturation, and annealing was allowed to occur at 50°C for 5 min. Reverse transcription was performed in a reaction volume of 20 μl at 50°C for 30 min after the addition of 1 U of avian myeloblastosis virus reverse transcriptase (Promega), 4 μl of 5× RT buffer, and 1 μl of a mixture containing a 10 mM concentration of each deoxynucleoside triphosphate. The mixture was inactivated by heating at 95°C for 5 min and placed immediately on ice. We then added 3 μl of 10× PCR buffer, 10 pmol of sense primer, 1.25 U of Taq polymerase (Quantum), and distilled water to a final volume of 100 μl and carried out PCR (30 cycles of denaturation at 95°C for 20 s, annealing at 45°C for 1 min, and elongation at 70°C for 1 min). The last elongation step was extended to 10 min. PCR products were quantified by electrophoresis on ethidium bromide-stained 1.5% agarose gels and were visualized under UV light.
Site-specific PCR.
Site-specific PCR was used as described by Guillot et al. to detect point mutations in the critical nucleotide positions implicated in the attenuation of OPV strains (position 480 for type 1, position 481 for type 2, and position 472 for type 3) (15). Briefly, the generic antisense primer UC53 (with a sequence corresponding to residues 577 to 595 of the serotype 1 Sabin strain) was used for reverse transcription. The targeted nucleotide position was then analyzed by PCR with the generic sense primer UG52 (with a sequence corresponding to residues 162 to 182 of the type 1 Sabin strain) and site-specific antisense primers differing by one residue at the 3′ end for identification of the critical position corresponding to the wild type or the attenuated type. An internal control was included in each reaction by adding the generic primer UC53. For each strain, two site-specific PCRs were carried out in parallel, with either the alternative wild-type or the attenuated-type primer. The PCR products were visualized by electrophoresis in 2% agarose gels.
Multiple RFLP analysis.
Three different genomic regions of the poliovirus genome, namely, VP3-VP1 (nucleotide positions 1913 to 2881 according to Sabin 1 numbering), VP1-2A (nucleotides 2870 to 3648), and 3D-3′UTR (nucleotides 6536 to 7441), were studied by RFLP analysis. These three regions were amplified by RT-PCR, using the oligonucleotide primer pairs UG24 and UC1, UG19 and UC13, and UG17 and UC10, respectively. All of these primers have been described in previous studies (2, 15). Reactions were performed as described above, except that UC10 and UG17 were used at a concentration of 50 pmol/μl for RT and PCRs. PCR products (18 μl of each reaction mix) were digested for 2 h with 10 U (each) of restriction enzymes at the appropriate temperature and in the appropriate buffer. For each PCR product, four restriction enzymes—DdeI, DpnII, RsaI, and HinfI—were used. Restriction profiles of the studied strains and of the reference OPV strains were visualized and compared by ethidium bromide staining after electrophoresis in 3% agarose gels at 5 V/cm for 2 h in Tris-acetate-EDTA buffer and visualization under UV light.
Each of the four restriction enzymes used recognizes a four-nucleotide sequence. Thus, a large number of putative sites (three nucleotides in the right positions) can give rise to a new restriction site following a single nucleotide substitution. For example, the cumulative numbers of putative sites (for the four enzymes) in the VP3-VP1 fragments of the Sabin 1, 2, and 3 strains are 195, 178, and 188, respectively. However, only one of the three possible substitutions can generate a new restriction site. Therefore, searches for new sites for these four enzymes correspond to checking for nucleotide substitutions at 65, 59, and 62 nucleotide positions, respectively. Moreover, these four enzymes recognize 12, 9, and 14 sites in the VP3-VP1 fragments of the three OPV strains. A single substitution occurring at one of the four nucleotide positions of a restriction site leads to the disappearance of that site. Therefore, searches for the disappearance of restriction sites in this fragment require the checking of 48, 36, and 56 nucleotide positions, respectively. In conclusion, comparative RFLP analysis of the VP3-VP1 fragment (or another fragment of similar length) of a studied strain and its reference OPV strain using these four restriction enzymes corresponds to the checking of about 100 nucleotide positions for possible substitutions.
Nucleotide sequence analysis.
Nucleotide sequencing of PCR products was performed with the same primers used for RT-PCR and with internal primers UG1 and UC11 (15). PCR products were sequenced following purification with a Qiaquick spin column purification kit (QIAGEN) directly after amplification. Sequencing reactions were performed with a BigDye Terminator cycle sequencing ready reaction kit (version 1.1) according to the recommendations of Applied Biosystems. Nucleotide sequences were aligned and compared using the Clustal W program (35).
RESULTS
We used a multiple RFLP method to investigate the possible long-term circulation of poliovirus vaccine strains in northwestern Russia by analyzing the genetic drift of OPV strains isolated during poliomyelitis surveillance activities.
The 39 studied strains were isolated from 23 children who could be classified into three groups according to epidemiological factors. Ten of these strains were isolated from patients with VAPP, 25 were isolated from patients with AFP, and 4 were isolated from healthy children living in close contact with children who had VAPP (Table 1). The 39 poliovirus strains studied were isolated from vaccinated children who had received their last dose of OPV more than 1 month before isolation (one of these children received an inactivated vaccine) or from nonvaccinated children. For most of the immunized children, poliovirus strains were isolated 1 or 2 months after administration of the last dose of OPV. However, for six of the immunized children, the interval between vaccination and poliovirus isolation was longer (3 to 8 months for five of these children and 2 years for the remaining child). Eleven of the 39 strains studied belonged to serotype 1, 13 strains belonged to serotype 2, and 15 strains belonged to serotype 3.
TABLE 1.
Poliovirus strains and epidemiological data for patients and healthy children
| Straina | Yr of isolation | Case classification | OPV history of the caseb | Time after vaccination (mo) |
|---|---|---|---|---|
| A3-VO225 | 1998 | AFP | 3 OPV | 1.5 |
| B2-SM227 | 1998 | AFP | >3 OPV | 1 |
| C2-SP242 | 1998 | AFP | >3 OPV | 6 |
| D1-AR260 | 1998 | VAPP | Nonvaccinated | |
| E3-SP328 | 1999 | AFP | >3 OPV | 8 |
| F1-SP335 | 1999 | AFP | 1 IPV | 3 |
| F3-SP335 | 1999 | AFP | 1 IPV | 3 |
| F1-SP336 | 1999 | AFP | 1 IPV | 3 |
| G1-SP403 | 1999 | AFP | >3 OPV | 3 |
| H1-SP411 | 1999 | AFP | >3 OPV | 24 |
| H1-SP412 | 1999 | AFP | >3 OPV | 24 |
| I2-SP471 | 1999 | AFP | 3 OPV | 1 |
| J1-SP474 | 1999 | AFP | 3 OPV | 1 |
| J2-SP474 | 1999 | AFP | 3 OPV | 1 |
| J3-SP474 | 1999 | AFP | 3 OPV | 1 |
| K1-SP476 | 1999 | AFP | >3 OPV | 1 |
| K1-SP477 | 1999 | AFP | >3 OPV | 1 |
| K2-SP477 | 1999 | AFP | >3 OPV | 1 |
| L3-AR488 | 1999 | AFP | >3 OPV | 1.5 |
| M3-KR494 | 1999 | AFP | >3 OPV | 1.5 |
| N1-PS525 | 1999 | AFP | >3 OPV | 2 |
| N2-PS525 | 1999 | AFP | >3 OPV | 2 |
| N3-PS525 | 1999 | AFP | >3 OPV | 2 |
| O2-KO532 | 1999 | AFP | >3 OPV | 5 |
| P3-PS573 | 1999 | AFP | >3 OPV | 1 |
| Q1-AR577 | 1999 | AFP | 1 OPV | 1.5 |
| R3-PS607 | 2000 | VAPP | 3 OPV | 2 |
| S3-VO610 | 2000 | VAPP | Nonvaccinated | |
| S3-VO611 | 2000 | VAPP | Nonvaccinated | |
| T2-SP681 | 2000 | VAPP | 1 OPV | 1.5 |
| T3-SP681 | 2000 | VAPP | 1 OPV | 1.5 |
| T2-SP682 | 2000 | VAPP | 1 OPV | 1.5 |
| T3-SP682 | 2000 | VAPP | 1 OPV | 1.5 |
| U2-MU876 | 2001 | VAPP | Nonvaccinated | |
| U2-MU877 | 2001 | VAPP | Nonvaccinated | |
| V2-MU905 | 2001 | Contact with VAPP patient | 2 OPV | 1 |
| V2-MU906 | 2001 | Contact with VAPP patient | 2 OPV | 1 |
| W3-MU911 | 2001 | Contact with VAPP patient | 2 OPV | 2 |
| W3-MU912 | 2001 | Contact with VAPP patient | 2 OPV | 2 |
In the name of each strain, the first letter corresponds to a laboratory code, the first number corresponds to the serotype of the strain, the two following letters indicate a given territory in the region, and the last numbers are the laboratory numbers.
IPV, inactivated vaccine.
Intratypic differentiation of these strains was carried out according to WHO recommendations, using ELISA and RT-PCR methods with cross-adsorbed antisera and specific primers, respectively. All strains reacted similarly to the corresponding OPV strains and were classified as Sabin-like on the basis of RT-PCR. ELISA demonstrated antigen modification in five of the strains. We also used a third method based on seroneutralization experiments with specific MAbs (Table 2) (9). Most strains reacted like OPV strains (Sabin-like). However, two strains were neutralized more efficiently by MAbs specific for wild-type strains than by MAbs specific for OPV strains (classified as non-Sabin-like). Four strains were not neutralized by any MAb (classified as nonneutralized).
TABLE 2.
Characteristics of poliovirus strains isolated from clinical samples
| Strain | Intratypic differentiation with MAbsa | RFLP profile(s)c
|
% Nucleotide sequence identity for VP3-VP1 and VP1-2Ae | ||
|---|---|---|---|---|---|
| VP3- VP1 | VP1- 2A | 3D-3′UTRd | |||
| Type 1 strains | |||||
| D1-AR260 | NSLb | 2 | 99.3 | ||
| F1-SP335 | SL | 100 | |||
| F1-SP336 | SL | 100 | |||
| G1-SP403 | SL | 99.9 | |||
| H1-SP411 | SL | 99.9 | |||
| H1-SP412 | SL | NS | |||
| J1-SP474 | SL | NS | |||
| K1-SP476 | SL | NS | |||
| K1-SP477 | SL | NS | |||
| N1-PS525 | SL | 99.9 | |||
| Q1-AR577 | NN | 1 | 99.7 | ||
| Type 2 strains | |||||
| B2-SM227 | SL | NS | |||
| C2-SP242 | NN | 100 | |||
| I2-SP471 | SL | NS | |||
| J2-SP474 | SL | 1 | 99.2 | ||
| K2-SP477 | SL | 1 | NS | ||
| N2-PS525 | SL | S3 | 99.8 | ||
| O2-KO532 | SL | S3 | 99.7 | ||
| T2-SP681 | NN | S2 and S1 | 99.7 | ||
| T2-SP682 | SL | 1 | 1 | S2, S1, and S3 | 99.7 |
| U2-MU876 | NN | S1 | 99.7 | ||
| U2-MU877 | SL | S1 | 99.8 | ||
| V2-MU905 | SL | 1 | S3 | NS | |
| V2-MU906 | SL | 1 | S3 | 99.5 | |
| Type 3 strains | |||||
| A3-VO225 | SL | 99.9 | |||
| E3-SP328 | SLb | 99.9 | |||
| F3-SP335 | SL | 100 | |||
| J3-SP474 | SL | 100 | |||
| L3-AR488 | SL | 100 | |||
| M3-KR494 | SL | 2 | 99.8 | ||
| N3-PS525 | SL | 1 | 99.8 | ||
| P3-PS573 | SLb | 2 | 99.7 | ||
| R3-PS607 | NSL | 1 | 1 | 98.6 | |
| S3-VO610 | SL | 99.8 | |||
| S3-VO611 | SL | 99.8 | |||
| T3-SP681 | SL | 2 | 99.7 | ||
| T3-SP682 | SL | 1 | 99.7 | ||
| W3-MU911 | SLb | 1 | S1 | NS | |
| W3-MU912 | SLb | 1 | S1 | 99.4 | |
NSL, non-Sabin-like; SL, Sabin-like; NN, nonneutralized (see Materials and Methods).
Antigenically modified according to ELISA.
All strains were analyzed, but only those with modified profiles are indicated, as follows: 1, modified for one of four enzymes; 2, modified for two of four enzymes.
Only profiles or mixtures of profiles corresponding to serotypes of OPV strains different from those determined by serotyping are indicated.
Percentages of nucleotide identity with the corresponding OPV reference strains are indicated. Nonidentity of >0.5% is shown in bold. NS, not sequenced.
We then applied the site-specific PCR method to check for possible point mutations that might affect the critical nucleotide positions in the 5′UTRs of genomes which are known to be involved in the attenuation of OPV strains (15). All strains studied had nucleotide substitutions at these positions (nucleotide position 480 for type 1, position 481 for type 2, and position 472 for type 3 strains).
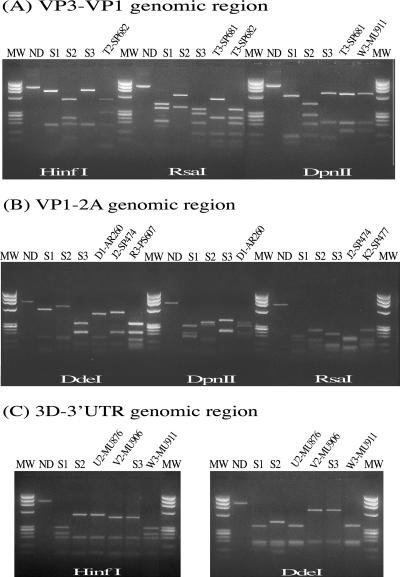
We analyzed the possible genetic drift of the strains, using a multiple RFLP method based on the comparative analysis of restriction profiles obtained with three RT-PCR amplicons and four restriction enzymes. Fragments VP3-VP1 (nucleotide positions 1913 to 2881) and VP1-2A (nucleotides 2870 to 3648) were located in the genomic regions encoding the capsid proteins VP3 and VP1 and the protease 2A. The third fragment, 3D-3′UTR (nucleotides 6536 to 7441), corresponds to the region of the genome encoding the 3D polymerase and to the 3′UTR at the 3′ end of the poliovirus genome. Each fragment was digested with the restriction enzymes DdeI, DpnII, RsaI, and HinfI. However, in most cases, the 3D-3′UTR fragment was digested by only two of these enzymes (DdeI and HinfI). The restriction profiles of the studied polioviruses were compared with those of the corresponding reference OPV strains to check for the appearance or disappearance of restriction sites. By this process, it was possible to check for possible nucleotide substitutions at approximately 100 nucleotide positions each in fragments VP3-VP1 and VP1-2A and at about 50 positions in the 3D-3′UTR fragment (see Materials and Methods for an explanation). Figure 1 shows RFLP profiles of some Sabin-derived isolates.
FIG. 1.
RFLP profiles of some Sabin strain-derived isolates. (A) VP3-VP1 genomic region, studied with restriction enzymes HinfI, RsaI, and DpnII. T2-SP682, T3-SP681, and T3-SP682, strains isolated from a VAPP patient; W3-MU911, strain isolated from a healthy contact with a VAPP patient. (B) VP1-2A genomic region, studied with restriction enzymes DdeI, DpnII, and RsaI. D1-AR260 and R3-PS607, strains isolated from VAPP patients; J2-SP474 and K2-SP477, strains isolated from AFP patients. (C) 3D-3′UTR genomic region, studied with restriction enzyme DdeI. U2-MU876, strain isolated from a VAPP patient; V2-MU906 and W3-MU911, strains isolated from healthy contacts with a VAPP patient. MW, molecular weight marker; ND, nondigested PCR product; S1, S2, and S3, Sabin type 1, 2, and 3 vaccine strains.
In analyses of the VP3-VP1 and VP1-2A fragments, 15 strains had profiles that were modified with respect to those of the reference OPV strains (Table 2). RFLP profiles of the 3D-3′UTR fragment showed that 10 strains had modified restriction profiles, indicating intertypic recombination events involving OPV strains of another serotype. Two of these isolates were actually a mixture of recombinant and nonrecombinant strains. Modified RFLP profiles did not appear to occur randomly. Instead, modifications appeared to depend on strain serotype, the fragments analyzed, and the restriction enzymes used (Table 3). For type 1 and 2 strains, modifications were most frequent in the VP1-2A fragment. Most of the profile modifications of type 2 strains were related to RsaI sites. RFLP analysis of the VP3-VP1 fragment with DpnII and RsaI led to the identification of the most strongly modified type 3 strains. Overall, 38.5% of the strains studied had restriction profiles displaying modifications in one or two of the genomic regions analyzed (VP3-VP1 and/or VP1-2A), and 25.6% of the strains had restriction profiles modified in the 3D-3′UTR genomic region, indicating intertypic recombination between OPV strains of different serotypes.
TABLE 3.
Fragments and enzymes implicated in modification of RFLP profiles
| Strain | RFLP profilea
|
|||||||
|---|---|---|---|---|---|---|---|---|
| VP3-VP1
|
VP1-2A
|
|||||||
| DpnII | HinfI | DdeI | RsaI | DpnII | HinfI | DdeI | RsaI | |
| Type 1 strains | ||||||||
| D1-AR260 | MOD | MOD | ||||||
| Q1-AR577 | MOD | |||||||
| Type 2 strains | ||||||||
| J2-SP474 | MOD | |||||||
| K2-SP477 | MOD | |||||||
| T2-SP682 | MOD | MOD | ||||||
| V2-MU905 | MOD | |||||||
| V2-MU906 | MOD | |||||||
| Type 3 strains | ||||||||
| M3-KR494 | MOD | MOD | ||||||
| N3-PS525 | MOD | |||||||
| P3-PS573 | MOD | MOD | ||||||
| R3-PS607 | MOD | MOD | ||||||
| T3-SP681 | MOD | MOD | ||||||
| T3-SP682 | MOD | |||||||
| W3-MU911 | MOD | |||||||
| W3-MU912 | MOD | |||||||
MOD, modified profile.
We determined the nucleotide sequences of the major parts of fragments VP3-VP1 (nucleotide positions 1977 to 2801) and VPI-2A (nucleotide positions 2921 to 3583) to confirm the results obtained with the multiple RFLP method and to determine precisely the level of genetic drift of the strains analyzed. Most of the strains with RFLP modifications (12 strains) were partially sequenced. Several unmodified strains (18 strains) were also sequenced as controls (Table 2). Five of the 12 sequenced strains with RFLP modifications had nucleotide substitution rates of >0.5% with respect to the reference OPV strain. These strains included one type 3 strain displaying 1.4% nucleotide divergence. For these five strains, we also determined the entire VP1 nucleotide sequence. The substitutions identified are shown in Table 4.
TABLE 4.
Nucleotide and amino acid substitutions in poliovirus vaccine mutant strains
| Strain | Case | Nucleotide substitution (nt position: substitution)
|
Amino acid substitutiona | |
|---|---|---|---|---|
| 5′UTR | VP3/VP1/2Aa | |||
| D1-AR260 PV1 | VAPP | 480:G→A | 1944: A→C | K401T |
| 2467: A→T | K575N | |||
| 2541: C→T | A600V | |||
| 2645: G→A | V635I | |||
| 2741: G→A | A667T | |||
| 2775: A→G | L678T | |||
| 2886: T→C | ||||
| 3328: A→G | ||||
| 3349: G→A | ||||
| 3367: C→T | ||||
| J2-SP474 PV2 | AFP | 481:A→G | 1980: G→T | E411D |
| 2007: G→A | ||||
| 2091: A→G | ||||
| 2523: T→C | ||||
| 2555: A→T | N603I | |||
| 2583: T→C | ||||
| 2625: G→A | ||||
| 2958: T→C | ||||
| 2986: A→G | K747E | |||
| 3014: C→T | S756F | |||
| 3243: C→T | ||||
| R3-PS607 PV3 | VAPP | 472: U→C | 2002: A→G | |
| 2014: C→T | ||||
| 2018: T→A | S426T | |||
| 2034: T→C | F431S | |||
| 2071: A→T | ||||
| 2089: T→C | ||||
| 2200: G→A | ||||
| 2210: T→C | ||||
| 2212: G→A | ||||
| 2388: G→A | S549N | |||
| 2476: G→A | ||||
| 2551: A→G | ||||
| 2636: G→A | A632T | |||
| 2790: T→C | M683T | |||
| 2812: T→C | ||||
| 2903: G→A | ||||
| 2950: C→T | ||||
| 3190: A→T | ||||
| 3211: T→C | ||||
| 3344: G→C | D868H | |||
| 3386: C→T | H882Y | |||
| 3517: A→G | ||||
| V2-MU906 PV2 | Contact with a VAPP patient | 481:A→G | 1986: C→T | |
| 2376: C→T | ||||
| 2494: A→G | M583V | |||
| 2958: T→C | ||||
| 3030: G→A | ||||
| 3258: G→A | ||||
| 3274: C→G | H843D | |||
| W3-MU912 PV3 | Contact with a VAPP patient | 472: U→C | 1991: G→A | D417N |
| 2034: T→C | F431S | |||
| 2446: C→T | ||||
| 2503: T→C | ||||
| 2516: C→A | L592I | |||
| 2572: C→T | ||||
| 2968: A→G | ||||
| 3088: G→A | ||||
| 3136: T→C | ||||
Nucleotide and amino acid substitutions in the capsid VP1 coding region are indicated in bold.
The mean substitution percentages for strains with modified or unmodified RFLP profiles are shown in Table 5. As expected, the percentage of substitutions was low (0.1%) for unmodified strains and significantly higher (0.5%) for modified strains. The mean percentage of nucleotide substitutions for the five strains with >0.5% substitutions was high (0.8%), indicating that these strains had circulated or multiplied for about 8 months according to the reported molecular clock of poliovirus strains, with about 1% nucleotide substitutions per year (3, 13, 24). We cannot rule out the possibility that some substitutions were not present in viruses in the original samples but appeared during the isolation process in cell cultures. Nevertheless, in this study all viruses were isolated according to the same procedures, and most of them showed only a few mutations or no mutation at all. The 18 vaccine strains with unmodified RFLP patterns showed zero to four substitutions in 1,486 sequenced nucleotides (mean substitution percentage, 0.1%) (Tables 2 and 5).
TABLE 5.
Mean percentages of nucleotide substitutions in strains with and without RFLP modifications
| RFLP profile | No. of sequenced strains | Mean % of substitutions |
|---|---|---|
| Nonmodified | 18 | 0.1 |
| Modified | ||
| All | 12 | 0.5 |
| Those with ≥0.5% substitutions | 5 | 0.8 |
DISCUSSION
Using the multiple RFLP method for analysis of the OPV strains isolated from clinical samples, we identified several mutant strains with modified restriction profiles. Some of them were intertypic recombinant strains. We also found that a large proportion of them had high percentages of nucleotide substitutions (>0.5%) with respect to the corresponding OPV reference strains. These strains included a type 3 VDPV with 1.4% nucleotide substitution, indicating the long-term multiplication and/or circulation of OPV strains.
The multiple RFLP method appears to detect OPV strains with significant genetic drift particularly efficiently. There was a strong correlation between the results for multiple RFLP analysis and the results of partial genomic sequencing. The sequenced strains with restriction profiles modified in the capsid genomic region had a higher percentage of nucleotide substitutions (0.5%) than the sequenced strains with unmodified profiles (0.1%). The five strains with >0.5% nucleotide divergence belonged to the group of strains with modified restriction profiles. Since RFLP analysis of the capsid genomic region corresponds to the checking of approximately 200 nucleotide positions, this method has a high probability of detecting all OPV strains displaying significant numbers of nucleotide substitutions. Although genomic sequencing remains essential for determining the precise genetic drift of vaccine strains with modified RFLP profiles, these results show that RFLP analysis is a potentially valuable tool for rapid initial screening, in particular in laboratories where sequencing capacity is not readily available.
RFLP analysis of the 3D-3′UTR fragment in the 3′-terminal part of the genome did not appear to be a useful means of detecting nucleotide substitutions. Indeed, most restriction profile modifications concerning this fragment detected in this study were associated with intertypic genetic recombination events between OPV strains of different serotypes. It has been shown that intertypic recombination is very frequent in children vaccinated with OPV and that this event occurs soon after vaccination (10). There is therefore no clear correlation between genetic recombination and the long-term circulation/multiplication of OPV strains. Nevertheless, four of the five poliomyelitis outbreaks involving VDPV were caused by strains resulting from recombination between an OPV strain and unidentified human enteroviruses, probably species C enteroviruses (18, 20, 31, 32, 39). Since the rapid detection of such dangerous by-products of the eradication program has become one of the priorities of poliomyelitis surveillance, RFLP analysis of the 3′-terminal part of the genomes of OPV strains isolated from clinical samples remains useful. Multiple RFLP analysis has already been shown to detect such recombinant VDPV in Madagascar (31). Our results suggest that analysis of the 3D-3′UTR fragment could already be limited to two restriction enzymes (such as HinfI and DdeI, which were used systematically in this study) rather than four. Analysis with these two enzymes alone is sufficient to detect recombination with OPV strains or with unidentified enteroviruses.
Detailed analysis of the RFLP results obtained in this study showed that modified RFLP profiles did not appear to occur randomly, instead depending on the strain serotype, the fragment analyzed, and the restriction enzymes used (Table 3). Most of the modifications detected for a given serotype concerning the VP3-VP1 and VP1-2A fragments were detected with one or two restriction enzymes. Moreover, only 5 of the 12 sequenced strains with modified profiles were found to have acquired large numbers of substitutions. This suggests that the multiple RFLP method could be simplified according to the serotype of the strains studied. The use of a single fragment and two enzymes—such as the VP3-VP1 fragment with DpnII and RsaI for type 3 strains—may be sufficient to detect most mutant strains (Table 3). However, analysis of a larger number of strains and results is required for the development of simplifications guaranteeing at least the systematic detection of all OPV strains with >1% nucleotide divergence (VDPV). Moreover, we cannot exclude the possibility that in the future, the systematic detection of strains with lower levels of nucleotide divergence—such as strains with 0.5% to 1.0% nucleotide divergence, indicating circulation/multiplication for more than 6 months—will be required for the final stages in the eradication program. Since the VDPV implicated in poliovirus outbreaks showed >1% nucleotide divergence, it may be possible to carry out surveillance for pre-VDPV strains as indicators of future potential problems.
The large proportion of strains with high percentages of nucleotide substitutions detected in this study was rather surprising. However, these results may be accounted for by the selection of 39 poliovirus strains from 97 strains on the basis of particular epidemiological parameters. The strains studied were isolated from nonvaccinated children or from vaccinated children some time after administration of the last dose of OPV (more than 1 month). Poliovirus excretion is observed over a period of 3 to 4 weeks for unvaccinated children and over a shorter period for children who have already been in contact with the virus (1). It would therefore seem likely that children excreting OPV strains for more than 1 month after the last dose of vaccine have recently acquired the virus by contact or that these children present immunological problems affecting viral clearance that may lead to chronic infection (16). Our choice of strains may therefore have favored the discovery of strains with significant genetic drift. The 58 OPV strains that we did not analyze were isolated sooner after the last dose of vaccine and are much more likely to resemble the OPV reference strains. If this proves to be the case, then the percentage of VDPV (>1% nucleotide divergence; 1 VDPV in 97 isolated strains) is close to 1%—a value consistent with the balance sheet obtained by systematic sequencing of isolated OPV strains from any source (6). However, the percentage of OPV strains with high levels of nucleotide divergence (>0.5%) remains high (5%), indicating that several strains have circulated for 6 to 16 months according to the molecular clock of poliovirus strains, with about 1% nucleotide substitutions per year (3, 13, 24).
Vaccine coverage of the pediatric population is traditionally very high (>95%) in the country where the studied strains were isolated. Thus, our results suggest that the prolonged persistence and circulation of OPV strains are not rare in certain populations of well-immunized children. Given the large number of vaccinated children, each unvaccinated child is very likely to come into contact with one or several recently vaccinated children excreting OPV strains. This was the case for one unvaccinated child who developed VAPP in this study. In this case, the child stayed in the hospital for more than a month, and paralysis began just after discharge from the hospital. Epidemiological investigations revealed that this child had been in close contact with several recently vaccinated children while in the hospital. We isolated poliovirus strains (type 2 and type 3 strains [V2-MU905/V2MU906 and W3-MU911/W3-MU912, respectively]) from two children who had been in recent contact with this VAPP patient. Unfortunately, none of these strains were similar to those isolated from the patient with VAPP (type 2 strains U2-MU876 and U2-MU877). However, both strain types isolated from contacts showed high percentages of nucleotide substitutions in the capsid VP1 region, i.e., 0.6% and 0.7% for the type 2 and type 3 strains, respectively. These strains also had substitutions at critical nucleotide positions in the 5′UTR, and the serotype 3 strain presented a substitution at another critical nucleotide position (position 2034) implicated in attenuation. The isolation of strains with high levels of nucleotide divergence from healthy children has important implications, as these strains can be dangerous for unvaccinated children.
The ability of OPV strains to circulate in a population with good vaccine coverage is presumably as little as that of homotypic wild poliovirus strains. Several studies of strains isolated from well-immunized human populations have shown that OPV strains may undergo several interhuman transmissions, leading to disease in unvaccinated individuals (7, 14, 34). Such OPV strains were isolated from immunocompetent patients with paralytic poliomyelitis and from a healthy contact of the VAPP patient from this study. VDPV strains have been isolated from sewage with no known source of excretion in countries with effective vaccine coverage (4, 17, 33). The description of a type 2 revertant poliovirus strain with only a few mutations isolated from a nonvaccinated immunodeficient patient with VAPP showed that transmissibility may be acquired very early in the evolution of vaccine strains of poliovirus (8). The spread of this poliovirus from the initial polio patient to several contacts illustrated how VDPV may emerge (8).
The circulation of OPV strains in countries with high vaccine coverage may cause future problems for the poliomyelitis eradication program. Moreover, the emergence of circulating and pathogenic VDPV in countries with low vaccine coverage and subsequent poliomyelitis outbreaks could jeopardize the final goal of poliomyelitis eradication, i.e., the elimination of pathogenic poliovirus strains. The risk will be even higher during the period following the cessation of poliomyelitis vaccination, when the increasing nonvaccinated population will be in contact with possible residual vaccine poliovirus strains. This threat has made it necessary to develop new, rapid, and simple surveillance methods for detecting mutant OPV strains. This study demonstrated that the multiple RFLP assay is an efficient tool for the screening of these strains. This method is simple and can be performed rapidly, fulfilling two essential properties for laboratories with no convenient access to sequencing facilities. Although genomic sequencing remains required for confirming and determining precisely the genetic drift of mutant strains, the multiple RFLP method allows rapid initial screening. It detected a large proportion of OPV mutants among the studied strains isolated in areas under the surveillance of the Saint-Petersburg Subnational Poliomyelitis Laboratory. It also rapidly detected the VDPV responsible for the outbreak of poliomyelitis in Madagascar in 2002 (31).
Acknowledgments
This study was partly supported by grants from the Direction of Foreign Affairs (AC02 Vaccin Polio Oral), the Transverse Research Programs (PTR120) of the Pasteur Institute, and the World Health Organization (OMS B3/181/125).
Footnotes
Published ahead of print on 6 September 2006.
REFERENCES
- 1.Alexander, J. P., Jr., H. E. Gary, Jr., and M. A. Pallansch. 1997. Duration of poliovirus excretion and its implications for acute flaccid paralysis surveillance: a review of the literature. J. Infect. Dis. 175:S176-S182. [DOI] [PubMed] [Google Scholar]
- 2.Balanant, J., S. Guillot, A. Candrea, F. Delpeyroux, and R. Crainic. 1991. The natural genomic variability of poliovirus analyzed by a restriction fragment length polymorphism assay. Virology 184:645-654. [DOI] [PubMed] [Google Scholar]
- 3.Bellmunt, A., G. May, R. Zell, P. Pring-Akerblom, W. Verhagen, and A. Heim. 1999. Evolution of poliovirus type I during 5.5 years of prolonged enteral replication in an immunodeficient patient. Virology 265:178-184. [DOI] [PubMed] [Google Scholar]
- 4.Blomqvist, S., C. Savolainen, P. Laine, P. Hirttio, E. Lamminsalo, E. Penttila, S. Joks, M. Roivainen, and T. Hovi. 2004. Characterization of a highly evolved vaccine-derived poliovirus type 3 isolated from sewage in Estonia. J. Virol. 78:4876-4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cammack, N., A. Phillips, G. Dunn, V. Patel, and P. D. Minor. 1988. Intertypic genomic rearrangements of poliovirus strains in vaccinees. Virology 67:507-514. [PubMed] [Google Scholar]
- 6.CDC. 2003. Laboratory surveillance for wild and vaccine-derived polioviruses, January 2002-June 2003. Morb. Mortal. Wkly. Rep. 52:913-916. [PubMed] [Google Scholar]
- 7.Cherkasova, E. A., E. A. Korotkova, M. L. Yakovenko, O. E. Ivanova, T. P. Eremeeva, K. M. Chumakov, and V. I. Agol. 2002. Long-term circulation of vaccine-derived poliovirus that causes paralytic disease. J. Virol. 76:6791-6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cherkasova, E. A., M. L. Yakovenko, G. V. Rezapkin, E. A. Korotkova, O. E. Ivanova, T. P. Eremeeva, L. I. Krasnoproshina, N. I. Romanenkova, N. R. Rozaeva, L. Sirota, V. I. Agol, and K. M. Chumakov. 2005. Spread of vaccine-derived poliovirus from a paralytic case in an immunodeficient child: an insight into the natural evolution of oral polio vaccine. J. Virol. 79:1062-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crainic, R., P. Couillin, B. Blondel, N. Cabau, A. Boué, and F. Horodniceanu. 1983. Natural variation of poliovirus neutralization epitope. Infect. Immun. 41:1217-1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cuervo, N., S. Guillot, N. Romanenkova, M. Combiescu, A. Aubert-Combiescu, M. Seghier, V. Caro, S. Guillot, R. Crainic, and F. Delpeyroux. 2001. Genomic features of intertypic recombinant Sabin strains excreted by primary vaccinees. J. Virol. 75:5740-5751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Friedrich, F., E. F. Da-Silva, and H. G. Schatzmayr. 1996. Type 2 poliovirus recombinants isolated from vaccine-associated cases and from healthy contacts in Brazil. Acta Virol. 40:27-33. [PubMed] [Google Scholar]
- 12.Furione, M., S. Guillot, D. Otelea, J. Balanant, A. Candrea, and R. Crainic. 1993. Polioviruses with natural recombinant genomes isolated from vaccine-associated paralytic poliomyelitis. Virology 196:199-208. [DOI] [PubMed] [Google Scholar]
- 13.Gavrilin, G. V., E. A. Cherkasova, G. Y. Lipskaya, O. M. Kew, and V. I. Agol. 2000. Evolution of circulating wild poliovirus and of vaccine-derived poliovirus in an immunodeficient patient: a unifying model. J. Virol. 74:7381-7390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Georgescu, M. M., F. Delpeyroux, M. Tardy-Panit, J. Balanant, M. Combiescu, A. A. Combiescu, S. Guillot, and R. Crainic. 1994. High diversity of poliovirus strains isolated from the central nervous system from patients with vaccine-associated paralytic poliomyelitis. J. Virol. 68:8089-8101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guillot, S., V. Caro, N. Cuervo, E. Korotkova, M. Combiescu, A. Persu, A. Aubert-Combiescu, F. Delpeyroux, and R. Crainic. 2000. Natural genetic exchanges between vaccine and wild poliovirus strains in humans. J. Virol. 74:8434-8443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Halsey, N. A., J. Pinto, F. Espinosa-Rosales, M. A. Faure-Fontenla, E. da Silva, A. J. Khan, A. D. Webster, P. Minor, G. Dunn, E. Asturias, H. Hussain, M. A. Pallansch, O. M. Kew, J. Winkelstein, and R. Sutter. 2004. Search for poliovirus carriers among people with primary immune deficiency diseases in the United States, Mexico, Brazil, and the United Kingdom. Bull. W. H. O. 82:3-8. [PMC free article] [PubMed] [Google Scholar]
- 17.Horie, H., H. Yoshida, K. Matsuura, M. Miyazawa, K. Wakabayashi, A. Nomoto, and S. Hashizume. 2002. Isolation of vaccine-derived type 1 polioviruses displaying similar properties to virulent wild strain Mahoney from sewage in Japan. J. Med. Virol. 68:445-451. [DOI] [PubMed] [Google Scholar]
- 18.Kew, O., V. Morris-Glasgow, M. Landaverde, C. Burns, J. Shaw, Z. Garib, J. Andre, E. Blackman, C. J. Freeman, J. Jorba, R. Sutter, G. Tambini, L. Venczel, C. Pedreira, F. Laender, H. Shimizu, T. Yoneyama, T. Miyamura, H. van der Avoort, M. S. Oberste, D. Kilpatrick, S. Cochi, M. Pallansch, and C. de Quadros. 2002. Outbreak of poliomyelitis in Hispaniola associated with circulating type 1 vaccine-derived poliovirus. Science 296:356-359. [DOI] [PubMed] [Google Scholar]
- 19.Kew, O. M., and B. K. Nottay. 1984. Evolution of the oral poliovaccine strains in humans occurs by both mutation and intramolecular recombination, p. 357-367. In R. Chanock and R. Lerner (ed.), Modern approaches to vaccines. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 20.Kew, O. M., R. W. Sutter, E. M. de Gourville, W. R. Dowdle, and M. A. Pallansch. 2005. Vaccine-derived polioviruses and the endgame strategy for global polio eradication. Annu. Rev. Microbiol. 59:587-635. [DOI] [PubMed] [Google Scholar]
- 21.Kew, O. M., R. W. Sutter, B. K. Nottay, M. J. McDonough, D. R. Prevots, L. Quick, and M. A. Pallansch. 1998. Prolonged replication of a type 1 vaccine-derived poliovirus in an immunodeficient patient. J. Clin. Microbiol. 36:2893-2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khetsuriani, N., D. R. Prevots, L. Quick, M. E. Elder, M. Pallansch, O. Kew, and R. W. Sutter. 2003. Persistence of vaccine-derived polioviruses among immunodeficient persons with vaccine-associated paralytic poliomyelitis. J. Infect. Dis. 188:1845-1852. [DOI] [PubMed] [Google Scholar]
- 23.Lipskaya, G. Y., A. R. Muzychenko, O. K. Kutitova, S. V. Maslova, M. Equestre, S. G. Drozdov, R. Perez Bercoff, and V. I. Agol. 1991. Frequent isolation of intertypic poliovirus recombinants with serotype 2 specificity from vaccine-associated polio cases. J. Med. Virol. 35:290-296. [DOI] [PubMed] [Google Scholar]
- 24.Liu, H. M., D. P. Zheng, L. B. Zhang, M. S. Oberste, O. M. Kew, and M. A. Pallansch. 2003. Serial recombination during circulation of type 1 wild-vaccine recombinant polioviruses in China. J. Virol. 77:10994-11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Macadam, A. J., C. Arnold, J. Howlett, A. John, S. Marsden, F. Taffs, P. Reeve, N. Hamada, K. Wareham, J. Almond, N. Cammack, and P. D. Minor. 1989. Reversion of the attenuated and temperature-sensitive phenotypes of the Sabin type 3 strain of poliovirus in vaccinees. Virology 172:408-414. [DOI] [PubMed] [Google Scholar]
- 26.MacLennan, C., G. Dunn, A. P. Huissoon, D. S. Kumararatne, J. Martin, P. O'Leary, R. A. Thompson, H. Osman, P. Wood, P. Minor, D. J. Wood, and D. Pillay. 2004. Failure to clear persistent vaccine-derived neurovirulent poliovirus infection in an immunodeficient man. Lancet 363:1509-1513. [DOI] [PubMed] [Google Scholar]
- 27.Martín, J., G. Dunn, R. Hull, V. Patel, and P. D. Minor. 2000. Evolution of the Sabin strain of type 3 poliovirus in an immunodeficient patient during the entire 637-day period of virus excretion. J. Virol. 74:3001-3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Minor, P. D. 2004. Polio eradication, cessation of vaccination and re-emergence of disease. Nat. Rev. Microbiol. 2:473-482. [DOI] [PubMed] [Google Scholar]
- 29.Nkowane, B. M., S. G. F. Wassilak, and W. A. Orenstein. 1987. Vaccine-associated paralytic poliomyelitis. United States: 1973 through 1984. JAMA 257:1335-1340. [PubMed] [Google Scholar]
- 30.Pipkin, P. A., D. J. Wood, V. R. Racaniello, and P. D. Minor. 1993. Characterisation of L cells expressing the human poliovirus receptor for the specific detection of polioviruses in vitro. J. Virol. Methods 41:333-340. [DOI] [PubMed] [Google Scholar]
- 31.Rousset, D., M. Rakoto-Andrianarivelo, R. Razafindratsimandresy, B. Randriamanalina, S. Guillot, J. Balanant, P. Mauclere, and F. Delpeyroux. 2003. Recombinant vaccine-derived poliovirus in Madagascar. Emerg. Infect. Dis. 9:885-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shimizu, H., B. Thorley, F. J. Paladin, K. A. Brussen, V. Stambos, L. Yuen, A. Utama, Y. Tano, M. Arita, H. Yoshida, T. Yoneyama, A. Benegas, S. Roesel, M. Pallansch, O. Kew, and T. Miyamura. 2004. Circulation of type 1 vaccine-derived poliovirus in the Philippines in 2001. J. Virol. 78:13512-13521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shulman, L. M., Y. Manor, R. Handsher, F. Delpeyroux, M. J. McDonough, T. Halmut, I. Silberstein, J. Alfandari, J. Quay, T. Fisher, J. Robinov, O. M. Kew, R. Crainic, and E. Mendelson. 2000. Molecular and antigenic characterization of a highly evolved derivative of the type 2 oral poliovaccine strain isolated from sewage in Israel. J. Clin. Microbiol. 38:3729-3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Strebel, P. M., A. Aubert-Combiescu, N. Ion-Nedelcu, S. Biberi-Moroeanu, M. Combiescu, R. W. Sutter, O. M. Kew, M. A. Pallansch, P. A. Patriarca, and S. L. Cochi. 1994. Paralytic poliomyelitis in Romania, 1984-1992. Evidence for a high risk of vaccine-associated disease and reintroduction of wild-virus infection. Am. J. Epidemiol. 140:1111-1124. [DOI] [PubMed] [Google Scholar]
- 35.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van der Avoort, H. G., B. P. Hull, T. Hovi, M. A. Pallansch, O. M. Kew, R. Crainic, D. J. Wood, M. N. Mulders, and A. M. van Loon. 1995. Comparative study of five methods for intratypic differentiation of polioviruses. J. Clin. Microbiol. 33:2562-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.WHO. 2001. Polio laboratory manual. WHO, Geneva, Switzerland.
- 38.Yang, C. F., L. De, B. P. Holloway, M. A. Pallansch, and O. M. Kew. 1991. Detection and identification of vaccine-related polioviruses by the polymerase chain reaction. Virus Res. 20:159-179. [DOI] [PubMed] [Google Scholar]
- 39.Yang, C. F., T. Naguib, S. J. Yang, E. Nasr, J. Jorba, N. Ahmed, R. Campagnoli, H. van der Avoort, H. Shimizu, T. Yoneyama, T. Miyamura, M. Pallansch, and O. Kew. 2003. Circulation of endemic type 2 vaccine-derived poliovirus in Egypt from 1983 to 1993. J. Virol. 77:8366-8377. [DOI] [PMC free article] [PubMed] [Google Scholar]