Abstract
Human enterovirus (HEV) infections can be asymptomatic or cause only mild illness; recent evidence may implicate HEV infection in type 1 diabetes mellitus and myocarditis. Here, we report the molecular characterization of HEV obtained in serial monthly collections from healthy Norwegian infants. A total of 1,255 fecal samples were collected from 113 healthy infants beginning at age 3 months and continuing to 28 months. The samples were analyzed for HEV nucleic acid by real-time PCR. Fifty-eight children (51.3%) had HEV infections. One hundred forty-five positive samples were typed directly by nucleotide sequencing of the VP1 region. HEV-A was detected most frequently, with an overall prevalence of 6.8%. HEV-B was present in 4.8% of the samples and HEV-C in only 0.2% of the samples. No poliovirus or HEV-D group viruses were detected. Twenty-two different serotypes were detected in the study period: the most common were EV71 (14.5%), CAV6 (10.5%), CAV4 (8.9%), E18 (8.9%), and CBV3 (7.3%). These findings suggest that the prevalence of HEV infections in general, and HEV-A infections in particular, has been underestimated in epidemiological studies based on virus culture.
The genus Enterovirus belongs to the family Picornaviridae. Although the majority of Human enterovirus (HEV) infections are asymptomatic, enterovirus infections can cause upper respiratory illness, febrile rash, aseptic meningitis, pleurodynia, encephalitis, acute flaccid paralysis, and neonatal sepsis-like disease (35). HEV may also be implicated in the pathogeneses of severe chronic diseases, including type 1 diabetes mellitus (33), myocarditis and congestive cardiomyopathy (20), and neuromuscular diseases (8).
HEVs contain a linear (7.4-kb) single-stranded RNA genome comprising a 5′ and a 3′ noncoding region and a single, long open reading frame coding for a polyprotein of about 2,200 amino acids. HEVs are classified, based on molecular and biological properties, into five species (19, 36): (i) Poliovirus (types 1 to 3), (ii) Human enterovirus A (HEV-A) (CAV2 to CAV8, CAV10, CAV12, CAV14, CAV16, and EV71), (iii) Human enterovirus B (HEV-B) (CAV9, CBV1 to CBV6, E17, E9, E11 to E21, E24 to E27, E29 to E33, and EV69), (iv) Human enterovirus C (HEV-C) (CAV1, CAV11, CAV13, CAV15, CAV17 to CAV22, and CAV24), and (v) Human enterovirus D (HEV-D) (EV68 and EV70). Several new serotypes (EV73 to EV78 and EV89 to EV91) were recently described (28, 29, 31, 32).
Neutralization tests of cultured virus may not be sufficient to identify some serotypes (26). Many coxsackievirus A strains can be isolated and propagated only in suckling mice (24). To address the need for a robust and universal typing system, many investigators have turned to the use of molecular methods (4, 30, 41). The introduction of molecular sequencing has extended the potential of virus surveillance and epidemiological studies by facilitating the identification of genotypes (14).
Most previous data on HEV circulation have been obtained from analysis of specimens from individuals with disease. Studies of enterovirus circulation in healthy populations antedate the advent of molecular technologies (12, 13, 16-18, 21, 23, 38, 39).
We have reported that 11.3% of fecal samples obtained prospectively from 113 healthy Norwegian infants were positive for enterovirus RNA (11). Here, we present the results of a comprehensive molecular characterization of these samples.
MATERIALS AND METHODS
Study design and subjects.
Participating children were enrolled in a prospective study (MIDIA) to investigate potential environmental triggers of type 1 diabetes mellitus. The MIDIA study recruits newborn infants at their first visit to health care centers. Candidates are screened for the HLA genotype conferring the highest risk of type 1 diabetes (HLA-DQB1*02-DQA1*05-DRB1*03/DQB1*0302-DQA1*03-DRB1*0401), which is carried by 2.7% of newborns (10). Over the period from September 2001 to October 2003, 113 of 116 infants identified as genetically susceptible were enrolled (60 males; 53 females). No more than one child in any nuclear family was enrolled. Beginning at 3 months of age and followed up to 28 months (for the first children recruited), monthly stool samples were obtained. Specimens were collected from September 2001 through November 2003; 96% (1,257/1,306) of the scheduled stool samples were received. Two samples were accidentally destroyed during handling. Eighty-nine percent of the stool samples were collected from children residing in the counties of Akershus (southeast Norway) and Hordaland (west coast), two counties separated by more than 400 km.
Processing of stool samples.
Total nucleic acids were extracted from 140 μl of 1,255 stool suspensions using the QIAamp Viral RNA Mini Kit (QIAGEN). HEV nucleic acid was detected and quantitated using a one-step real-time reverse transcription (RT)-PCR targeting the 5′ untranscribed region (11).
Molecular typing.
HEV serotypes of samples positive in real-time PCR were determined by VP1 sequencing of RNA (4, 44). In instances where samples could not be serotyped by this approach (39 of 145 samples), the RNA polymerase region was amplified and sequenced (4). Based on the RNA polymerase sequence, serotype- or species-specific VP1 primers were designed (Table 1) (4, 44). In total, 14 additional nesting primer sets (localized inside the sequence product of the first amplification) were designed, facilitating serotype assignment of 91.7% of the enterovirus-positive samples.
TABLE 1.
Oligonucleotide primers used for VP1 characterization of enteroviruses
| Primer | 5′-3′ sequencea | Enterovirus specificity | Positionsb | Usec |
|---|---|---|---|---|
| ENV71SP-F1 | GCACAGGTYTCNGTNCCRTTYATGTC | HEV-A | 2987-3012 | PCR/seq. |
| ENV71SP-R1 | CATGCCCTGACRTGYTTCATYCTCAT | HEV-A | 3207-3182 | PCR/seq. |
| VP1-2S-CAV16 | GCAGTACATGTATGTCCCSCCMGGSG | HEV-A | 2878-2903 | PCR |
| VP1-2A-CAV16 | TCGCACCCCTGGGCRGTGGTGGA | HEV-A | 3507-3485 | PCR |
| VP1-2S-CAV2 | CAGTACATGTAYGTCCCNCCYGGRG | HEV-A | 2879-2903 | PCR/seq. |
| VP1-2A-CAV2 | CCGGTCTGACAATTACATCGAGC | HEV-A | 3537-3515 | PCR/seq. |
| VP1-2S-CAV3 | CAGTACATGTAIGTTCCACCTGGTG | HEV-A | 2879-2903 | PCR/seq. |
| VP1-2A-CAV3 | CCCGTCTGGCAGTTGCATCGAGC | HEV-A | 3537-3515 | PCR/seq. |
| VP1-2S-CAV4 | CARTACATGTATGTGCCACCYGGRG | HEV-A | 2879-2903 | PCR |
| VP1-2A-CAV4 | CCTGTTTGGCAATTACAGCGGGC | HEV-A | 3537-3515 | PCR |
| VP1-2S-CAV6 | CAGTACATGTAYGTRCCRCCRGGTG | HEV-A | 2879-2903 | PCR |
| VP1-2A-CAV6 | CCAGTCTGGCAGTTACATCGAGC | HEV-A | 3537-3515 | PCR |
| VP1-2S-CAV10 | CAGTATATGTATGTNCCTCCNGGYG | HEV-A | 2879-2903 | PCR |
| VP1-2A-CAV10 | CCTGTCTGACAGTTGCACCGAGC | HEV-A | 3537-3515 | PCR |
| VP1-2S-CAV12 | CAGTACATGTTIGTGCCICCTGGTG | HEV-A | 2879-2903 | PCR |
| VP1-2A-CAV12 | CCAGTCTGACAATTGCATCGAGC | HEV-A | 3537-3515 | PCR/seq. |
| VP1-2S-ENV71 | CARTAYATGTTTGTNCCSCCYGG | EV71 | 2879-2903 | PCR |
| VP1-2A-ENV71 | TCACAACCYTGRGCRGTGGTAGA | EV71 | 3507-3485 | PCR |
| VP1-CAV6-F | CGGTGTTCGCAAAATTGAGT | CAV6 | 2958-2977 | PCR |
| VP1-CAV6-R | TCACATCCTTGAGCAGTAGTGG | CAV6 | 3507-3486 | PCR/seq. |
| VP1-CAV10-F | GCCCCTAAACCGACTGGTAG | CAV10 | 2903-2922 | PCR |
| VP1-CAV10-R | ACCCCTGTGCAGTGGTAGAG | CAV10 | 3503-3484 | PCR |
| VP1-HEV71-F | CCAAGCCAGACTCCAGAGAA | EV71 | 2907-2926 | PCR/seq. |
| VP1-HEV71-R | ATTACAGCGGGCAATTGTGT | EV71 | 3526-3507 | PCR/seq. |
| VP1-CBV3-F | CGGTGCCAGATAAGGTTGAC | CBV3 | 2907-2926 | PCR/seq. |
| VP1-CBV3-R | TCTGGCTATTGTATCGCATCC | CBV3 | 3520-3500 | PCR |
| VP1-EV18-F | GGCCAAGGTGGATAGTTACG | EV18 | 2914-2933 | PCR |
| VP1-EV18-R | ACACCTGGCGATGGTATCAC | EV18 | 3523-3504 | PCR |
I, inosine; Y, C or T; W, A or T; R, A or G; K, G or T.
Relative to the genome of coxsackievirus A6 strain Gdula (GenBank accession number AY421764).
seq., sequencing.
First-round VP1 amplification products obtained by the method of Casas et al. (4) or separate random-hexamer-primed RT uncoupled from the first round of PCR were used as templates with the new sense and antisense primers. PCR was carried out in a total volume of 20 μl with 10 pmol of each primer (Table 1), 1 to 2 μl of the RT-PCR products or cDNA, 2 μl 10× PCR buffer II (Applied Biosystems), 2.5 mmol MgCl2, 0.5 mmol deoxynucleoside triphosphates, and 2.5 U of Taq polymerase (Perkin-Elmer). The thermal-cycling program consisted of 5 min of denaturation at 94°C, followed by 40 cycles of 30 s of denaturation at 94°C, 30 s of annealing at 50 to 55°C, and 1 min of extension at 72°C.
Nucleotide sequence determination and analysis.
Direct product sequencing was performed using the amplification primers. Both strands were sequenced by automatic methods (BigDye version 3.1 and ABI PRISM 310 Genetic Analyzer; Applied Biosystems, Foster City, CA). In instances where sequences could not be obtained directly, the products were cloned into a pGEM-T-Easy vector (Promega) and five clones from each sample were sequenced on both strands.
Raw sequence data were analyzed with Sequencher (version 4.2; Gene Codes Corporation, Ann Arbor, MI) to obtain the final consensus sequence. Ambiguous nucleotides were resolved by resequencing them.
Phylogenetic analysis.
The VP1 capsid coding sequences determined in this study were included in a phylogenetic analysis with reference strains of all enterovirus serotypes. Multiple sequence alignments were performed with ClustalX version 1.82 (42). Phylogenetic analysis was performed using the Kimura two-parameter model as a model of nucleotide substitution and the neighbor-joining method to reconstruct the phylogenetic tree (MEGA program version 3.0 [22]). The statistical significance of the phylogenies constructed was estimated by bootstrap analysis with 1,000 pseudoreplicate data sets.
Infection episodes and coinfections.
A new infection episode was defined as detection of a new genotype after a monthly sample was negative for that genotype. Coinfection was defined as the simultaneous presence of two or more different genotypes in the same stool sample.
Statistical analysis.
The software packages used were Stata version 9.0 (Stata Corporation, College Station, TX) and SPSS version 12.0.1 (SPSS, Chicago, IL). The overall prevalence for each species over the whole study period was computed with 95% confidence intervals (CI). The confidence intervals for prevalence were computed based on standard errors estimated by bootstrapping, using each child as a unit to provide a simple adjustment for dependence between repeated infections (15). Relationships of sex, year, season, and geographic location with prevalence for the main HEV species were assessed.
Associations of viral load and duration of infection, sex, age, and enterovirus species were assessed in linear-regression models, using the viral load of the first positive sample in an infection period. The potential dependence in data from repeated measurements for each child was handled in the regression models by generalized estimating equations using the xtgee procedure in STATA (Stata cross-sectional time-series reference manual, release 8, StataCorp, Stata Press, College Station, TX).
The cumulative incidence of first infection during follow-up for individual children at different ages was estimated using Kaplan-Meier survival analysis. Differences between groups were tested using the log rank test. A P value of 0.05 or less was considered to be statistically significant.
Nucleotide sequence accession numbers.
The 3′-end VP1 sequences reported here were deposited in the GenBank sequence database under accession no. DQ317159 to DQ317293. The RNA polymerase sequences were reported under accession no. DQ315510 to DQ315562.
RESULTS
Molecular typing and phylogenetic analysis.
The HEV type was identified in 92% of the samples (133/145) by VP1 sequencing and in 6.2% (9 cases) by RNA polymerase sequencing. In total, 97.9% (142/145) of the real-time PCR-positive samples could be assigned to the species level. The VP1 sequencing and phylogenetic analysis allowed further subtyping. The three samples that were not identified had low viral loads. The positive samples represented 124 discrete infection episodes. A total of 22 different serotypes were detected (Table 2). The most prevalent serotypes were EV71 (14.5%), CAV6 (10.5%), CAV4 (8.9%), E18 (8.9%), and CBV3 (7.3%). Whereas CBV3 and E18 were detected throughout the study period, CAV4, CAV5, CAV6, CAV10, and CBV1 were detected only during short intervals. CAV16, E5, E11, E25, E30, E3, E13, and a CAV19-like strain were represented in only one or two infection episodes. The circulation of specific serotypes was not restricted to a single county or municipality.
TABLE 2.
Enterovirus serotypes and species detected in feces specimens taken from healthy children aged 3 to 28 months in Norwaya
| Species | Serotype | No. of episodes | % (n = 124) |
|---|---|---|---|
| HEV-A | CAV2 | 3 | 2.4 |
| HEV-A | CAV4 | 11 | 8.9 |
| HEV-A | CAV5 | 7 | 5.6 |
| HEV-A | CAV6 | 13 | 10.5 |
| HEV-A | CAV10 | 7 | 5.6 |
| HEV-A | CAV14 | 6 | 4.8 |
| HEV-A | CAV16 | 1 | 0.8 |
| HEV-A | HEV71 | 18 | 14.5 |
| HEV-A | HEV-Ab | 4 | 3.2 |
| HEV-B | CAV9 | 4 | 3.2 |
| HEV-B | CBV1 | 5 | 4.0 |
| HEV-B | CBV3 | 9 | 7.3 |
| HEV-B | CBV4 | 3 | 2.4 |
| HEV-B | CBV5 | 4 | 3.2 |
| HEV-B | EV3 | 2 | 1.6 |
| HEV-B | EV5 | 1 | 0.8 |
| HEV-B | EV9 | 3 | 2.4 |
| HEV-B | EV11 | 1 | 0.8 |
| HEV-B | EV13 | 2 | 1.6 |
| HEV-B | EV18 | 11 | 8.9 |
| HEV-B | EV25 | 1 | 0.8 |
| HEV-B | EV30 | 1 | 0.8 |
| HEV-B | HEV-Bb | 2 | 1.6 |
| HEV-C | CAV19 | 2 | 1.6 |
| Untyped | HEV | 3 | 2.4 |
In the period from September 2001 to November 2003. The frequencies of different serotypes are reported as numbers of infection episodes.
Species were identified by pairwise comparison of RNA polymerase sequences.
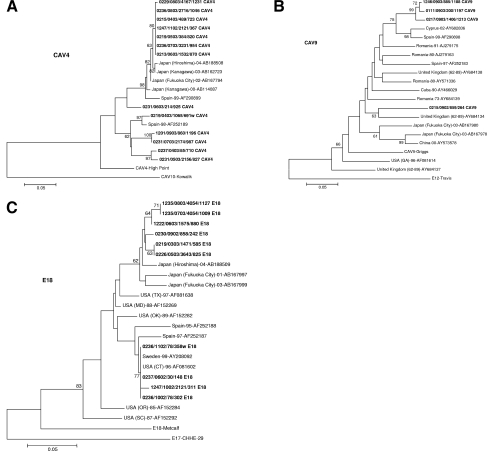
Individual serotypes of the VP1 sequences were compared phylogenetically with all corresponding sequences of the respective serotypes in GenBank. Three serotypes (of the 22 analyzed) segregated in different clusters (CAV4, CAV9, and E18 [Fig. 1A to C, respectively]), indicating that there have been at least two introductions of genotypes into the population for each of these serotypes. Two distinct clusters were observed for the CAV4-positive samples (Fig. 1A). Seven CAV4 sequences clustered together with sequences from Spain (1999) or Japan (2000 to 2004); six sequences segregated with a strain that circulated in Spain in 1998 (Fig. 1A). Some CAV9 sequences (Fig. 1B) were clustered with isolates from Cyprus (2002) and Spain (1999); one clustered with an unrelated cluster closer to an isolate from the United Kingdom (1962 to 1989). E18 sequences clustered with sequences isolated in Japan (2001 to 2004) or Spain (1995 to 1997), the United States (1996), and Sweden (1999) (Fig. 1C). Although more than one strain was found to be cocirculating in certain periods, as seen for CAV4 (Fig. 1A), the two clusters of CAV9 and E18 seem to have been introduced to the cohort at different times (with one exception) (Fig. 1B and C). The circulation of subgroups for each of the serotypes did not appear to be geographically restricted (Fig. 1A to C).
FIG. 1.
Rooted phylogenetic trees of CAV4 (A), CAV9 (B), and E18 (C) 3′-end VP1 sequences and available reference sequences and prototypes from GenBank for each of the serotypes. The evolutionary distances were calculated using the Kimura two-parameter model as a model of nucleotide substitution and the neighbor-joining method to reconstruct the phylogenetic tree (MEGA version 3.0 software package). The bootstrap values (the percentage of 1,000 pseudoreplicate data sets) supporting each cluster are shown at the nodes; for clarity, only values of >60% are shown. The outgroup taxa are the prototype strains of the closest serotypes. Sequence names for field strains (in boldface) are constructed as follows: municipality number (starting with 1, in Hordaland county; starting with 0, in Akershus county), month and year of sampling, child identifier, sample number, and serotype. The reference sequences have GenBank accession numbers. The scale bar represents the genetic distance (the number of nucleotide substitutions per site).
Prevalence and epidemiology of HEV-A and HEV-B infections.
HEV was detected in 145 of 1,255 samples (11.6% of samples; 95% CI, 9.8% to 13.5%). HEV-A was detected most frequently, with an overall prevalence of 6.8% (95% CI, 5.0% to 8.6%); HEV-B was present in 4.8% of the samples (95% CI, 3.2% to 6.4%). HEV-C was present in only three samples (0.2%). No poliovirus or HEV-D group viruses were detected. Of 124 infection episodes, 70 (57%) were classified as HEV-A; 49 (40%) were classified as HEV-B (Table 2). HEV-B infections were more prevalent than HEV-A infections during the interval from March through August 2002 (2.9% to 4.4% versus 1.4% to 2.6%); however, during the interval from December 2002 through November 2003, HEV-A infections were more prevalent than HEV-B infections (3.9% to 10.4% versus 1.6% to 6.4%). From September through November 2002, HEV-A and HEV-B prevalence rates were similar (7.8%). The highest prevalence of HEV-A was observed from June through August 2003 (10.4%); the highest prevalence of HEV-B was observed from September through November 2002 (7.8%). The characteristic seasonal variation of HEV infections, with peak incidence in late summer and autumn, was seen for both HEV-A and HEV-B (data not shown).
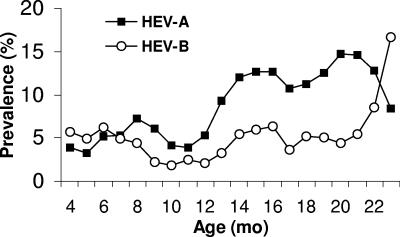
Figure 2 shows the age-specific prevalences of HEV-A and HEV-B infections for children aged 3 to 23 months. The 3-month moving averages of the age-specific prevalences of positive samples are plotted against the ages of the children in months. In both species, the highest prevalence was in the second year of life.
FIG. 2.
Three-month moving average of the age-specific prevalences of HEV-A- and HEV-B-positive samples for ages 3 to 23 months (n = 1,237 person-months). The curve was truncated at 24 months, due to few observations (18 stool samples) from 24 to 28 months of age. A logistic regression model was applied to adjust for the potential mutual confounding of age and season or year. Adjusting for season or year indicated that confounding by these factors was minimal or did not change the effect of age in the logistic-regression model.
The probability of having at least one HEV infection by the age of 12 months was approximately 40%; 90% of the children had been infected by the age of 2 years (see Fig. S1 in the supplemental material). There were no significant differences in prevalence of HEV-A or HEV-B between boys and girls or between the two main counties included in the study (data not shown).
Prolonged excretion, viral load, and coinfections.
In 28 of the 124 infection episodes, the same viral serotype was observed in two or more consecutive samples: 23 cases of two consecutive months, 4 cases of three consecutive months, and 1 case of four consecutive months. Prolonged duration of infection was associated with a higher viral load in the first sample, but not with sex, viral species, or season. The initial viral load was on average eightfold higher in infections that persisted for at least 2 months than in single-month episodes (P = 0.001). A trend toward shorter duration of infection with increasing age was not statistically significant (P = 0.078).
Stool samples from 10 children showed evidence of coinfection with more than one HEV. Eight samples contained two serotypes; two samples had three serotypes. In six samples, the coinfections were limited to HEV-A; in three cases, there was coinfection with HEV-A and HEV-B; in one case, there was coinfection with HEV-A and HEV-C. The viral loads in the coinfected samples were not significantly different than in samples containing only a single serotype.
DISCUSSION
HEV infections were prospectively assessed in stools of healthy children by using a molecular typing strategy. HEVs were detected in 11.6% of the stool samples; 98% of the HEV-positive samples were identified to the species level. HEV-A species were detected in 6.8% of the samples; HEV-B was detected in 4.8% of the samples. Although these findings contrast with recent studies of HEV epidemiology based on culture and serology that reported only infrequent HEV-A infections in symptomatic (2, 5-7, 9, 25, 27, 37, 43) or healthy (13, 16, 21, 23, 38) children, they are consistent with earlier studies in which suckling mice were employed for in vivo assays and HEV-A infections were observed to be relatively frequent in healthy children (12, 17, 39). In a 1958 and 1959 study of stool samples obtained from 2,084 healthy children less than 5 years of age in London, the prevalence rates for HEV-A (CAV2, -4, -5, -6, -8, -10, and -12) and HEV-B (CBV3 and -4) isolates were 5.4% and 1.2%, respectively (17). In a study of clinical samples (primarily stools) obtained in 1966 and 1967 from 625 healthy children in Australia, the prevalence rates for HEV-A (CAV2, -3, -4, -8, and -10) and HEV-B (CBV2, -4, and -5 and CAV9) isolates were 12.1% and 4.3%, respectively (12). The Australian samples were collected in a single nursery; thus, the extent to which they represent the prevalence in the general population is uncertain. Finally, a Malaysian study of stool samples collected in 1971 and 1972 from healthy children less than 7 years of age found prevalences for HEV-A of 5.3% and for HEV-B of 3.4% (39). No species identification was reported.
We speculate that differences in the prevalences of individual HEV serotypes between our study and others reflect methodological differences. Whereas in vitro culture methods appear to be more sensitive for HEV-B, in vivo culture methods are more sensitive for HEV-A. To our knowledge, there are no previous studies reporting the prevalence of HEV-A in clinical samples using molecular methods. Our data indicate similar or equal sensitivities for both HEV-A and HEV-B. Thus, the findings reported here may represent accurate levels of circulating HEV serotypes.
Our analysis of seasonality confirmed the established pattern for HEV infections, where the highest prevalence in the Northern Hemisphere occurs between July and November (11). This observation held for both HEV-A and HEV-B (data not shown).
Analysis of age-specific frequencies of HEV-A and HEV-B showed an apparent trough between the ages of 9 and 12 months, with a distinct increase after 12 months of age (Fig. 2). These findings are consistent with those of Gamble et al. (17), in which the frequencies of both HEV-A and HEV-B infections were lower in the first year of life.
Phylogenetic analysis of the C-terminal domain of the VP1 gene has facilitated the differentiation of circulating serotypes and the association of specific HEV genotypes with disease (1, 3, 34, 40). Some serotypes are associated with global epidemics (1). During the period from 2001 to 2003, E13 and E18 were reported as the more common serotypes causing outbreaks of aseptic meningitis in the United States and elsewhere (6, 37). Most of the infections were E18 (8.9% versus 1.6%). Our VP1 sequence analysis revealed the circulation of several strains of the same serotype (Fig. 1).
HEV coinfections were commonly observed in the Norwegian infants followed in the present study. Although coinfections have been described in the developing world (23), they are only infrequently reported in children in industrialized or temperate areas (13).
We found no association between sex and either the frequency or duration of infection. These findings are in contrast with those reported for HEV-associated diseases, where infections were more frequent in males than in females, particularly for severe diseases, such as meningitis and carditis (35).
Our results suggest that the prevalence of HEV-A infections has been underestimated in recent studies that have relied on cell culture for detection. The diagnosis of HEV-A infections could be considerably improved by the use of molecular methods that facilitate detection and genotypic analysis.
Supplementary Material
Acknowledgments
This study was supported by grants from the Norwegian Research Council (155300/320), the Ministry of Education of the Czech Republic (grant no. MSM0021620814), and the National Institutes of Health (AI55466 and AI51202). The MIDIA study is supported by the Norwegian Research Council (135893/330 and 156477/730).
We are grateful to the health care nurses and laboratory technicians in the MIDIA project and the laboratory technicians at the Department of Virology, Norwegian Institute of Public Health; to Trond Rasmussen for excellent data management and IT solutions; to Håkon Gjessing and Anders Skrondal for help with advanced statistical methods; and to Marian Rewers for sharing insights and for support.
Footnotes
Published ahead of print on 30 August 2006.
Supplemental material for this article may be found at http://jcm.asm.org.
REFERENCES
- 1.Avellon, A., I. Casas, G. Trallero, C. Perez, A. Tenorio, and G. Palacios. 2003. Molecular analysis of echovirus 13 isolates and aseptic meningitis, Spain. Emerg. Infect. Dis. 9:934-941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bahri, O., D. Rezig, B. B. Nejma-Oueslati, A. B. Yahia, J. B. Sassi, N. Hogga, A. Sadraoui, and H. Triki. 2005. Enteroviruses in Tunisia: virological surveillance over 12 years (1992-2003). J. Med. Microbiol. 54:63-69. [DOI] [PubMed] [Google Scholar]
- 3.Brown, B. A., M. S. Oberste, J. P. Alexander, Jr., M. L. Kennett, and M. A. Pallansch. 1999. Molecular epidemiology and evolution of enterovirus 71 strains isolated from 1970 to 1998. J. Virol. 73:9969-9975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Casas, I., G. F. Palacios, G. Trallero, D. Cisterna, M. C. Freire, and A. Tenorio. 2001. Molecular characterization of human enteroviruses in clinical samples: comparison between VP2, VP1, and RNA polymerase regions using RT nested PCR assays and direct sequencing of products. J. Med. Virol. 65:138-148. [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. 2006. Enterovirus surveillance—United States, 2002-2004. Morb. Mortal. Wkly. Rep. 55:153-156. [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. 2002. Enterovirus surveillance—United States, 2000-2001. Morb. Mortal. Wkly. Rep. 51:1047-1049. [PubMed] [Google Scholar]
- 7.Centers for Disease Control and Prevention. 2003. Outbreaks of aseptic meningitis associated with echoviruses 9 and 30 and preliminary surveillance reports on enterovirus activity—United States, 2003. Morb. Mortal. Wkly. Rep. 52:761-764. [PubMed] [Google Scholar]
- 8.Chia, J. K. 2005. The role of enterovirus in chronic fatigue syndrome. J. Clin. Pathol. 58:1126-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christensen, A., and S. A. Nordbø. 2003. Enterovirus infections diagnosed in middle Norway during the period 1992-2001. Tidsskr. Nor. Laegeforen. 123:3180-3183. (In Norwegian.) [PubMed] [Google Scholar]
- 10.Cinek, O., E. Wilkinson, L. Paltiel, O. D. Saugstad, P. Magnus, and K. S. Rønningen. 2000. Screening for the IDDM high-risk genotype. A rapid microtitre plate method using serum as source of DNA. Tissue Antigens 56:344-349. [DOI] [PubMed] [Google Scholar]
- 11.Cinek, O., E. Witsø, S. Jeansson, T. Rasmussen, P. Drevinek, T. Wetlesen, J. Vavrinec, B. Grinde, and K. S. Rønningen. 2006. Longitudinal observation of enterovirus and adenovirus in stool samples from Norwegian infants with the highest genetic risk of type 1 diabetes. J. Clin. Virol. 35:33-40. [DOI] [PubMed] [Google Scholar]
- 12.Cook, I., B. C. Allan, and S. Welham. 1969. Coxsackieviruses in normal children. Med. J. Aust. 2:789-792. [DOI] [PubMed] [Google Scholar]
- 13.Cooney, M. K., C. E. Hall, and J. P. Fox. 1972. The Seattle virus watch. 3. Evaluation of isolation methods and summary of infections detected by virus isolations. Am. J. Epidemiol. 96:286-305. [DOI] [PubMed] [Google Scholar]
- 14.Dalakas, M. C. 1995. Enteroviruses and human neuromuscular diseases, p. 387-398. In H. A. Rotbart (ed.), Human enterovirus infections. ASM Press, Washington, D.C.
- 15.Efron, B., and R. J. Tibshirani. 1993. An introduction to the bootstrap. Chapman & Hall, New York, N.Y.
- 16.Froeschle, J. E., P. M. Feorino, and H. M. Gelfand. 1966. A continuing surveillance of enterovirus infection in healthy children in six United States cities. II. Surveillance enterovirus isolates 1960-1963 and comparison with enterovirus isolates from cases of acute central nervous system disease. Am. J. Epidemiol. 83:455-469. [DOI] [PubMed] [Google Scholar]
- 17.Gamble, D. R. 1962. Isolation of Coxsackie viruses from normal children aged 0-5 years. Br. Med. J. 5270:16-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gelfand, H. M., A. H. Holguin, G. E. Marchetti, and P. M. Feorino. 1963. A continuing surveillance of enterovirus infections in healthy children in six United States cities. I. Viruses isolated during 1960 and 1961. Am. J. Hyg. 78:358-375. [DOI] [PubMed] [Google Scholar]
- 19.Hyypiä, T., T. Hovi, N. J. Knowles, and G. Stanway. 1997. Classification of enteroviruses based on molecular and biological properties. J. Gen. Virol. 78:1-11. [DOI] [PubMed] [Google Scholar]
- 20.Kearney, M. T., J. M. Cotton, P. J. Richardson, and A. M. Shah. 2001. Viral myocarditis and dilated cardiomyopathy: mechanisms, manifestations, and management. Postgrad. Med. J. 77:4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kogon, A., I. Spigland, T. E. Frothingham, L. Elveback, C. Williams, C. E. Hall, and J. P. Fox. 1969. The virus watch program: a continuing surveillance of viral infections in metropolitan New York families. VII. Observations on viral excretion, seroimmunity, intrafamilial spread and illness association in coxsackie and echovirus infections. Am. J. Epidemiol. 89:51-61. [DOI] [PubMed] [Google Scholar]
- 22.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 23.Kuramitsu, M., C. Kuroiwa, H. Yoshida, M. Miyoshi, J. Okumura, H. Shimizu, L. Narantuya, and D. Bat-Ochir. 2005. Non-polio enterovirus isolation among families in Ulaanbaatar and Tov province, Mongolia: prevalence, intrafamilial spread, and risk factors for infection. Epidemiol. Infect. 133:1131-1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lipson, S. M., R. Walderman, P. Costello, and K. Szabo. 1988. Sensitivity of rhabdomyosarcoma and guinea pig embryo cell cultures to field isolates of difficult-to-cultivate group A coxsackieviruses. J. Clin. Microbiol. 26:1298-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maguire, H. C., P. Atkinson, M. Sharland, and J. Bendig. 1999. Enterovirus infections in England and Wales: laboratory surveillance data: 1975 to 1994. Commun. Dis. Public Health 2:122-125. [PubMed] [Google Scholar]
- 26.Muir, P., U. Kammerer, K. Korn, M. N. Mulders, T. Pöyry, B. Weissbrich, R. Kandolf, G. M. Cleator, A. M. van Loon, et al. 1998. Molecular typing of enteroviruses: current status and future requirements. Clin. Microbiol. Rev. 11:202-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nairn, C., and G. B. Clements. 1999. A study of enterovirus isolations in Glasgow from 1977 to 1997. J. Med. Virol. 58:304-312. [PubMed] [Google Scholar]
- 28.Norder, H., L. Bjerregaard, L. Magnius, B. Lina, M. Aymard, and J. J. Chomel. 2003. Sequencing of ‘untypable’ enteroviruses reveals two new types, EV-77 and EV-78, within human enterovirus type B and substitutions in the BC loop of the VP1 protein for known types. J. Gen. Virol. 84:827-836. [DOI] [PubMed] [Google Scholar]
- 29.Oberste, M., D. Schnurr, K. Maher, S. al Busaidy, and M. Pallansch. 2001. Molecular identification of new picornaviruses and characterization of a proposed enterovirus 73 serotype. J. Gen. Virol. 82:409-416. [DOI] [PubMed] [Google Scholar]
- 30.Oberste, M. S., K. Maher, D. R. Kilpatrick, and M. A. Pallansch. 1999. Molecular evolution of the human enteroviruses: correlation of serotype with VP1 sequence and application to picornavirus classification. J. Virol. 73:1941-1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oberste, M. S., K. Maher, S. M. Michele, G. Belliot, M. Uddin, and M. A. Pallansch. 2005. Enteroviruses 76, 89, 90 and 91 represent a novel group within the species Human enterovirus A. J. Gen. Virol. 86:445-451. [DOI] [PubMed] [Google Scholar]
- 32.Oberste, M. S., S. M. Michele, K. Maher, D. Schnurr, D. Cisterna, N. Junttila, M. Uddin, J. J. Chomel, C. S. Lau, W. Ridha, S. al Busaidy, H. Norder, L. O. Magnius, and M. A. Pallansch. 2004. Molecular identification and characterization of two proposed new enterovirus serotypes, EV74 and EV75. J. Gen. Virol. 85:3205-3212. [DOI] [PubMed] [Google Scholar]
- 33.Oberste, M. S., and M. A. Pallansch. 2003. Establishing evidence for enterovirus infection in chronic disease. Ann. N. Y. Acad. Sci. 1005:23-31. [DOI] [PubMed] [Google Scholar]
- 34.Palacios, G., I. Casas, D. Cisterna, G. Trallero, A. Tenorio, and C. Freire. 2002. Molecular epidemiology of echovirus 30: temporal circulation and prevalence of single lineages. J. Virol. 76:4940-4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pallansch, M. A., and R. P. Roos. 2001. Enteroviruses: polioviruses, coxsackieviruses, echoviruses, and newer enteroviruses, p. 723-775. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, and B. Roizman, and S. E. Straus (ed.), Fields virology. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 36.Pöyry, T., L. Kinnunen, T. Hyypiä, B. Brown, C. Horsnell, T. Hovi, and G. Stanway. 1996. Genetic and phylogenetic clustering of enteroviruses. J. Gen. Virol. 77:1699-1717. [DOI] [PubMed] [Google Scholar]
- 37.Sedmak, G., D. Bina, and J. MacDonald. 2003. Assessment of an enterovirus sewage surveillance system by comparison of clinical isolates with sewage isolates from Milwaukee, Wisconsin, collected August 1994 to December 2002. Appl. Environ. Microbiol. 69:7181-7187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spigland, I., J. P. Fox, L. R. Elveback, F. E. Wassermann, A. Ketler, C. D. Brandt, and A. Kogon. 1966. The Virus Watch program: a continuing surveillance of viral infections in metropolitan New York families. II. Laboratory methods and preliminary report on infections revealed by virus isolation. Am. J. Epidemiol. 83:413-435. [DOI] [PubMed] [Google Scholar]
- 39.Tan, D. S., and S. K. Lam. 1978. Enterovirus survey before and after poliomyelitis vaccination in Kuala Lumpur, Malaysia. Southeast Asian J. Trop. Med. Public Health 9:301-307. [PubMed] [Google Scholar]
- 40.Thoelen, I., P. Lemey, I. Van Der Donck, K. Beuselinck, A. M. Lindberg, and M. Van Ranst. 2003. Molecular typing and epidemiology of enteroviruses identified from an outbreak of aseptic meningitis in Belgium during the summer of 2000. J. Med. Virol. 70:420-429. [DOI] [PubMed] [Google Scholar]
- 41.Thoelen, I., E. Moes, P. Lemey, S. Mostmans, E. Wollants, A. M. Lindberg, A. M. Vandamme, and M. Van Ranst. 2004. Analysis of the serotype and genotype correlation of VP1 and the 5′ noncoding region in an epidemiological survey of the human enterovirus B species. J. Clin. Microbiol. 42:963-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trallero, G., I. Casas, A. Tenorio, J. E. Echevarria, A. Castellanos, A. Lozano, and P. P. Brena. 2000. Enteroviruses in Spain: virological and epidemiological studies over 10 years (1988-97). Epidemiol. Infect. 124:497-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Welch, J., K. Maclaran, T. Jordan, and P. Simmonds. 2003. Frequency, viral loads, and serotype identification of enterovirus infections in Scottish blood donors. Transfusion 43:1060-1066. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.