Abstract
Eight optochin-susceptible (Opts) alpha-hemolytic (viridans) streptococcus isolates were characterized at the molecular level. These isolates showed phenotypic characteristics typical of both viridans streptococci and Streptococcus pneumoniae. Comparison of the sequence of housekeeping genes from these isolates with those of S. pneumoniae, Streptococcus mitis, Streptococcus oralis, and Streptococcus pseudopneumoniae suggested that the Opts isolates corresponded to streptococci of the mitis group. Besides, the Opts streptococci were negative by a Gen-Probe AccuProbe pneumococcus test and hybridized with specific pneumococcal probes (lytA and ply) but also with ant, a gene not present in most S. pneumoniae strains. Moreover, the isolates were insoluble in 1% sodium deoxycholate but completely dissolved in 0.1% deoxycholate. Sequence analysis of the lytA gene revealed that the Opts streptococci carried lytA alleles characteristic of those present in nonpneumococcal streptococci of the mitis group. The determination of the partial nucleotide sequence embracing the atp operon encoding the FoF1 H+-ATPase indicated that the optochin susceptibility of the isolates was due to the acquisition of atpC, atpA, and part of atpB from S. pneumoniae by horizontal gene transfer.
Streptococcus pneumoniae (the pneumococcus) is an important cause of morbidity and mortality worldwide, remaining a major etiological agent of community-acquired pneumonia, meningitis, and acute otitis media (6). Three phenotypic characteristics are used for identification of the pneumococcus among alpha-hemolytic (viridans) streptococci: optochin (Opt) susceptibility, bile (sodium deoxycholate [Doc]) solubility, and reaction with type-specific antisera (21). Although their colony morphologies can be very similar, the alpha-hemolytic streptococci of the mitis group (SMG), such as Streptococcus mitis or Streptococcus oralis, are classically Opt resistant (Optr), insoluble in 1% Doc, and nontypeable (21).
In most countries, the Opt susceptibility test remains the only clinical test carried out for the identification of the pneumococcus, although Doc solubility is also currently inspected in many U.S. laboratories. It is well known that the typical Opt susceptibility of pneumococci is due to the characteristics of the Fo complex of its FoF1 H+-ATPase (12), an enzyme essential for the viability of this organism (13). The primary roles of this enzyme are to create a proton gradient with the energy provided by ATP hydrolysis and to maintain the intracellular pH via proton extrusion (24). Hydrolysis of ATP on the cytoplasmic F1 sector (α, β, δ, ξ, and γ subunits) drives proton transport through the Fo cytoplasmic membrane sector (a, b, and c subunits) by long-range conformational changes (1, 34, 36). The activity of the FoF1 ATPase of S. pneumoniae is pH inducible and is regulated at the level of initiation of transcription (24).
Resistance to Opt or other amino-alcohol antimalarial drugs (e.g., quinine or mefloquine) in pneumococcus has been reported as a consequence of point mutations that change amino acid residues located in either one of the two transmembrane α-helices of the c subunit or one of the two last α-helices of the a subunit (8, 12, 25, 28, 33). This fact suggests that the above-mentioned α-helices of these subunits interact and that the mutated residues are important for the structure of the Fo complex and, consequently, for proton translocation. Although several S. pneumoniae Optr isolates have been reported previously (19, 27, 32, 33, 39), to the best of our knowledge there are only three reports describing Opts SMG (5, 23, 26). Only a single Opts SMG strain has been characterized as having a recombinant structure in the genes encoding the FoF1 H+-ATPase, that is, it had acquired atpC, atpA, and part of atpB from S. pneumoniae (23).
A new member of the mitis group (Streptococcus pseudopneumoniae) has been described recently (3). Isolates of this species exhibit an Optr phenotype when they are incubated under an atmosphere of increased CO2, but Opt susceptibility increases in the presence of O2 (3). For this reason, this previous report recommended the use of a CO2-enriched atmosphere for this test to prevent misidentification of SMG Opt variants as truly pneumococci. The same authors described S. pseudopneumoniae as Doc insoluble, although solubility in 0.1% Doc has been documented recently (20). It has also been found that some SMG isolates, including S. pseudopneumoniae strains, carry atypical alleles of the lytA gene (encoding the major pneumococcal autolysin LytA) and that most of them are insoluble in 1% Doc but dissolve completely when 0.1% Doc is used (20). This behavior is in agreement with the finding that 1% Doc (but not 0.1% Doc) inhibited the activity of the LytA-like amidases from SMG, whereas the LytA enzyme synthesized by typical pneumococci was not affected by the detergent (30).
In this work we have gained more insight into the knowledge of the Opts phenotype in SMG by characterizing eight Opts SMG strains isolated during 2002 at the Spanish Pneumococcal Reference Laboratory and have determined a partial sequence of the atp operon from two S. pseudopneumoniae strains.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and determination of optochin sensitivity.
The bacterial strains used in this work are listed in Table 1. Unless otherwise stated, streptococci were grown in Todd-Hewitt broth supplemented with 0.5% yeast extract at 37°C without shaking. For determination of Opt sensitivity, each isolate was cultured onto Mueller-Hinton agar plates (Difco) supplemented with 5% defibrinated sheep blood (BA plates). Several colonies were suspended in liquid culture, and 107 CFU was applied to BA plates in which 5-μg Opt disks of 6 mm (Becton Dickinson Microbiology Systems) were placed. After overnight incubation at 37°C in either 5% CO2 or O2 atmosphere, inhibition zones around the disk were measured. MICs were determined by the agar dilution method as recommended by the Clinical and Laboratory Standards Institute (29), except that plates were incubated in either 5% CO2 or ambient atmosphere. The MIC was defined as the lowest concentration of drug that inhibited visible growth. S. pneumoniae ATCC 6303, S. pneumoniae R6, S. mitis NCTC 12261T, S. oralis NCTC 11427T, and two S. pseudopneumoniae strains (CCUG 49455T and CCUG 48465) were used as controls. Opt was purchased from Sigma Chemical, St. Louis, Mo.
TABLE 1.
Relevant characteristics of the strains used in this study
| Strain | Origin(s) and patient condition(s)a | Organism(s) identified by API (% identification) | Opt susceptibility
|
Doc phenotype | APc,d | Presence ofc:
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Zone size (mm) in:
|
MIC (μg/ml) in:
|
Phenotype (CO2/O2)b | ||||||||||
| CO2 | O2 | CO2 | O2 | lytA | ply | ant | ||||||
| SMG 578 | Nose, sinusitis | S. oralis(98.5) | 15 | 22 | 1.5 | 0.75 | S/S | +/− | − | + | + | + |
| SMG 1237 | BAA, pneumonia, HIV+ | S. oralis(84.4) | 18 | 21 | 1.5 | 0.37 | S/S | +/− | − | + | + | + |
| SMG 1504 | LRT | S. oralis(99.0) | 20 | 23 | 1.5 | 0.75 | S/S | +/− | − | + | + | + |
| SMG 1956 | Sputum, fever | S. oralis(94.9) | 17 | 24 | 1.5 | 0.75 | S/S | +/− | − | + | + | + |
| SMG 2410 | Pus, abscess | S. oralis(89.9) | 15 | 20 | 1.5 | 0.75 | S/S | +/− | − | + | + | + |
| SMG 2859 | Ascites | S. oralis(82.4) | 18 | 20 | 1.5 | 0.75 | S/S | +/− | − | + | + | + |
| SMG 3137 | BAA, pneumonia | S. oralis(53.4), S. pneumoniae (32.6) | 16 | 20 | 1.5 | 0.75 | S/S | +/− | − | + | + | + |
| SMG 3198 | Sputum, bronchitis | S. oralis(93.5) | 12 | 18 | 3 | 0.75 | IR/S | +/− | − | + | + | + |
| S. mitis NCTC 12261T | 6 | 6 | 96 | 48 | R/R | − | − | − | − | + | ||
| S. oralis NCTC 11427T | 6 | 6 | 96 | 48 | R/R | − | − | − | − | + | ||
| S. pneumoniae R6 | 12 | 19 | 3 | 1.5 | IR/S | + | + | + | + | − | ||
| S. pneumoniaeATCC 6303 | 16 | 22 | 1.5 | 0.75 | S/S | + | + | + | + | − | ||
| S. pseudopneumoniaeCCUG 49455T | 10 | 20 | 6 | 1.5 | IR/S | +/− | + | + | + | − | ||
| S. pseudopneumoniaeCCUG 48465 | 12 | 21 | 6 | 0.75 | IR/S | +/− | + | + | + | − | ||
BAA, bronchoalveolar aspirate; HIV+, human immunodeficiency virus positive; LRT, lower respiratory tract.
Opt susceptibility phenotypes were categorized as follows: resistant (R), MIC of ≥6 μg/ml or zone size diameter of <10 mm; intermediate resistant (IR), MIC of 3 to 6 μg/ml or zone size diameter of 10 to 14 mm; susceptible (S), MIC of ≤1.5 μg/ml or zone size diameter of >14 mm.
+, positive; −, negative.
AP, AccuProbe.
Deoxycholate solubility tests.
Aliquots (0.5 ml) of exponentially growing cultures received 50 μl of 1 M potassium phosphate buffer (pH 8.0) and 50 μl of a 10% or 1% Doc solution in water. The mixtures were incubated for up to 15 min at 37°C. Lysis was followed by decrease of the turbidity. The strains that lysed with 1% Doc were designated Doc+. This was the case for true pneumococcal isolates. However, those SMG strains that lysed with 0.1% Doc but not with 1% Doc were designated Doc+/−.
Gen-Probe (AccuProbe).
For detection of the rRNA genes, an AccuProbe S. pneumoniae culture identification test (Gen-Probe, San Diego, Calif.) was used according to the manufacturer's instructions with four colonies from an overnight culture on 5% BA plates.
PCR amplification and DNA sequence determination and analysis.
PCR amplifications were performed using 0.5 to 1 U of Thermus thermophilus thermostable DNA polymerase (Biotools), 0.1 μg of chromosomal DNA, 1 μM (each) of the synthetic oligonucleotide primers, and 0.2 mM of each deoxynucleoside triphosphate in a final volume of 50 μl in the buffer recommended by the manufacturers. Amplification was achieved with an initial cycle of 1 to 5 min of denaturation at 94°C, 25 to 30 cycles of 1 min of denaturation at 94°C, 1 min of annealing at 55°C, and 1 to 4 min of polymerase extension at 72°C, with a final 8-min extension at 72°C and slow cooling at 4°C. PCR products were purified using MicroSpin S400 HR columns (Amersham-Pharmacia-Biotech) and sequenced on both strands with an Applied Biosystems Prism 377 DNA sequencer. The following atp operon regions were amplified. Fragments of about 3 kb from SMG isolates 1504, 1237, 1956, 2859, and 3137 were amplified with oligonucleotides pepti101 (23) and atpδ110 (24), located in spr1284 and in the gene of the delta subunit of the FoF1 H+-ATPase, respectively. Fragments of 1,935 bp from isolates 578, 2410, and 3198 were amplified with oligonucleotides pepti368 (5′-CCAGGTTTCCGTCATTTTGAAAC-3′) and atpBEND (5′-GGCTTCTCCTAGCTGATCGATATAC-3′), located in spr1284 and in atpB, respectively. To amplify the atpCAB region from S. pseudopneumoniae strains, PCR fragments were obtained by using oligonucleotides atpWO (23) and atpBEND. The upstream atpC regions from those strains were amplified with oligonucleotides spr1370-196 (5′-ATTCCTTGCCTTGCTCTCAGCAG-3′) and spr1368END2 (5′-GACACAATTTCTAGGTTATAACTGG-3′), located in spr1370 and downstream spr1368, respectively, yielding fragments of about 3 kb. PCR fragments were sequenced with the same oligonucleotides and with several internal oligonucleotides. In addition, direct sequencing of chromosomal DNAs by use of oligonucleotide atpCorf2 (24) was performed. PCR amplification and sequencing of the lytA alleles from SMG strains were carried out using conditions and oligonucleotide primers described elsewhere (20). Multilocus sequence typing (MLST) was carried out exactly as described elsewhere (11) with the tools provided by the MLST website (http://www.mlst.net) for the molecular characterization of pneumococci. Multiple sequence alignments were created with PILEUP (9) or CLUSTAL W (38). Pairwise evolutionary distances (PEDs) (estimated number of substitutions per 100 bases) were determined using the Distances program (9) with the Kimura two-parameter distance correction (18). Sequence comparisons were carried out by running the BLAST program (2) with data from the EMBL/UniProtKB database and preliminary genomic data for S. pneumoniae 670 and the type strain of S. mitis (The Institute for Genomic Research website at http://www.tigr.org) and for other pneumococcal strains (The Sanger Institute; http://www.sanger.ac.uk/Projects/S_pneumoniae).
Southern blot identification of strains.
Restriction fragments carrying lytA and ply (pneumolysin) DNA probes and PCR products carrying the ant probe (a homolog of genes encoding aminoglycoside adenylyltransferases) were obtained as described previously (14, 23). Probes were labeled with a Phototope-Star detection kit (New England Biolabs). Southern blotting and hybridization were carried out by following the manufacturer's instructions.
Nucleotide sequence accession numbers.
The sequences of the atpCAB genes reported here have been deposited in the EMBL/GenBank/DDBJ databases. The lytA alleles have been assigned accession numbers AM113498, AM113499, AM113501, AM113502, and AM1113505, the housekeeping fragments are accession numbers DQ659935 to DQ659964 and DQ665241 to DQ665250, and the atp genes and their surrounding regions are accession numbers DQ659925 to DQ659934.
RESULTS
Phenotypic characteristics of Opts SMG.
The Spanish Pneumococcal Reference Laboratory receives pneumococcal isolates for typing purposes and antibiotic resistance surveillance. Isolates are routinely confirmed to be S. pneumoniae isolates by means of the Opt susceptibility and Doc solubility tests. Among the isolates received during 2002, 2.4% (78 out of 3,240) showed any discrepancy in those tests and did not show hybridization with AccuProbe. Among the 78 AccuProbe-negative isolates, 11 exhibited a clear Opt inhibition zone (≥12 mm) in the presence of 5% CO2. Eight of these 11 isolates were chosen for further studies (Table 1). Phenotypic characterization of these Opts SMG isolates by use of a Rapid ID 32 STREP kit (bioMerieux, La Balme les Grottes, France) system classified them as S. oralis isolates. This was not completely unexpected since it is documented that phenotypic characterization is of limited value for identification of many species of nonhemolytic streptococci and particularly of SMG (16).
As previously reported (3), S. pseudopneumoniae showed Opt susceptibility (zones of inhibition of ≥14 mm) with Opt disks when the test was carried out in ambient air and Opt intermediate resistance in a CO2-enriched atmosphere. Unexpectedly, a similar reaction was also observed for S. pneumoniae R6 (Table 1), making it difficult to evaluate the significance of the results. As it remains unclear whether incubation in CO2, as recommended in the Manual of Clinical Microbiology (35), or ambient atmosphere, as recommended by the manufacturers of Opt test disks, is optimal for the identification of pneumococci, we decided to establish the Opt susceptibilities of various strains by following the standard procedures used to determine the MIC. The Opt MICs were always higher in the presence of 5% CO2 than in ambient air. The lower Opt susceptibility in an atmosphere of increased CO2 may be related to the activation of the promoter of the atp operon in response to acid, which would produce a twofold increase in the amount of the FoF1 H+-ATPase (24), increasing the Opt MIC by titration.
The eight isolates showed a Doc+/− phenotype, that is, they did not lyse in the presence of 1% Doc but were solubilized when 0.1% Doc was used (not shown). Furthermore, they autolyzed after a prolonged incubation at 37°C, confirming previous results obtained with strains 578 and 1504, which synthesize a defective but functional LytA autolysin (20). These results taken together strongly suggested the presence of an atypical lytA allele not only in strains 578 and 1504 but also in the other SMG strains (see below).
Genetic relatedness of the Opts SMG isolates.
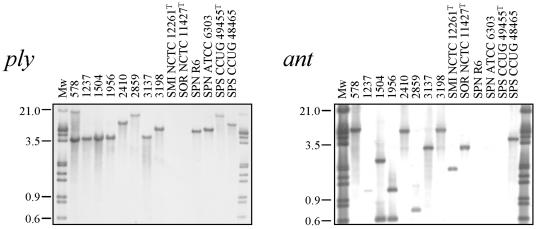
The eight Opts Doc+/− AccuProbe-negative SMG isolates were inspected to determine the presence of pneumococcal-specific genes, such as ply or lytA (not shown). Southern blot hybridization experiments showed that all eight strains harbored both genes (Fig. 1 and unpublished observations). Quite surprisingly, however, they also hybridized with the ant probe, a gene that is not normally present in S. pneumoniae but that is found in S. mitis and S. oralis (4).
FIG. 1.
Southern blot hybridization of streptococcal isolates by hybridization with ply- and ant-specific probes. Chromosomal DNAs were cleaved with ClaI (A) and EcoRV plus NcoI (B), and the fragments were separated in 1% agarose gels. The gel was blotted, and the blot was probed with biotinylated DNA as follows: an insert of plasmid pJCP191 containing the ply gene and a PCR fragment from S. pneumoniae 3870 containing most of the ant gene. Far-left and far-right lanes, biotinylated DNA ladder. Numbers at left are molecular size markers in kilodaltons. SMI, S. mitis; SOR, S. oralis; SPN, S. pneumoniae; SPS, S. pseudopneumoniae.
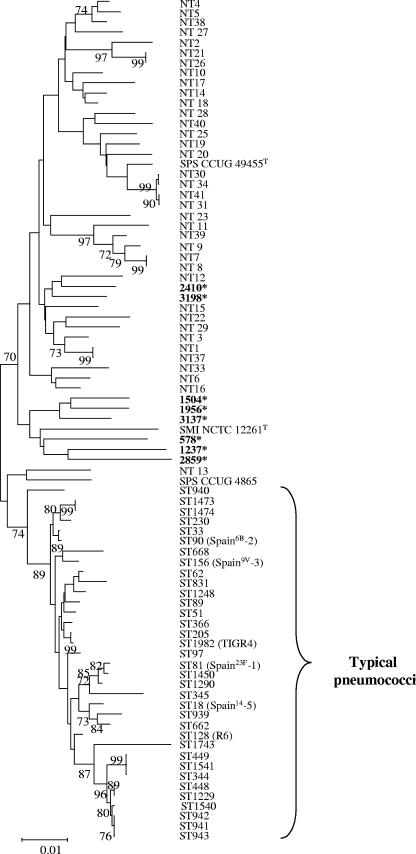
Taking into account these results, the genetic relationship between the Opts SMG strains and other strains was investigated by using MLST as previously suggested (15). Partial sequences of gdh, gki, recP, and spi were determined from the Opts SMG and S. pseudopneumoniae strains. The concatenated allelic profiles were compared to those present at the pneumococcal MLST website (15) and to that of S. mitis NCTC 12261T (The Institute for Genomic Research; http://www.tigr.org). The eight Opts SMG isolates grouped with the nontypeable, nonpneumococcal isolates, as did S. pseudopneumoniae and S. mitis type strains. Six out of eight Opts SMG isolates formed a clade with S. mitis NCTC 12261T (Fig. 2).
FIG. 2.
Dendrogram of genetic relationships between the SMG isolates examined in this study (shown in boldface type and labeled with an asterisk) and other streptococcal isolates. The dendrogram was constructed from housekeeping gene sequence data by using the neighbor-joining method. Only bootstrap confidence values of ≥70% are shown. NT and ST denote nontypeable SMG and sequence type, respectively (11). The scale represents the number of nucleotide substitutions per site. SPS, S. pseudopneumoniae; SMI, S. mitis.
Genetic analysis of lytA alleles carried by Opts SMG.
The lytA alleles from the isolates studied here were PCR amplified and sequenced, except those from isolates 578 and 1504 and the two S. pseudopneumoniae strains that had been already reported (20). Three new, different lytA alleles were found (data not shown). Strains 1237 and 2859 carried the same lytA allele, whereas alleles from strains 3137 and 2410 differed from each other and from those included in the EMBL database. The lytA alleles from strains 1956 and 3198 were identical, respectively, to those present in Opts SMG isolates 1504 and 3072 previously reported (20). All sequences matched the signatures characteristic of lytA alleles from nonpneumococcal SMG isolates, including the 6-bp deletion located near the 3′ end of the gene (20), and explained why the Opts SMG isolates studied here showed a Doc+/− phenotype (Table 1).
Susceptibilities of isolates to optochin and organization and sequence of the atp chromosomal region.
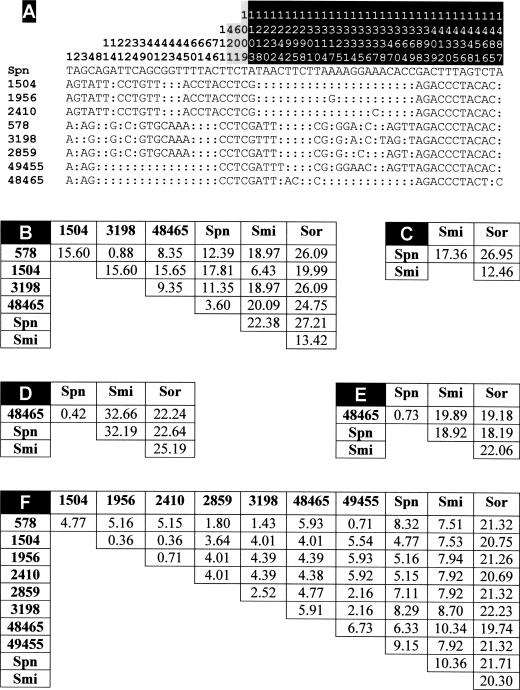
Nucleotide sequences of 1,499-bp fragments containing atpC, atpA, part of atpB, and 114 bp located upstream of atpC from the 10 SMG strains, including the two strains of S. pseudopneumoniae, were determined. The sequences from strains 578 and 1504 were identical to those from strains 1237 and 3137, respectively. Besides, all of the isolates showed identical sequences from nucleotide positions 77 to 1217, which included the atpC and atpA genes as well as the first 138 bp of atpB (Fig. 3A). Moreover, sequence comparisons with those included in the EMBL database (21 March 2006, last date accessed) also showed that the atpC gene of the SMG strains was identical to that of strain R6 (EMBL accession numbers Z26851, AE008506, and AF368465) and to 15 other entries from pneumococcal strains. Moreover, we also found that the atpA alleles of R6 and the Opts SMG strains differed at only three positions and always at the third position of the corresponding codon: T-to-C transitions at codons 24 and 220 (positions 421 and 1009 in Fig. 3A) and a C-to-T transition at codon 84 (position 601 in Fig. 3A). None of these differences produced changes in the predicted primary sequence of the ATPase a subunit.
FIG. 3.
Polymorphism in the atpC, atpA, and atpB genes of Opts SMG. (A) The nucleotides present at each polymorphic site are shown for S. pneumoniae R6, but for the other strains, only nucleotides that differ from those in R6 are shown. Nucleotide positions at the 1,499-bp fragments are indicated vertically above the sequences. Nucleotide positions located upstream of atpC, in atpA, or in atpB are indicated on a white, gray, or black background, respectively. Colons indicate nucleotides identical to those of strain R6. SMG strains are identified by numbering corresponding to that in Table 1. (B to F) Pairwise comparison of the nucleotide sequences located upstream of atpC (nucleotide positions 1 to 114) (B), in atpC (nucleotide positions 115 to 315) (C), in atpA (nucleotide positions 350 to 1066) (D), in the conserved part of atpB (nucleotide positions 1080 to 1217) (E), and in the most divergent region of atpB (nucleotide positions 1218 to 1499) (F). Matrices of PEDs between aligned sequences are shown. Abbreviations: Spn, S. pneumoniae R6; Smi, type strain of S. mitis; Sor, type strain of S. oralis.
Upstream of atpC (nucleotide positions 1 to 114) and downstream of nucleotide 138 of atpB (nucleotide positions 1218 to 1499), the sequences of SMG strains clearly diverged from that of the pneumococcal strain R6 (Fig. 3A). Moreover, sequence divergence in those regions was found among the Opts SMG strains, being more evident upstream of atpC (PEDs up to 15.6%) (Fig. 3B) than at the 3′ end of atpB (PEDs lower than 6%) (Fig. 3F). Besides, evolutionary divergence ranged between 3.6% (for the S. pseudopneumoniae strains) and 17.8% (for strains 1504, 1956, 2410, and 3137) compared with the S. pneumoniae R6 strain (Fig. 3B). It should be noted that, in this region, strain 1504 and relatives were much more evolutionarily related to the type strain of S. mitis (PED of 6.43%) than to S. pneumoniae, although other Opts SMG strains clearly diverged (PEDs higher than 11%) from both S. pneumoniae R6 and the type strain of S. mitis (Fig. 3B). The type strain of S. oralis was only distantly related to the Opts SMG strains (PEDs higher than 19%). Taking together all these results, it is assumed that the Opts SMG strains acquired their atpC and atpA genes from S. pneumoniae via horizontal transfer.
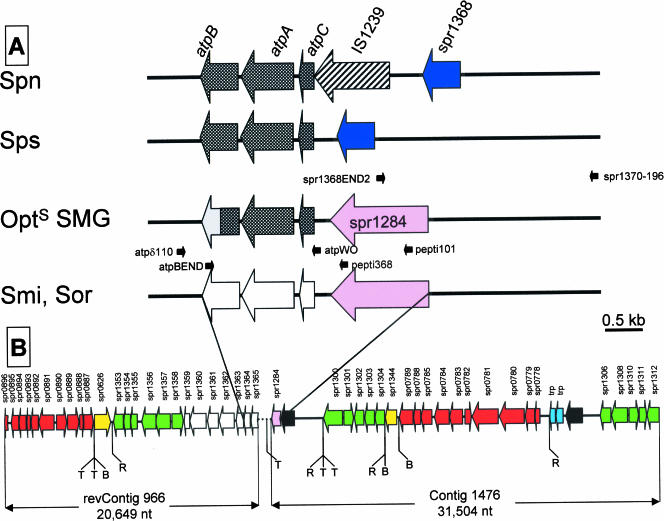
Early reports had suggested that an insertion sequence (IS) was located immediately upstream of the S. pneumoniae atp operon (12). Nucleotide sequencing has since confirmed the presence in this region of the pneumococcal chromosome of a copy of IS1239 (spr1367, or SP1515) in every strain tested, namely, R6 (17), TIGR4 (37), G54 (10), 670 (http://www.tigr.org), and Spain23F-1, OXC14, and INV104B (http://www.sanger.ac.uk/Projects/S_pneumoniae) (data not shown). On the other hand, Martín-Galiano et al. recently reported that, in the type strains of S. mitis and S. oralis, atpC is preceded by a gene highly similar to S. pneumoniae spr1284, or SP1429, putatively encoding a protease (23). In S. pneumoniae, however, atpC and spr1284 are located approximately 65 kb apart (23, 37). Moreover, both genes are apparently arranged in an inverted position in type strains of S. mitis and S. oralis with respect to S. pneumoniae (where spr1284 is located downstream of atpC). PCR amplification and nucleotide sequencing with appropriate oligonucleotide primers showed that, with the significant exception of the S. pseudopneumoniae strains, the gene spr1284 is located immediately upstream of atpC in the Opts SMG strains (Fig. 4). In S. pseudopneumoniae strains, the gene spr1368 is located immediately upstream of atpC, which is equivalent to the gene organization in S. pneumoniae isolates but lacking the spr1367 copy of IS1239 (Fig. 4).
FIG. 4.
Genetic structure of the atp region and its surrounding regions in SMG. Big arrows indicate the genes and their direction of transcription, using as a reference the S. pneumoniae genome (17, 37). The oligonucleotides used in PCR experiments are indicated by small black arrows. (A) Crosshatched and open arrows correspond to the atp genes of S. pneumoniae (Spn)/S. pseudopneumoniae (Sps) or S. mitis (Smi)/S. oralis (Sor), respectively. The atpB gene from Opts SMG is represented as a doubly shaded arrow, where the gray color indicates sequence divergence from either S. pneumoniae or S. mitis genes. (B) Partial genomic map of the DNA regions flanking the S. mitis type strain operon. The DNA region linking contig 966, whose complementary and inverted sequence is represented (revContig 966), and contig 1476 is represented as a dotted line; it has been sequenced previously (23), and it corresponds to the S. mitis/S. oralis scheme shown in panel A. Genes are named according to their S. pneumoniae R6 homologues. Yellow and pink arrows indicate genes that are translocated compared with their location and orientation in S. pneumoniae. Red arrows indicate gene or gene clusters that are inverted. S. mitis genes showing synteny with those of S. pneumoniae are indicated by green or open (for the atp genes) arrows. Light-blue and filled arrows represent transposase genes and genes lacking any significant similarity with those included in the databases, respectively. T, B, and R indicate the locations of transposase genes/IS-like elements and BOX or RUP repeats, respectively, as annotated in the S. pneumoniae R6 genome sequence (17). nt, nucleotides.
DISCUSSION
The chromosomal organization of the atp operon region in S. pneumoniae, S. mitis, S. pseudopneumoniae, and Opts SMG (Fig. 4) and the nucleotide sequences of the atpC-atpA-atpB regions strongly suggest a recombinational origin for the eight Opts isolates analyzed in this work. This recombination resulted in the acquisition of a region of about 1.3 kb that included the complete atpC and atpA genes and 200 to 300 bp of atpB from S. pneumoniae. The same region was involved in the interchanges yielding the Optr S. pneumoniae strain M222 (12) and the Opts S. mitis isolate 1162/99 (23), suggesting that the reconstitution of a functional FoF1 H+-ATPase requires a b subunit with an N terminus compatible with the a subunit and a C terminus compatible with the δ subunit. Besides, the gene organization upstream of atpC appears to be quite variable depending on the particular species and/or isolate studied. The comparison of the genomic regions of S. pneumoniae and S. mitis embracing the atp operon revealed the existence of significant differences between both species, with an obvious loss of synteny (Fig. 4B). Most of these differences were related to genomic rearrangements, suggesting frequent events of inversion and/or translocation of either individual genes or clusters of genes. It has been proposed previously that the observed genomic plasticity of the pneumococcal genome (7) may be related, at least in part, to the abundant presence of ISs and repeat elements, such as those named BOX (22) and RUP (repeat unit of pneumococcus) (31). As shown in Fig. 4B, transposase genes, RUPs, and/or BOX elements were very frequently placed at the boundaries of the translocated/inverted DNA regions. We propose that one such genomic rearrangement might be responsible for the differences observed in the genes located upstream of atpC in the closely related Optr/Opts SMG. It is not known if there is any advantage for selecting for Opts SMG, but the involvement of the FoF1 H+-ATPase in the maintenance of the intracellular pH and of the membrane potential (24) suggests a relation between Opt susceptibility and the adaptation of S. pneumoniae to the different habitats in which it causes infection.
The eight Opts SMG isolates examined in this study represent a kind of streptococcus that could not be classified as S. pneumoniae on the basis of being AccuProbe negative or from their location in the phylogenetic tree constructed with housekeeping genes (Fig. 2). However, those isolates carry at least two pneumococcal virulence determinants (lytA and ply), and it was found that all of them perform an atypical bile solubility behavior, according to the genetic characteristics of their lytA alleles. At least seven of them were associated with disease, and all were penicillin resistant. These data and the description of three SMG isolates from the United Kingdom associated with respiratory diseases and also carrying lytA and ply (40) reflect the genetic interchange that takes place among SMG and between SMG and S. pneumoniae in nature.
Currently, most clinical laboratories depend on the Opt susceptibility test for S. pneumoniae identification. Accordingly, the eight Opts SMG isolates described in this work were identified as S. pneumoniae isolates by the clinical laboratories that had sent them to the reference laboratory. Since misidentification of Opts SMG isolates as S. pneumoniae isolates may have significant implications for the management of patients, the performance of at least two tests for the identification of S. pneumoniae, i.e., Opt susceptibility and bile solubility, should be required. Nevertheless, the results presented here, together with other previous reports of Opt variant and/or Doc variant phenotypes in SMG strains, reflect the importance of describing accurately the conditions used for both tests, that is, CO2 atmosphere composition and percentage of Doc, respectively.
Acknowledgments
We thank M. J. Ferrándiz for the critical reading of the manuscript. The technical assistance of Alicia Rodriguez-Bernabé and E. Cano is greatly acknowledged.
L.B. and A.H.-M. received fellowships from Instituto de Salud Carlos III. This study was supported by grants BIO2005-02189 and BMC2003-00074 from the Dirección General de Investigación Científica y Técnica.
Sequencing of S. mitis NCTC 12261T was accomplished with support from the National Institute of Dental and Craniofacial Research.
Footnotes
Published ahead of print on 13 September 2006.
REFERENCES
- 1.Abrahams, J. P., A. G. W. Leslie, and J. E. Walker. 1994. Structure at 2.8 Å resolution of F1-ATPase from bovine heart mitochondria. Nature 370:621-628. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arbique, J. C., C. Poyart, P. Trieu-Cuot, G. Quesne, M. da Glória S. Carvalho, A. G. Steigerwalt, R. E. Morey, D. Jackson, R. J. Davidson, and R. R. Facklam. 2004. Accuracy of phenotypic and genotypic testing for identification of Streptococcus pneumoniae and description of Streptococcus pseudopneumoniae sp. nov. J. Clin. Microbiol. 42:4686-4696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balsalobre, L., M. J. Ferrándiz, J. Liñares, F. Tubau, and A. G. de la Campa. 2003. Viridans group streptococci are donors in horizontal transfer of topoisomerase IV genes to Streptococcus pneumoniae. Antimicrob. Agents Chemother. 47:2072-2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borek, A. P., D. C. Dressel, J. Hussong, and L. R. Peterson. 1997. Evolving clinical problems with Streptococcus pneumoniae: increasing resistance to antimicrobial agents, and failure of traditional optochin identification in Chicago, Illinois, between 1993 and 1996. Diagn. Microbiol. Infect. Dis. 29:209-214. [DOI] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. 1997. Prevention of pneumococcal disease: recommendations of the Advisory Committee on Immunization Practices (ACIP). Morb. Mortal. Wkly. Rep. Recomm. Rep. 46:1-24. [Google Scholar]
- 7.Claverys, J. P., M. Prudhomme, I. Mortier-Barriere, and B. Martin. 2000. Adaptation to the environment: Streptococcus pneumoniae, a paradigm for recombination-mediated genetic plasticity? Mol. Microbiol. 35:251-259. [DOI] [PubMed] [Google Scholar]
- 8.Cogné, N., J. Claverys, F. Denis, and C. Martin. 2000. A novel mutation in the α-helix 1 of the C subunit of the F1/Fo ATPase responsible for optochin resistance of a Streptococcus pneumoniae clinical isolate. Diagn. Microbiol. Infect. Dis. 38:119-121. [DOI] [PubMed] [Google Scholar]
- 9.Devereux, J., P. Haeberli, and O. Smithies. 1984. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 12:387-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dopazo, J., A. Mendoza, J. Herrero, F. Caldara, Y. Humbert, L. Friedli, M. Guerrier, E. Grand-Schenk, C. Gandin, M. de Francesco, A. Polissi, G. Buell, G. Feger, E. García, M. Peitsch, and J. F. García-Bustos. 2001. Annotated draft genomic sequence from a Streptococcus pneumoniae type 19F clinical isolate. Microb. Drug Resist. 7:99-125. [DOI] [PubMed] [Google Scholar]
- 11.Enright, M. C., and B. G. Spratt. 1998. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology 144:3049-3060. [DOI] [PubMed] [Google Scholar]
- 12.Fenoll, A., R. Muñoz, E. García, and A. G. de la Campa. 1994. Molecular basis of the optochin-sensitive phenotype of pneumococcus: characterization of the genes encoding the Fo complex of the Streptococcus pneumoniae and Streptococcus oralis H+-ATPases. Mol. Microbiol. 12:587-598. [DOI] [PubMed] [Google Scholar]
- 13.Ferrándiz, M. J., and A. G. de la Campa. 2002. The membrane-associated FoF1 ATPase is essential for the viability of Streptococcus pneumoniae. FEMS Microbiol. Lett. 212:133-138. [DOI] [PubMed] [Google Scholar]
- 14.Ferrándiz, M. J., A. Fenoll, J. Liñares, and A. G. de la Campa. 2000. Horizontal transfer of parC and gyrA in fluoroquinolone-resistant clinical isolates of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 44:840-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanage, W. P., T. Kaijalainen, E. Herva, A. Saukkoriipi, R. Syrjanen, and B. G. Spratt. 2005. Using multilocus sequence data to define the pneumococcus. J. Bacteriol. 187:6223-6230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoshino, T., T. Fujiwara, and M. Kilian. 2005. Use of phylogenetic and phenotypic analyses to identify nonhemolytic streptococci isolated from bacteremic patients. J. Clin. Microbiol. 43:6073-6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoskins, J., W. E. Alborn, Jr., J. Arnold, L. C. Blaszczak, S. Burgett, B. S. DeHoff, S. T. Estrem, L. Fritz, D.-J. Fu, W. Fuller, C. Geringer, R. Gilmour, J. S. Glass, H. Khoja, A. R. Kraft, R. E. Lagace, D. J. LeBlanc, L. N. Lee, E. J. Lefkowitz, J. Lu, P. Matsushima, S. M. McAhren, M. McHenney, K. McLeaster, C. W. Mundy, T. I. Nicas, F. H. Norris, M. O'Gara, R. B. Peery, G. T. Robertson, P. Rockey, P.-M. Sun, M. E. Winkler, Y. Yang, M. Young-Bellido, G. Zhao, C. A. Zook, R. H. Baltz, S. R. Jaskunas, P. R. Rosteck, P. L. Skatrud, and J. I. Glass. 2001. Genome of the bacterium Streptococcus pneumoniae strain R6. J. Bacteriol. 183:5709-5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111-1120. [DOI] [PubMed] [Google Scholar]
- 19.Kontiainen, S., and A. Sivonen. 1987. Optochin resistance in Streptococcus pneumoniae strains isolated from blood and middle ear fluid. Eur. J. Clin. Microbiol. 6:422-424. [DOI] [PubMed] [Google Scholar]
- 20.Llull, D., R. López, and E. García. 2006. Characteristic signatures of the lytA gene provide a rapid and reliable diagnosis of Streptococcus pneumoniae infections. J. Clin. Microbiol. 44:1250-1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lund, E., and J. Herichsen. 1978. Laboratory diagnosis, serology and epidemiology of Streptococcus pneumoniae. Methods Microbiol. 12:241-262. [Google Scholar]
- 22.Martin, B., O. Humbert, M. Camara, E. Guenzi, J. Walker, T. Mitchell, P. Andrew, M. Prudhomme, G. Alloing, R. Hakenbeck, D. A. Morrison, G. J. Boulnois, and J. P. Claverys. 1992. A highly conserved repeated DNA element located in the chromosome of Streptococcus pneumoniae. Nucleic Acids Res. 20:3479-3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martín-Galiano, A. J., L. Balsalobre, A. Fenoll, and A. G. de la Campa. 2003. Genetic characterization of optochin-susceptible viridans group streptococci. Antimicrob. Agents Chemother. 47:3187-3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martín-Galiano, A. J., M. J. Ferrándiz, and A. G. de la Campa. 2001. The promoter of the operon encoding the FoF1 ATPase of Streptococcus pneumoniae is inducible by pH. Mol. Microbiol. 41:1327-1338. [DOI] [PubMed] [Google Scholar]
- 25.Martín-Galiano, A. J., B. Gorgojo, C. M. Kunin, and A. G. de la Campa. 2002. Mefloquine and new related compounds target the Fo complex of the FoF1 H+-ATPase of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 46:1680-1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mundy, L. S., E. N. Janoff, K. E. Schwebke, C. J. Shanholtzer, and K. E. Willard. 1998. Ambiguity in the identification of Streptococcus pneumoniae. Optochin, bile solubility, quellung, and the AccuProbe DNA probe tests. Am. J. Clin. Pathol. 109:55-61. [DOI] [PubMed] [Google Scholar]
- 27.Muñoz, R., A. Fenoll, D. Vicioso, and J. Casal. 1990. Optochin-resistant variants of Streptococcus pneumoniae. Diagn. Microbiol. Infect. Dis. 13:63-66. [DOI] [PubMed] [Google Scholar]
- 28.Muñoz, R., E. García, and A. G. de la Campa. 1996. Quinine specifically inhibits the proteolipid subunit of the FoF1 H+-ATPase of Streptococcus pneumoniae. J. Bacteriol. 178:2455-2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.National Committee for Clinical Laboratory Standards. 2004. Performance standards for antimicrobial susceptibility testing. Fourteenth informational supplement. NCCLS document M100-S14. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 30.Obregón, V., P. García, E. García, A. Fenoll, R. López, and J. L. García. 2002. Molecular peculiarities of the lytA gene isolated from clinical pneumococcal strains that are bile insoluble. J. Clin. Microbiol. 40:2545-2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oggioni, M. R., and J. P. Claverys. 1999. Repeated extragenic sequences in prokaryotic genomes: a proposal for the origin and dynamics of the RUP element in Streptococcus pneumoniae. Microbiology 145:2647-2653. [DOI] [PubMed] [Google Scholar]
- 32.Phillips, G., R. Barker, and O. Brogan. 1988. Optochin-resistant Streptococcus pneumoniae. Lancet ii:281. [DOI] [PubMed] [Google Scholar]
- 33.Pikis, A., J. M. Campos, W. J. Rodriguez, and J. M. Keith. 2001. Optochin resistance in Streptococcus pneumoniae: mechanism, significance, and clinical implications. J. Infect. Dis. 184:582-590. [DOI] [PubMed] [Google Scholar]
- 34.Rastogi, V. K., and M. E. Girvin. 1999. Structural changes linked to proton translocation by subunit c of the ATP synthase. Nature 402:263-268. [DOI] [PubMed] [Google Scholar]
- 35.Ruoff, K. L., R. A. Whiley, and D. Beighton. 2003. Streptococcus, p. 405-421. In P. R. Murray, E. J. Baron, J. H. Jorgensen, M. A. Pfaller, and R. H. Yolken (ed.), Manual of clinical microbiology, 8th ed. ASM Press, Washington, D.C.
- 36.Stock, D., A. G. W. Leslie, and J. E. Walker. 1999. Molecular architecture of the rotary motor in ATP synthase. Science 286:1700-1705. [DOI] [PubMed] [Google Scholar]
- 37.Tettelin, H., K. E. Nelson, I. T. Paulsen, J. A. Eisen, T. D. Read, S. Peterson, J. Heidelberg, R. T. DeBoy, D. H. Haft, R. J. Dodson, A. S. Durkin, M. Gwinn, J. F. Kolonay, W. C. Nelson, J. D. Peterson, L. A. Umayam, O. White, S. L. Salzberg, M. R. Lewis, D. Radune, E. Holtzapple, H. Khouri, A. M. Wolf, T. R. Utterback, C. L. Hansen, L. A. McDonald, T. V. Feldblyum, S. Angiuoli, T. Dickinson, E. K. Hickey, I. E. Holt, B. J. Loftus, F. Yang, H. O. Smith, J. C. Venter, B. A. Dougherty, D. A. Morrison, S. K. Hollingshead, and C. M. Fraser. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293:498-506. [DOI] [PubMed] [Google Scholar]
- 38.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsai, H. Y., P. R. Hsueh, L. J. Teng, P. I. Lee, L. M. Huang, C. Y. Lee, and K. T. Luh. 2000. Bacteremic pneumonia caused by a single clone of Streptococcus pneumoniae with different optochin susceptibilities. J. Clin. Microbiol. 38:458-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Whatmore, A. M., A. Efstratiou, A. P. Pickerill, K. Broughton, G. Woodard, D. Sturgeon, R. George, and C. G. Dowson. 2000. Genetic relationships between clinical isolates of Streptococcus pneumoniae, Streptococcus oralis, and Streptococcus mitis: characterization of “atypical” pneumococci and organisms allied to S. mitis harboring S. pneumoniae virulence factor-encoding genes. Infect. Immun. 68:1374-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]