Abstract
Tropheryma whipplei, the agent of Whipple's disease, is a gram-positive rod-shaped bacterium that belongs to the group of actinobacteria. In order to produce monoclonal antibodies (MAbs) against this bacterium, we inoculated mice with two different strains, Slow2 and Endo5. We produced 13 and 10 MAbs against Slow2 and Endo5, respectively. Nine of the Slow2 MAbs and seven of the Endo5 MAbs recognized a 58-kDa epitope. In addition, three other Endo5 MAbs detected a unique 84-kDa epitope. These MAbs were species specific, as they did not react with a selection of 22 different bacterial species, but they were not strain specific, as they did react with six other strains of T. whipplei. Two-dimensional gel electrophoresis (2-DE) was combined with mass spectrometry (MS) to identify the 58-kDa and 84-kDa epitopes recognized by MAbs. After trypsin in-gel digestion of the spot, the 58-kDa protein was identified as an ATP synthase F1 complex beta chain, whereas the 84-kDa protein was identified as a polyribonucleotide nucleotidyltransferase by MS with matrix-assisted laser desorption ionization-time of flight. In an in vitro model, one of these MAbs allowed good detection of T. whipplei in stool samples, contrary to a rabbit polyclonal antibody, which led to high fluorescent background. In the prospective studies, the produced MAb will be tested for detection of T. whipplei in clinical samples, and the gene coding for identified 58-kDa and 84-kDa antigens will be tentatively cloned and then tested for its use in a diagnostic enzyme-linked immunosorbent assay for Whipple's disease.
Whipple's disease is a multisystemic bacterial infection which may involve any organ system in the body. This disease is known mainly as a chronic pathology involving the intestine. Malabsorption, diarrhea, weight loss, and eventually association with adenopathies and polyarthritis correspond to the classical symptoms of Whipple's disease (4, 7, 17, 22). Occasionally, it is also associated with cardiac manifestations, such as myocarditis, pericarditis, and endocarditis, or central nervous system involvement (21, 31, 34, 38). Diagnosis of infection is usually based on classical histopathological examination of a duodenal biopsy specimen showing infiltration by large macrophages that contain periodic acid-Schiff-positive, non-acid-fast bacteria (1). The determination of the nucleotide sequence of the 16S rRNA gene of Tropheryma whipplei (32), the agent of Whipple's disease (14, 40), and then its isolation by cell culture provided the basis for the development of species-specific diagnostic PCR systems (27, 39). These PCR-based diagnostic methods have become standards for the diagnosis of Whipple's disease. Using a shell vial cell culture system, we first isolated the Whipple's disease bacterium from the cardiac valve of a patient with Whipple's disease-related endocarditis and successfully established a stable culture (28). Since then, the isolation methods were improved and allowed us and others to isolate more T. whipplei strains (20). We first developed a specific microimmunofluorescence (MIF) assay with Labteck slide-grown bacteria (28). This technique presents several major drawbacks, most important being loss of antigenicity of T. whipplei isolates after several subcultures. Considering the fact that Whipple's disease is rare, a sensitive screening test not requiring invasive specimens as a tool for patient follow-up under antibiotic treatment would be extremely helpful. The need for standardization of diagnostic antigens is a strong rationale for the development of new serodiagnostic reagents. However, the immunodominant antigens of T. whipplei during infection are not well characterized. As a result, the ability of a single or multiple selected proteins to serve as an alternative to purified whole bacteria as antigens for serological diagnostic tests is untested.
In a previous study, we produced some monoclonal antibodies against the Twist-Marseille strain of T. whipplei (16). For unknown reasons and even with several subcloning attempts, hybridomas producing monoclonal antibodies (MAbs) were progressively lost. Moreover, since the separation based on a single physicochemical property is not sufficient, the immunodominant epitopes of the strain were not identified and characterized by general Western immunoblotting. In contrast, two-dimensional gel electrophoresis (2-DE) blotting is a technique that combines two physicochemical properties, pI and molecular mass. In this technique, the experimental conditions can be optimized according to the proteins of interest (25). It is possible to separate the components from each other only on combining two techniques, isoelectric focusing (IEF) and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Therefore, the combination of the high-resolution electrophoresis (2-DE) with subsequent transfer onto a protein-binding membrane (blotting), immunological detection, and mass spectrometry (MS) is a powerful tool to identify and characterize immunodominant epitopes of T. whipplei.
In the present study, we first produced the monoclonal antibodies against the Slow2 and Endo5 strains of T. whippleiand then identified and characterized the recognized epitopes with 2-DE blotting and MS.
MATERIALS AND METHODS
Preparation of antigen.
T. whipplei strain Slow2-Marseille, which was grown previously in 30 ml of minimal essential medium according to Raoult et al. (28), was cultured on HEL cell monolayers in 150-cm2 cell culture flasks. HEL cells infected with bacteria were harvested from 40 150-cm2 flasks into 40 ml of phosphate-buffered saline (PBS). Trypsin (Gibco) was added at a final concentration of 5 mg ml−1, and the suspension was incubated at 30°C for 45 min. The suspension was then subjected to sonication (three times for 1 min, each time on ice), after which the unlysed cells were removed by centrifugation at 100 × g for 15 min. The supernatant was layered onto a 25% (wt/vol) sucrose solution in PBS. After centrifugation at 9,000 × g for 30 min at 4°C, the pellet containing the bacteria was resuspended in 2 ml of PBS and carefully layered onto a 25 to 45% (wt/vol) Renografin step gradient (in PBS). This gradient was subjected to centrifugation at 130,000 × g for 1 h at 5°C. The bacteria were then harvested from the interface of the 25 to 45% Renografin gradient and washed twice in PBS. For SDS-PAGE, the bacteria were resuspended in sterile distilled water at a final concentration of 1 mg ml−1. Another T. whipplei strain, Endo5, was cultured in axenic liquid medium as previously described (33) and washed twice in PBS.
Production of MAbs.
The monoclonal antibodies (MAbs) were produced by inoculation of 6- to 8-week-old immunocompetent BALB/c mice with a total of 0.1 mg of purified strain Slow2-Marseille and Endo5 with CpG adjuvant, respectively, as described previously (9, 15). The isotypes of the MAbs were determined with an ImmunoType Mouse Monoclonal Antibody Isotyping kit with antisera to mouse immunoglobulin M (IgM), IgA, IgG1, IgG2a, IgG2b, and IgG3 (Sigma Chemical Co.). The specificities of the MAbs were tested by Western immunoblotting.
Specificity assay.
Cross-reaction was determined by MIF assay (26). The MAbs produced were tested against antigens from six other T. whipplei strains isolated in our laboratory (Twist, Dig7, Endo7, Dig9, Neuro1, and Neuro2) and 22 diverse bacterial strains that were also isolated in our laboratory from clinical samples, including Actinomyces meyeri, Actinomyces viscosus, Actinomyces pyogenes, Nocardia asteroides, Propionibacterium acnes, Mycobacterium marinum, Mycobacterium avium, Bacillus cereus, Listeria monocytogenes, the Corynebacterium ANF group, Corynebacterium striatum, Streptococcus bovis, Streptococcus agalactiae (group B streptococcus), Clostridium perfringens, Clostridium bifermentans, Fusobacterium necrophorum, Escherichia coli, Yersinia enterocolitica, Shigella sonnei, Shigella flexneri, Salmonella enterica, and Campylobacter jejuni.
SDS-PAGE and Western blot study.
SDS-PAGE and Western blotting were performed according to the method originally developed by Laemmli (13). Antigens were treated with proteinase K or boiled. Heat denaturation was performed by boiling the antigens at 100°C for 10 min. For Western blotting, the strips were incubated with diluted supernatants of MAbs (1:10 dilution) and polyclonal mouse T. whipplei antisera diluted in PBS (1:100 dilution) at room temperature for 1 h and then were washed three times with PBS-Tween.
Preparation of crude extracts for 2-DE.
The bacterial suspension was precipitated by using a PlusOne 2-D Clean-Up kit (Amersham Biosciences, Uppsala, Sweden) and resuspended directly in rehydration solution {7 M urea, 2 M thiourea, 4% (wt/vol) 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate)}. The protein content of the solution was determined using a commercially available protein assay system that incorporated bovine serum albumin (BSA) as a standard (Bio-Rad, Hercules, CA) (2).
2-DE blotting.
Immobiline DryStrips (13 cm, pH 4 to 7; Amersham Biosciences, Uppsala, Sweden) were rehydrated overnight with 250 μg of proteins in rehydration solution supplemented with 2% (vol/vol) immobilin pH gradient (IPG) buffer (pH 4 to 7) (Amersham Biosciences). IEF was carried out according to the manufacturer's protocol (Multiphor II system; Amersham Biosciences). Prior to electrophoresis in the second dimension, the strips had been equilibrated for 15 min in 10 ml of equilibration buffer (30% [vol/vol] glycerol, 2% [wt/vol] SDS, 6 M urea, 50 mM Tris-HCl, bromophenol blue, pH 8.8) containing 65 mM of dithiothreitol. This step was repeated once again using 10 ml of equilibration buffer supplemented with 100 mM of iodoacetamide. The strips so treated were then embedded in 0.5% agarose, and the proteins were resolved by 9 to 16% gradient SDS-PAGE (Bio-Rad Protean II xi chamber). Electrophoresis was performed at the constant voltage of 250 V until the bromophenol blue reached the end of the gel. The molecular weight (Mr) was determined by running standard protein markers (LMW; Bio-Rad, Hercules, CA). Gels were then processed either for silver staining (23) or for immunoblotting (10). For immunoblotting, the proteins were transferred onto nitrocellulose membranes (Trans-blot Transfer Medium; Pure Nitrocellulose Membrane, 0.45 μm; Bio-Rad) by using a semidry transfer unit (Hoefer TE 77; Amersham Biosciences).
Digestion peptides and MALDI mass spectrometry analysis.
The protein spots excised from silver-stained gels were destained and subjected to in-gel digestion with trypsin (sequencing grade-modified porcine trypsin; Promega, Madison, WI) (35). The peptides obtained from protein digestion were dissolved in 10 to 20 μl of 0.1% trifluoroacetic acid (TFA). The peptide mixture was then analyzed using an Ettan pro matrix-assisted laser desorption ionization (MALDI) spectrometer (Amersham Biosciences) in positive ion reflector mode. For this, the sample (0.3 μl) of peptide mixture was cocrystallized in the presence of 0.5% TFA onto the MALDI target with an equal amount of matrix solution (3 mg/ml of α-cyano-4-hydroxycinnamic acid in 50% acetonitrile). Alternatively, the peptide mixtures derived from protein digestion were desalted and concentrated using zip tips (Millipore, Bedford, MA) and deposited onto the MALDI target by elution with the matrix solution. Proteins were identified and assigned a number by Profound (ProteoMetrics, LLC, New York, NY) and Mascot (Matrix Science Ltd., London, United Kingdom) software for comprehensive sequence databases (24, 36).
Immunofluorescence detection of T. whipplei in stool samples.
A healthy individual's stool sample, in which T. whipplei PCR detection was negative by previously described techniques (30), was selected. This stool sample was diluted in PBS (20%, wt/vol) and mixed well. Four hundred microliters of this sample was suspended in sterile distilled water and submitted to 1-min sedimentation. Supernatant was removed and aliquoted into two parts. To one part a suspension of T. whipplei strain Endo5 suspended in PBS was added in order to obtain a concentration of 104 T. whipplei cells per ml of dilution. Two microliters from each part was deposited onto glass slides, air dried, and fixed with methanol for 5 min. Slides were stored at 4°C before use. For immunofluorescense assay, slides were saturated by incubation with PBS-5% BSA at 37°C for 30 min and then washed twice with PBS-0.1% Tween for 10 min and once with sterile distilled water for 5 min. Samples were incubated either with WS5H4 mouse monoclonal antibody or with rabbit polyclonal serum at its respective 1:100 or 1:400 dilution in PBS-3% BSA-0.1% Tween for 30 min at 37°C. After a washing step as described above, bound antibodies were revealed with fluorescein isothiocyanate-conjugated IgG goat anti-mouse or anti-rabbit antibody (Immunotech, Marseille, France) diluted 1:2,000 in PBS-3% BSA-0.1% Tween-0.2% Evans blue (BioMerieux, Marcy l'Etoile, France). For image scanning, slides were mounted with Fluoprep (BioMerieux, Marcy l'Etoile, France) after subsequent washing procedures and examined under an Olympus BX-51 epifluorescence microscope at ×100 magnification. In order to see the specificity of MAb, we also tested 15 stool samples prepared as mentioned above using WS5H4 mouse monoclonal antibody and compared the results to those obtained with rabbit polyclonal serum.
RESULTS
SDS-PAGE and Western blotting of T. whipplei.
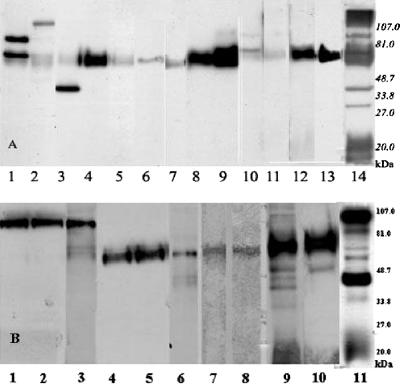
We obtained 13 MAbs against strain Slow2 and 10 MAbs against strain Endo5 (Table 1). Of these, 16 MAbs reacted with a 58-kDa antigen and 3 MAbs with an 84-kDa antigen. The MAbs WS6F5, WS1F6, and 6C3 recognized 134-, 65-, and 47-kDa antigens, respectively, whereas MAb 7H3 reacted simultaneously with 105- and 65-kDa protein bands (Fig. 1).
TABLE 1.
Hybridomas obtained from inoculation with Slow2 and Endo5 strainsa
| Hybridoma | Isotype | Strain from which hybridoma was obtained | Size (kDa) of recognized antigens from:
|
Epitope | |
|---|---|---|---|---|---|
| 1-D WB | 2-D WB | ||||
| WS3E5 | IgG1 | Slow2 | 58 | 58 | ATP synthase F1 complex β chain |
| WS3F9 | IgG1 | Slow2 | 58 | 58 | ATP synthase F1 complex β chain |
| WS1F6 | IgG1 | Slow2 | 65 | NI | NI |
| WS4D11 | IgG1 | Slow2 | 58 | 58 | ATP synthase F1 complex β chain |
| WS2A3 | IgG1 | Slow2 | 58 | 58 | ATP synthase F1 complex β chain |
| WS5D1 | IgG1 | Slow2 | 58 | 58 | ATP synthase F1 complex β chain |
| WS7H3 | IgG1 | Slow2 | 65-105 | Smear | NI |
| WS6F5 | IgG1 | Slow2 | 134 | Smear | NI |
| WS5H4 | IgG1 | Slow2 | 58 | 58 | ATP synthase F1 complex β chain |
| WS7G2 | IgG1 | Slow2 | 58 | 58 | ATP synthase F1 complex β chain |
| WS6C3 | IgG1 | Slow2 | 47 | Smear | NI |
| WS1C6 | IgM | Endo5 | 58 | 58 | ATP synthase F1 complex β chain |
| WS5E5 | IgM | Endo5 | 58 | 58 | ATP synthase F1 complex β chain |
| WE7F6 | IgG1 | Endo5 | 58 | 58 | ATP synthase F1 complex β chain |
| WE10D11 | IgG1 | Endo5 | 58 | 58 | ATP synthase F1 complex β chain |
| WE11H11 | IgG1 | Endo5 | 58 | 58 | ATP synthase F1 complex β chain |
| WE11G10 | IgG1 | Endo5 | 58 | 58 | ATP synthase F1 complex β chain |
| WE8D5 | IgG1 | Endo5 | 58 | 58 | ATP synthase F1 complex β chain |
| WE9C1 | IgG1 | Endo5 | 58 | 58 | ATP synthase F1 complex β chain |
| WE9D4 | IgG1 | Endo5 | 58 | 58 | ATP synthase F1 complex β chain |
| WE8H5 | IgG2a | Endo5 | 84 | 84 | Polyribonucleotide nucleotidyltransferase |
| WE11B10 | IgG2a | Endo5 | 84 | 84 | Polyribonucleotide nucleotidyltransferase |
| WE11F10 | IgG2a | Endo5 | 84 | 84 | Polyribonucleotide nucleotidyltransferase |
NI, not identified; 1-D WB, one-dimensional Western blot; 2-D WB, two-dimensional Western blot; smear, it was not possible to identify an antigen clearly.
FIG. 1.
Immunoblots of antigens of T. whipplei with mono- and polyclonal antibodies. (A) T. whipplei Slow2 with its monoclonal antibodies. Lane 1, MAb WS7H3; lane 2, MAb WS6F5; lane 3, MAb WS6C3; lane 4, MAb 7G2; lane 5, MAb WS5E5; lane 6, WS2A3; lane 7, MAb 3E5; lane 8, MAb WS5D1; lane 9, MAb 3F9; lane 10, MAb WS1F6; lane 11, MAb WS1C6; lane 12, MAb WS4D11; 13, MAb WS5H4; and lane 14, polyclonal mouse antiserum. (B) T. whipplei Endo5 with its monoclonal antibodies. Lane 1, MAb WE11F10; lane 2, MAb WE11B10; lane 3, MAb WE8H5; lane 4, MAb 11G10; lane 5, MAb WE8D5; lane 6, MAb WE9D4; lane 7, MAb WE7F6; lane 8, MAb WE9C1; lane 9, MAb WE11H11; lane 10, MAb WE10D11; and lane 11, polyclonal mouse antiserum.
2-DE and Western blotting.
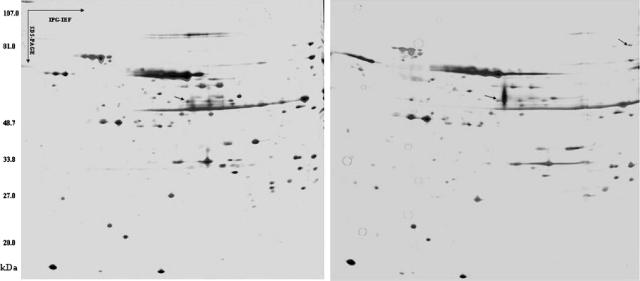
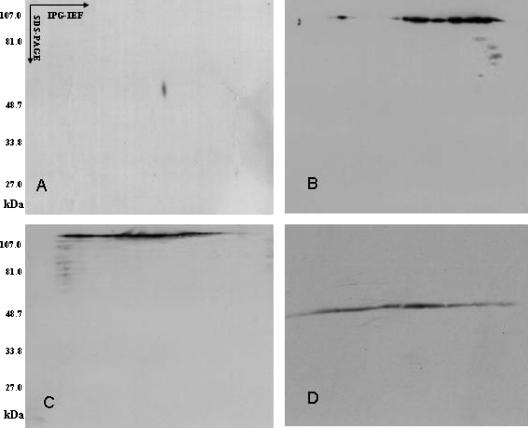
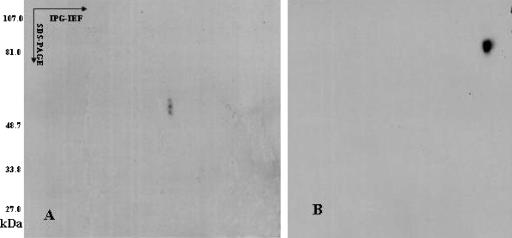
To identify the epitopes recognized by the MAbs, Slow2 and Endo5 strain extracts were subjected to 2-DE and subsequent Western blotting analysis. Figure 2 shows the typical electrophoregram of Slow2 and Endo5 extract components, which were obtained under the same experimental conditions, such as molecular masses (Mr) of 20 to 107 and pI 4.5 to 5.5, visualized by silver staining. After 2-DE, the proteins were transferred onto a nitrocellulose membrane, which was subsequently incubated with the MAbs. On Western blotting with MAbs WS3E5, WS3F9, WS1F6, WS4D11, WS2A3, WS5D1, WS1C6, WS5E5, WS5H4, and WS7G2, only one immunoreactive spot was detected at a 58-kDa protein (Fig. 3A). MAbs WS6C3 and WS6F5 recognized the protein smear of about 47 to 48 kDa and 134 kDa, respectively, and the WS7H3 MAb detected many spots ranging from 47 to 105 kDa (Fig. 3B to D). It was thus extremely difficult to pick up spots except one that belong to WS7G2. In addition, the MAbs WE11F10, WE11B10, and 8H5 detected an epitope of an 84-kDa protein, whereas other MAbs of Endo5 recognized the same spot that was noticed in the case of Slow2 at 58 kDa (Fig. 4).
FIG. 2.
Two-dimensional gel of T. whipplei extract with silver staining (the first one is for Slow2, the second one is for Endo5). Proteins were resolved in the first dimension over a pI gradient of 4.5 to 5.5, followed by a second-dimension separation by SDS-PAGE in a 10% acrylamide gel. The prominent spots at 60 kDa and 84 kDa (arrow) were cored from the gel and submitted for analysis by mass spectrometry. These spots corresponded to the 2-DE blotting.
FIG. 3.
Two-dimensional Western blot showing the reactivity of MAbs with Slow2 proteins. The monoclonal antibodies against Slow2 were from the supernatant of hybridoma 7G2 (A), 7H3 (B), 6F5 (C), and 6C3 (D). The 7G2 antibody bound only one spot at 60 kDa and pI 5.1.
FIG. 4.
Two-dimensional Western blot showing the reactivity of MAbs with Endo5 proteins. The monoclonal antibodies against Endo5 were from the supernatant of hybridoma 11G10 (A) and 11F10 (B). The 11F10 antibody bound a unique spot at 84 kDa and pI 5.3. The MAb WE11G10 recognized the same spot as WS7G2.
Identification of spots.
The spots recognized by MAbs at 58 and 84 kDa were excised, digested with trypsin, and subjected to peptide sequencing by MALDI-time of flight. Proteins were identified using the SwissProt database with Mascot search engine (www.matrixscience.com). In the identification of the 58-kDa spot, 16 peptides were obtained by mass spectrometry and matched the ATP synthase F1 complex beta chain of T. whipplei strain Twist. The molecular mass and pI of this protein were recorded as 52.5 kDa and 5.1, respectively. On the other hand, 17 peptides were obtained from an 84-kDa spot and matched the polyribonucleotide nucleotidyltransferase of T. whipplei strain Twist, the molecular mass of which was found to be 81 kDa.
Specificity.
The results of MIF assay showed that MAbs did not react either with HEL cells or with any of the 22 diverse bacterial strains tested. All MAbs reacted with the six other T. whipplei strains tested.
Immunofluorescence detection of T. whipplei in stool samples.
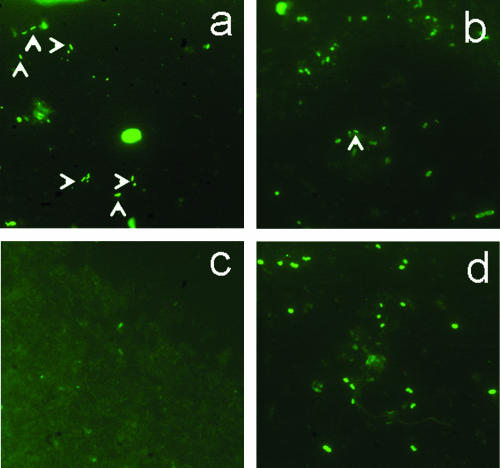
For the 15 stool samples tested using polyclonal rabbit serum, high fluorescent background and numerous fluorescent bacteria were observed, whereas no fluorescent bacteria were observed with WS5H4 MAb. The same observation was made with the contaminated sample (Fig. 5c and d). Bacteria with typical T. whipplei morphology were detected by MAb. On the contrary, the rabbit polyclonal serum reacted with many bacteria, most of which have no morphological features of T. whipplei (Fig. 5a and b).
FIG. 5.
Immunofluorescence detection of T. whipplei by using WS5H4 MAb (a) and rabbit polyclonal serum (b) in artificially contaminated stools (c) and in a negative stool control (d). Arrows indicate bacilli with typical T. whipplei morphology.
DISCUSSION
Since the clinical diagnosis of Whipple's disease is difficult and the isolation of the causative agent is time-consuming, the diagnosis of the disease is mainly based on the results of pathology and specific DNA detection. Although the serological diagnosis was encouraging in laboratory tests, this technique presents several drawbacks that render its routine use difficult (28). The identification and characterization of the immunodominant antigens could have important repercussions for developing novel diagnostic, prophylactic, and therapeutic techniques for Whipple's disease. Moreover, the sequencing of epitope polypeptide will provide the foundation for cloning and expression of recombinant antigen to be used in an enzyme-linked immunosorbent assay.
The monoclonal antibody technique has proven to be a powerful tool in studying the antigenicity and virulence of microorganisms (41). In the present work, we generated 13 and 10 MAbs that were as efficient as mouse polyclonal antibodies in recognizing T. whipplei strains Slow2 and Endo5, respectively, by the MIF assay. These MAbs were demonstrated to be specific, because they did not react either with 22 other pathogenic, phylogenetically closely related gram-positive bacteria, with common gastrointestinal pathogenic bacteria, or with bacterial species that have been shown to be cross-reactive with the Whipple's disease bacillus, such as Streptococcusagalactiae and Shigella flexneri (6, 11). The 58- and 84-kDa antigens appeared to be the immunodominant antigens, because most MAbs were found to have strong reactivity to these antigens. The MAb 7H3 recognized two protein bands, such as 105 and 65 kDa, in which the same epitope was probably present. Three antigens of 134, 65, and 47 kDa were recognized by only one MAb (Fig. 1).
In a previous work, we had produced some monoclonal antibodies against T. whipplei strain Twist-Marseille. However, the immunodominant epitopes of the strain had not been identified and characterized, because the proteins were not separated well by SDS-PAGE. Results of the two-dimensional blotting indicated that MAbs WS7G2 and WE11F10 were directed against only one epitope located at 58 and 84 kDa, respectively, which were reproduced several times by other MAbs (WS4D11, WS5D1, WS1C6, WS5E5, WS5H4, WE11G10, WE11B10, and WE8H5). However, other MAbs (such as 6F5, 6C3, and 7H3) recognized either a smear band or many spots. These results made it difficult for us to pick up the spots for MS. Therefore, only the 58- and 84-kDa antigenic spots were further analyzed by MS with MALDI-time of flight. These proteins were identified, respectively, as an ATP synthase F1 complex beta chain with a 168 score and 46% sequence coverage and a polyribonucleotide nucleotidyltransferase with a 130 score and 29% sequence coverage, which matched T. whipplei strain Twist isolated in Europe (29) and another strain from the United States (19) in the Mascot database, respectively. These results obtained from two strains from different geographical regions suggest that these epitopes are common to all T. whipplei strains and were confirmed by testing the corresponding MAbs to six other unrelated T. whipplei strains. Interestingly, the sizes estimated by SDS-PAGE were higher compared to the molecular mass determined by MALDI-MS, by which the molecular masses recorded were as low as 52 and 81 kDa. This can be explained in general by the fact that SDS-PAGE gives only a rough estimation of molecular mass.
The data presented in this paper demonstrate that 2-DE combined with MS constitutes a sensitive and powerful technique to identify the epitope of T. whipplei recognized by MAbs. The produced MAbs may be useful for better detection of T. whipplei in tissues or stools, and the 58- and 84-kDa antigens recognized by our MAbs are good candidates for the development of an enzyme-linked immunosorbent assay using the recombinant antigens. In a previous work, we used serological proteomic approaches for the identification of candidate antigens in Whipple's disease (12). The 58- and 84-kDa antigens identified herein were not detected. That does not mean that these antigens are not immunogenic for humans, because many proteins are present in the same area as these antigens, and sera of patients and controls recognize many protein spots. Only production of the 58- and 84-kDa antigens for testing with patients and control sera will enable us to address this issue (these tests are currently in progress).
Recently, the presence of T. whipplei in stool samples of patients with Whipple's disease was reported (8), and an isolate was obtained from stool samples (30). The MAb WS5H4, which recognizes the 58-kDa epitope, was demonstrated in this study to be an efficient means to detect T. whipplei in stool samples, contrary to rabbit polyclonal serum, which cross-reacts with many other bacteria. In the future, this MAb will be used in our laboratory prospectively in combination with PCR amplification for the detection of T. whipplei in stool samples. This approach could help to differentiate, contrary to PCR, true digestive Whipple's disease from simple carriage without using an invasive procedure (3, 5, 18, 37).
Acknowledgments
This project was funded under the 5th Framework Programme of the European Commission (Ref. QLG1-CT-2002-01049).
Footnotes
Published ahead of print on 20 September 2006.
REFERENCES
- 1.Black-Schaffer, B. 1949. The tinctoral demonstration of a glycoprotein in Whipple's disease. Proc. Soc. Exp. Biol. Med. 72:225-227. [DOI] [PubMed] [Google Scholar]
- 2.Bradford, M. M. 1976. A rapid and sensitive for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 3.Dutly, F. 2000. Tropheryma whippelii DNA in saliva of patients without Whipple's disease. Infection 28:219-222. [DOI] [PubMed] [Google Scholar]
- 4.Dutly, F., and M. Altwegg. 2001. Whipple's disease and “Tropheryma whippelii”. Clin. Microbiol. Rev. 14:561-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ehrbar, H. U., P. Bauerfeind, F. Dutly, H. R. Koelz, and M. Altwegg. 1999. PCR-positive tests for Tropheryma whippelii in patients without Whipple's disease. Lancet 353:2214. [DOI] [PubMed] [Google Scholar]
- 6.Evans, D., and M. H. Ali. 1985. Immunohistochemistry in the diagnosis of Whipple's disease. J. Clin. Pathol. 38:372-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fenollar, F., and D. Raoult. 2001. Whipple's disease. Clin. Diagn. Lab. Immunol. 8:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gross, M., C. Jung, and W. G. Zoller. 1999. Detection of Tropheryma whipplei (Whipple's disease) in faeces. Ital. J. Gastroenterol. Hepatol. 31:70-72. [PubMed] [Google Scholar]
- 9.Harlow, E., and D. Lane. 1988. Monoclonal antibodies and growing hybridomas, p. 139-282. In E. Harlow and D. Lane (ed.), Antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 10.Kaufmann, H., J. E. Bailey, and M. Fussenegger. 2001. Use of antibodies for detection of phosphorylated proteins separated by two-dimensional gel electrophoresis. Proteomics 1:194-199. [DOI] [PubMed] [Google Scholar]
- 11.Kirkpatrick, P. M., Jr., S. P. Kent, A. Milhas, and P. Pritchett. 1978. Whipple's disease: case report with immunological studies. Gastroenterology 75:297-301. [PubMed] [Google Scholar]
- 12.Kowalczewska, M., F. Fenollar, D. Lafitte, and D. Raoult. 1996. Identification of candidate antigen in Whipple's disease using a serological proteomic approach. Proteomics 6:3294-3305. [DOI] [PubMed] [Google Scholar]
- 13.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 227:680-685. [DOI] [PubMed] [Google Scholar]
- 14.La Scola, B., F. Fenollar, P. E. Fournier, M. Altwegg, M. N. Mallet, and D. Raoult. 2001. Description of Tropheryma whipplei gen. nov. sp. nov., the Whipple' s disease bacillus. Int. J. Syst. Evol. Microbiol. 51:1471-1479. [DOI] [PubMed] [Google Scholar]
- 15.Liang, Z., and D. Raoult. 2000. Species-specific monoclonal antibodies for rapid identification of Bartonella quintana. Clin. Diagn. Lab. Immunol. 7:21-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liang, Z., B. La Scola, and D. Raoult. 2002. Monoclonal antibodies to immunodominant epitope of Tropheryma whipplei. Clin. Diagn. Lab. Immunol. 9:156-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maiwald, M., and D. A. Relman. 2001. Whipple's disease and Tropheryma whippelii: secrets slowly revealed. Clin. Infect. Dis. 32:457-463. [DOI] [PubMed] [Google Scholar]
- 18.Maiwald, M., F. Schuhmacher, H. J. Ditton, and A. Von Herbay. 1999. Environmental occurrence of the Whipple's disease bacterium (Tropheryma whipplei). Appl. Environ. Microbiol. 64:760-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maiwald, M., P. W. Lepp, and D. A. Relman. 2003. Analysis of conserved non-rRNA genes of Tropheryma whipplei. Syst. Appl. Microbiol. 26:3-12. [DOI] [PubMed] [Google Scholar]
- 20.Maiwald, M., A. von Herbay, D. N. Fredricks, C. C. Ouverney, J. C. Kosek, and D. A. Relman. 2003. Cultivation of Tropheryma whipplei from cerebrospinal fluid. J. Infect. Dis. 188:801-808. [DOI] [PubMed] [Google Scholar]
- 21.Maizel, H., J. M. Ruffin, and W. O. Dobbins. 1970. Whipple's disease: a review of 19 patients from one hospital and a review of the literature since 1950. Medicine 49:175-205. [PubMed] [Google Scholar]
- 22.Misbah, S., and N. Mapstone. 2000. Whipple's disease revisited. J. Clin. Pathol. 53:750-755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nesterenko, M. V., M. Tilley, and S. J. Upton. 1994. A simple modification of Blum's silver stain method allows for 30 minute detection of proteins in polyacrylamide gels. J. Biochem. Biophys. Methods 28:239-242. [DOI] [PubMed] [Google Scholar]
- 24.Perkins, D. N., D. J. Pappin, D. M. Creasy, and J. S. Cottrell. 1999. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20:3551-3567. [DOI] [PubMed] [Google Scholar]
- 25.Petersen, A. 2003. Two-dimensional electrophoresis replica blotting: a valuable technique for the immunological and biochemical characterization of single components of complex extracts. Proteomics 3:1206-1214. [DOI] [PubMed] [Google Scholar]
- 26.Philip, R. N., E. A. Casper, W. Burgdorfer, R. K. Gerloff, L. B. Hughes, and E. J. Bell. 1978. Serologic typing of rickettsiae of the spotted fever group by microimmunofluorescence. J. Immunol. 121:1961-1968. [PubMed] [Google Scholar]
- 27.Ramzan, N. N., E. Loftus, Jr., L. J. Burgart, M. Rooney, K. P. Batts, R. H. Wiesner, D. N. Fredricks, D. A. Relman, and H. H. Persing. 1997. Diagnosis and monitoring of Whipple disease by polymerase chain reaction. Ann. Intern. Med. 126:520-527. [DOI] [PubMed] [Google Scholar]
- 28.Raoult, D., M. L. Birg, B. La Scola, P. E. Fournier, M. Enea, H. Lepidi, V. Roux, J. C. Piette, F. Vandenesch, D. Vital-Durand, and T. J. Marrie. 2000. Cultivation of the bacillus of Whipple's disease. N. Engl. J. Med. 342:620-625. [DOI] [PubMed] [Google Scholar]
- 29.Raoult, D., H. Ogata, S. Audic, C. Robert, K. Suhre, M. Drancourt, and J. M. Claverie. 2003. Tropheryma whipplei twist: a human pathogenic actinobacteria with a reduced genome. Genome Res. 13:1800-1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raoult, D., F. Fenollar, and M. L. Birg. 2006. Culture of Whipple's disease bacterium from the stools. N. Engl. J. Med. 355:1503-1505. [DOI] [PubMed] [Google Scholar]
- 31.Ratliff, N. B., J. T. McMahon, T. J. Naab, and D. M. Cosgrove. 1984. Whipple's disease in the porcine leaflets of a Carpentier-Edwards prosthetic mitral valve. N. Engl. J. Med. 311:902-903. [DOI] [PubMed] [Google Scholar]
- 32.Relman, D. A., T. M. Schmidt, R. P. MacDermott, and S. Falkow. 1992. Identification of the uncultured bacillus of Whipple's disease. N. Engl. J. Med. 327:293-301. [DOI] [PubMed] [Google Scholar]
- 33.Renesto, P., N. Crapoulet, H. Ogata, B. La Scola, G. Vestris, J. M. Claverie, and D. Raoult. 2003. Genome-based design of a cell-free culture medium for Tropheryma whipplei. Lancet 362:447-449. [DOI] [PubMed] [Google Scholar]
- 34.Rickman, L. S., W. R. Freeman, W. R. Green, S. Feldman, J. Sullivan, V. Russack, and D. A. Relman. 1995. Brief report: uveitis caused by Tropheryma whipplei (Whipple's bacillus). N. Engl. J. Med. 322:363-366. [DOI] [PubMed] [Google Scholar]
- 35.Schocker, F., and W. M. Becker. 2001. Optimization of electrophoresis for the identification of low molecular mass allergens in hazelnuts. J. Chromatogr. B. Biomed. Sci. Appl. 756:105-111. [DOI] [PubMed] [Google Scholar]
- 36.Shevchenko, A., M. Wilm, O. Vorm, and M. Mann. 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68:850-858. [DOI] [PubMed] [Google Scholar]
- 37.Street, S., H. D. Donoghue, and G. H. Neild. 1999. Tropheryma whipplei DNA in saliva of healthy people. Lancet 354:1178-1179. [DOI] [PubMed] [Google Scholar]
- 38.Vital-Durand, D., C. Lecomte, P. Cathebras, H. Rousset, and P. Godeau. 1997. Whipple disease. Clinical review of 52 cases. Medicine 76:170-184. [DOI] [PubMed] [Google Scholar]
- 39.Von Herbay, A., H. J. Ditton, and M. Maiwald. 1996. Diagnostic application of a polymerase chain reaction assay for the Whipple's disease bacterium to intestinal biopsies. Gastroenterology 110:1735-1743. [DOI] [PubMed] [Google Scholar]
- 40.Wilson, K. H., R. Blitchington, R. Frothingham, and J. A. P. Wilson. 1991. Phylogeny of the Whipple's-disease-associated bacterium. Lancet 338:474-475. [DOI] [PubMed] [Google Scholar]
- 41.Xu, W., and D. Raoult. 1997. Production of monoclonal antibodies against Rickettsia massiliae and their use in antigenic and epidemiological studies. J. Clin. Microbiol. 35:1715-1721. [DOI] [PMC free article] [PubMed] [Google Scholar]