Abstract
Enterococcus faecalis isolates of porcine origin were screened for the presence of a previously identified pathogenicity island (PAI). By using the esp gene as a genetic marker for the presence of this PAI, 9 esp-positive and 10 esp-negative isolates of porcine origin were investigated by use of a designed oligonucleotide array. The results indicated the clustering of esp-positive strains by multilocus sequence typing (MLST), but surprisingly, all strains investigated contained parts of the PAI. None of the strains of animal origin investigated belonged to previously identified MLST complex 2, where most isolates from patients cluster. Five of the nine esp-positive E. faecalis isolates of animal origin belonged to the same PAI complex as human isolate MMH594 but differed in their sequence types, which strongly indicates the horizontal transfer of the PAI between enterococci of porcine and human origin.
The esp gene is significantly associated with infection-derived isolates of Enterococcus faecalis (10) and E. faecium (13), contributes to the persistence of urinary tract infections in the bladder (9), and enhances biofilm formation by E. faecalis (11, 12). Recently, the esp gene has been shown to be part of a large pathogenicity island (PAI) harboring multiple virulence factors in E. faecalis (8) and E. faecium (4). These data suggest that esp may serve as a marker for the presence of the PAI and as a means of identifying virulent lineages of enterococci. The esp gene not only is detected in isolates of human origin but also can be detected in enterococcal isolates from both production and companion animals (2, 3; A. M. Hammerum and L. B. Jensen, Letter, Antimicrob. Agents Chemother. 40:4396, 2002).
In a previous study, 75 selected Enterococcus faecalis isolates of porcine origin from 2001 were screened for the presence of the esp gene by PCR. Six isolates (isolates D1 to D6) were found to be positive (Hammerum and Jensen, letter). These isolates, together with newly identified esp-positive isolates (isolates D27, D34, and D38) and an additional 10 esp-negative isolates (D26, D28 to D33, and D35 to D37) representing randomly selected isolates with different resistance profiles, were included in this study. All E. faecalis isolates were obtained from pig feces in 2001 as part of the Danish Integrated Antimicrobial Resistance and Research Programme. Only one E. faecalis isolate was collected from a pig on each of the sampled farms (1). The E. faecalis isolates should therefore be epidemiologically unrelated. To test this hypothesis, pulsed-field gel electrophoresis was performed with all esp-positive strains. Only two of these isolates (isolates D4 and D5) showed highly similar profiles (data not shown).
All isolates were analyzed by the newly described multilocus sequence typing (MLST) scheme for E. faecalis (7; http://www.mlst.net). The allelic profiles of the E. faecalis strains were obtained by sequencing internal fragments of seven housekeeping genes, gdh, gyd, pstS, gki, aroE, xpt, and yqiL (http://www.mlst.net); and the sequence types (STs) of all isolates were determined.
Among the nine esp-positive isolates of porcine origin, five belonged to ST16, two belonged to ST40, and one isolate each belonged to ST26 and ST47 (Table 1). Isolates belonging to ST16 have previously been isolated from nonhospitalized humans and animals. Isolates belonging to ST40 have previously been isolated from a hospitalized patient, a food handler from Spain, and a seal from The Netherlands (7). esp-positive human isolate MMH594 has ST6, which belongs to MLST clonal complex 2. This complex comprises hospital-derived isolates exclusively (7). None of the animal isolates tested belonged to this complex. The esp-negative isolates were genetically more diverse than the esp-positive isolates. Two esp-negative isolates each were ST40 (isolates D32 and D37) and ST99 (isolates D33 and D35), while the other esp-negative isolates were of genetically unrelated STs (Table 1). Two STs, ST40 and ST16, were represented by both esp-positive and esp-negative isolates.
TABLE 1.
Allelic profiles and STs of the E. faecalis isolates
| Strain no. | Origin | esp statusa | ST | Allelic profile
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| gdh | gyd | pstS | gki | aroE | xpt | yqiL | ||||
| OG1 | Human | Neg | 1 | 3 | 1 | 16 | 1 | 1 | 1 | 1 |
| MMH594 | Human | Pos | 6 | 12 | 7 | 3 | 7 | 6 | 1 | 5 |
| D2 | Porcine | Pos | 16 | 5 | 1 | 1 | 3 | 7 | 7 | 6 |
| D4 | Porcine | Pos | 16 | 5 | 1 | 1 | 3 | 7 | 7 | 6 |
| D5 | Porcine | Pos | 16 | 5 | 1 | 1 | 3 | 7 | 7 | 6 |
| D6 | Porcine | Pos | 16 | 5 | 1 | 1 | 3 | 7 | 7 | 6 |
| D29 | Porcine | Neg | 16 | 5 | 1 | 1 | 3 | 7 | 7 | 6 |
| D34 | Porcine | Pos | 16 | 5 | 1 | 1 | 3 | 7 | 7 | 6 |
| D38 | Porcine | Pos | 26 | 3 | 1 | 11 | 12 | 9 | 14 | 7 |
| D1 | Porcine | Pos | 40 | 3 | 6 | 23 | 12 | 9 | 10 | 7 |
| D27 | Porcine | Pos | 40 | 3 | 6 | 23 | 12 | 9 | 10 | 7 |
| D37 | Porcine | Neg | 40 | 3 | 6 | 23 | 12 | 9 | 10 | 7 |
| D32 | Porcine | Neg | 40 | 3 | 6 | 23 | 12 | 9 | 10 | 7 |
| D3 | Porcine | Pos | 47 | 19 | 1 | 24 | 22 | 19 | 17 | 14 |
| D26 | Porcine | Neg | 63 | 21 | 2 | 1 | 3 | 7 | 1 | 6 |
| D28 | Porcine | Neg | 96 | 27 | 12 | 35 | 32 | 6 | 2 | 23 |
| D30 | Porcine | Neg | 97 | 2 | 7 | 11 | 21 | 3 | 4 | 2 |
| D31 | Porcine | Neg | 98 | 25 | 2 | 15 | 12 | 23 | 18 | 26 |
| D33 | Porcine | Neg | 99 | 21 | 9 | 1 | 3 | 7 | 1 | 29 |
| D35 | Porcine | Neg | 99 | 21 | 9 | 1 | 3 | 7 | 1 | 29 |
| D36 | Porcine | Neg | 100 | 34 | 2 | 17 | 37 | 29 | 23 | 6 |
Neg, negative; Pos, positive.
All isolates were then screened for the presence of genes colocalized with the esp gene in the previously identified PAI by using a custom array. The custom oligonucleotide array was designed to accommodate all of the 129 open reading frames (ORFs) represented on the E. faecalis PAI as duplicate spots of 50-mer oligonucleotides. Each computer-generated oligomer represented a unique region of each of the PAI genes, was amino modified, and was arrayed on pan epoxy glass slides by using an Affymetrix 417 arrayer (MWG Biotech, Ebersberg, Germany). Total genomic DNA was isolated as described earlier (10) and labeled with fluorescent dyes, and hybridization experiments were performed with a mixture of two target preparations applied to each array. The processed microarray slides were read in the green (Cy3) and red (Cy5) channels of a ScanArray Express scanner (PE Life Sciences, Boston, MA). DNA from prototype PAI-positive human isolate MMH594 (8) and DNA from PAI-negative strain OG1RF (5), a well-characterized laboratory strain, were included as controls for the microarray hybridization. To confirm that the PAI genes were in fact organized in a contiguous manner and that the positive hybridization on the microarray was not indicative of the random distribution of genes throughout the genome, we performed overlapping long-range PCR throughout the length of the PAI, as described previously (8). Using isolate D34 as a prototype for the esp-positive isolates, we verified that the PAI genes were in fact contiguous both in esp-positive and in esp-negative strains. Furthermore, amplification across the left end of the PAI insertion site revealed that the PAI in the isolates tested was inserted at the same site where it is inserted in prototype human isolate MMH594, revealing the 10-bp duplication at the target site (8).
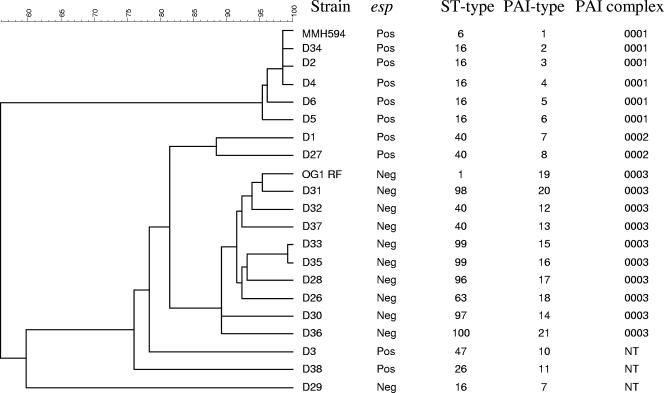
The pathochip hybridization results revealed a high degree of plasticity within the PAI region of the genomes of the enterococci isolated from pigs (Fig. 1). Surprisingly, all isolates, including the esp-negative isolates, contained fragments of the PAI. Interestingly, if single-linkage clustering (nearest-neighbor clustering), based on presence and absence of PAI genes, was used, the isolates tested could be grouped into two major complexes (PAI complexes 1 and 3) of 6 and 10 isolates, respectively, and a minor complex (PAI complex 2) of two isolates when an arbitrarily set cutoff of 85% similarity for grouping of the complexes was used (Fig. 2). Of the nine esp-positive E. faecalis isolates tested, five (55%) isolates, all of which were ST16, clustered in PAI complex 1, indicating that the PAI of porcine origin highly resembles the previously published PAI of strain MMH594 (8). The remaining 4 esp-positive isolates as well as the 11 esp-negative isolates contained a PAI that differed considerably from the published MMH594 PAI (8). Furthermore, these isolates, with the exception of strain D29, were of different STs and were not evolutionarily linked to the ST16 isolates. Previous studies by MLST based on seven housekeeping genes have defined distinct clonal complexes (CCs) of E. faecalis adapted to the hospital environment (CC2 and CC9). Of the isolates included in this study, only human isolate MMH594, which is ST6, grouped in one of these complexes, CC2. Porcine isolates of ST16 belonged to a previously reported prominent singleton clone represented by 11 isolates from hospitalized patients; 3 isolates from nonhospitalized individuals; and 3 isolates from animals, not including pigs, recovered from Spain and The Netherlands (7). Of the other STs reported in this study, four (ST1, ST26, ST40, and ST47) were found previously, although they were found infrequently and were not associated with pigs, whereas six STs (ST63, ST96, ST97, ST98, ST99, and ST100) represented novel STs (7).
FIG. 1.
Comparison of esp-positive E. faecalis isolates (isolates D3 through D38) and esp-negative E. faecalis isolates (isolates D36 through D35) (x axis) from pigs for the presence (gray) or the absence (black) of PAI genes (y axis) (4). MMH594 and OG1RF were included as controls for PAI-positive and PAI-negative strains, respectively.
FIG. 2.
Clustering of the tested E. faecalis strains by using single-linkage clustering (nearest-neighbor clustering), based on the presence and the absence of the individual genes composing the PAI. An arbitrarily set cutoff of 85% similarity for the grouping of complexes was used. NT, nontypeable.
The results presented here indicate the clonal expansion of most of the esp-positive E. faecalis isolates of porcine origin, with five of the nine isolates belonging to MLST group ST16. These five isolates all contained PAIs belonging to PAI complex 1. In addition, the finding of highly similar PAIs among genetically diverse E. faecalis isolates (like PAI complex 1 in isolates with ST6 and ST16, which do not belong to the same clonal complex, and PAI complex 3 in isolates with eight different STs that group in two different clonal complexes and five singleton clonal complexes) indicates the horizontal transfer of PAI gene sequences. Horizontal transfer of the esp gene has already been demonstrated in E. faecalis (6), and recent unpublished work has revealed that this is accompanied by the transfer of large regions of the E. faecalis PAI. Our finding that different PAI variants could be grouped on the basis of the presence and the absence of ORFs suggests that the insertions and the deletions of PAI ORFs driving the evolution of the PAI do not occur randomly but occur between defined clusters. Ongoing work suggests that large, contiguous regions of the PAI may be lost or gained as an evolutionary process, but the mechanisms defining these remain to be characterized (N. Shankar, unpublished data). Whether the plasticity of the E. faecalis PAI may favor adaptation to different ecological niches, like the guts of humans and animals, is unknown and remains to be further investigated.
The identification of highly homologous PAIs among enterococci isolated from production animals could indicate the presence of a nonhuman reservoir of the E. faecalis PAI implicated in virulence. The existence of a nonhuman reservoir of enterococcal virulence genes is undesirable and calls for careful monitoring and further research to gain more insight into the impact of the horizontal transfer of these genes from animals to humans, directly or via the food chain, and its influence on the establishment of pathogenic E. faecalis strains capable of causing infections in humans.
Acknowledgments
This work was funded, in part, through a grant from the Oklahoma Center for the Advancement of Science and Technology to N.S. This work was a part of the Danish Integrated Antimicrobial Resistance and Research Programme and was funded by the Danish Ministry of Family and Consumer Affairs and the Danish Ministry of the Interior and Health.
We thank Michael S. Gilmore for help with the microarray experiments.
Footnotes
Published ahead of print on 6 September 2006.
REFERENCES
- 1.Danish Integrated Antimicrobial Resistance and Research Programme. 2002. Consumption of antimicrobial agents and occurrence of antimicrobial resistance in bacteria from food animals, food and humans in Denmark. Danish Zoonosis Centre, Copenhagen, Denmark.
- 2.Garcia-Migura, L., E. Pleydell, S. Barnes, R. H. Davies, and E. Liebana. 2005. Characterization of vancomycin-resistant Enterococcus faecium isolates from broiler poultry and pig farms in England and Wales. J. Clin. Microbiol. 43:3283-3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harada, T., N. Tsuji, K. Otsuki, and T. Murase. 2005. Detection of the esp gene in high-level gentamicin resistant Enterococcus faecalis strains from pet animals in Japan. Vet. Microbiol. 106:139-143. [DOI] [PubMed] [Google Scholar]
- 4.Leavis, H., J. Top, N. Shankar, K. Borgen, M. Bonten, J. van Embden, and R. J. L. Willems. 2004. Novel putative enterococcal pathogenicity island linked to the esp virulence gene of Enterococcus faecium and associated with epidemicity. J. Bacteriol. 186:672-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murray, B. E., K. V. Singh, R. P. Ross, J. D. Heath, G. M. Dunny, and G. M. Weinstock. 1993. Generation of restriction map of Enterococcus faecalis OG1 and investigation of growth requirements and regions encoding biosynthetic function. J. Bacteriol. 175:5216-5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oancea, C., I. Klare, W. Witte, and G. Werner. 2004. Conjugative transfer of the virulence gene esp, among isolates of Enterococcus faecium and Enterococcus faecalis. J. Antimicrob. Chemother. 54:232-235. [DOI] [PubMed] [Google Scholar]
- 7.Ruiz-Garbajosa, P., M. J. M. Bonten, D. A. Robinson, J. Top, S. R. Nallapareddy, C. Torres, T. M. Coque, R. Cantón, F. Baquero, B. E. Murray, R. del Campo, and R. J. L. Willems. 2006. Multilocus sequence typing scheme for Enterococcus faecalis reveals hospital-adapted genetic complexes in a background of high rates of recombination. J. Clin. Microbiol. 44:2220-2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shankar, N., A. S. Baghdayan, and M. S. Gilmore. 2002. Modulation of virulence within a pathogenicity island in vancomycin-resistant Enterococcus faecalis. Nature 417:746-750. [DOI] [PubMed] [Google Scholar]
- 9.Shankar, N., C. V. Lockatell, A. S. Baghdayan, C. Drachenberg, M. S. Gilmore, and D. E. Johnson. 2001. Role of Enterococcus faecalis surface protein Esp in the pathogenesis of ascending urinary tract infection. Infect. Immun. 69:4366-4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shankar, V., A. S. Baghdayan, M. M. Huycke, G. Lindahl, and M. S. Gilmore. 1999. Infection-derived Enterococcus faecalis strains are enriched in esp, a gene encoding a novel surface protein. Infect. Immun. 67:193-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tendolkar, P. M., A. S. Baghdayan, M. S. Gilmore, and N. Shankar. 2004. Enterococcal surface protein, Esp, enhances biofilm formation by Enterococcus faecalis. Infect. Immun. 72:6032-6039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Toledo-Arana, A., J. Valle, C. Solano, M. J. Arrizubieta, C. Cucarella, M. Lamata, B. Amorena, J. Leiva, J. R. Penades, and I. Lasa. 2001. The enterococcal surface protein, Esp, is involved in Enterococcus faecalis biofilm formation. Appl. Environ. Microbiol. 67:4538-4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Willems, R. J., W. Homan, J. Top, M. van Santen-Verheuvel, D. Tribe, X. Manzioros, C. Gaillard, C. M. Vandenbroucke-Grauls, E. M. Mascini, E. van Kregten, J. D. van Embden, and M. J. Bonten. 2001. Variant esp gene as a marker of a distinct genetic lineage of vancomycin-resistant Enterococcus faecium spreading in hospitals. Lancet 357:853-855. [DOI] [PubMed] [Google Scholar]