Abstract
Regions of difference (RDs) have been described in clinical isolates of Mycobacterium tuberculosis, but the potential epidemiological and clinical relevance of the genotypes of these RDs remains to be investigated. We screened a population-based sample of 648 isolates for the deletion of five RDs, designated RD105, RD181, RD142, RD150, and RD239, using microarray-based hybridization, PCR, and DNA sequencing and assessed the associations between the RD deletions and the clinical characteristics of the patients using chi-square analysis and multivariate logistic regression model. Of the 648 isolates, 18 (2.8%) had the RD239 deletion and 39 (6.0%) had the RD105 deletion. The deletions of RD142, RD150, and RD181 subdivided the isolates with the RD105 deletion into four groups comprising a group with concurrent deletions of RD105, RD181, and RD142 (n = 13); a group with concurrent deletions of RD105, RD181, and RD150 (n = 5); a group with concurrent deletions of RD105 and RD181 (n = 13); and a group with a deletion of RD105 only (n = 8). Extrathoracic tuberculosis is statistically significantly associated with infection with the isolates with concurrent deletions of RD105, RD181, and RD142 (adjusted odds ratio [OR] = 3.05; 95% confidence interval [CI] = 1.58, 5.90) and the isolates with concurrent deletions of RD105, RD181, and RD150 (adjusted OR = 11.09; 95% CI = 4.27, 28.80), after controlling for the previously identified risk factors for extrathoracic tuberculosis (human immunodeficiency virus serostatus, race, gender, and the genotype of the plcD gene). These two combinations of RD deletions have the potential for predicting the clinical presentation of M. tuberculosis infection in the human host.
Although the overall genome of Mycobacterium tuberculosis is relatively more conserved than the genomes of many other bacterial species (9, 12), large sequence polymorphisms (LSPs) of M. tuberculosis consisting of insertions and deletions have been identified by comparison of complete genome sequences between CDC1551, a clinical strain that was found to be highly transmissible in humans (22), and H37Rv, a laboratory strain (9, 10). CDC1551 has been found to induce a more rapid and robust host immune response in vivo and in vitro than H37Rv (16).
In addition, LSPs among M. tuberculosis clinical isolates have been reported. Among 100 clinical isolates, 68 regions of difference (RDs) of M. tuberculosis have been identified by DNA microarray analysis and sequencing (21). Some of the RDs had frequencies of deletion of more than 20% (21). Among these RDs, the RD105 deletion was exclusively found in all strains in a genetically related group, named the Beijing/W lineage (20); thus, the RD105 deletion is suggested to be a marker of Beijing/W lineage strains (20). In addition, RD142, RD150, and RD181 have been found to subdivide the Beijing/W lineage strains into four subgroups, a group with concurrent deletions of RD105, RD181, and RD142; a group with concurrent deletions of RD105, RD181, and RD150; a group with concurrent deletions of RD105 and RD181; and a group with only the RD105 deletion (20). The Beijing/W lineage includes a large number of M. tuberculosis strains circulating around the world. Strains from the Beijing/W lineage have been associated with global transmission and drug resistance (3). Three principal genetic groups, based on the single-nucleotide polymorphisms of the katG gene codon 463 and the gyrA gene codon 95, have been used to describe the divergence of M. tuberculosis complex strains (9, 10, 19). The Beijing/W lineage strains of M. tuberculosis were classified into group 1 (9, 10). Given the reported association between the deletions of the four RDs and the Beijing/W lineage, exploration of the relationship between the three genetic groups and the RD deletions with a population-based sample may provide new insight into the evolution of M. tuberculosis.
Despite the report of the relationship between a given set of RD deletions and a family of M. tuberculosis clinical strains, whether these RDs have epidemiological and clinical significance beyond their associations with the Beijing/W strains remain to be investigated. Current tuberculosis (TB) control strategies assume that all clinical strains of M. tuberculosis are equally transmissible and virulent in humans, but if different RD genotypes account for different biological attributes of M. tuberculosis strains regarding virulence and transmissibility, alternative TB control strategies may be in order. To gain a better understanding of the clinical and epidemiological relevance of the presence or absence of the RDs and to explore the usefulness of these RDs as markers for a particular lineage of subpopulations, we screened a clinically and epidemiologically well characterized population-based collection of clinical isolates for the presence or absence of the four RDs associated with the Beijing/W lineage and RD239, which was included because of its high frequency of deletion among clinical isolates reported previously (21); we also analyzed the association between the RD genotypes and the clinical and epidemiological characteristics of the patients.
MATERIALS AND METHODS
Study isolates.
This study included 648 M. tuberculosis isolates from 648 patients diagnosed with culture-confirmed TB in Arkansas between 1 January 1996 and 31 December 2000; these represent 91.9% of the culture-confirmed cases detected during the study period. Genomic DNA was extracted from Lowenstein-Jensen slant cultures by standard procedures (18). The distributions of the demographics, social behaviors, and clinical characteristics of the patients were compared for the 648 available cases and the 56 unavailable cases. No statistically significant differences in the distributions of the variables studied were found between the study sample and the excluded cases. Previously, the isolates were genotyped by IS6110-based restriction fragment length polymorphism (IS6110 RFLP) analysis. Isolates with six IS6110-hybridizing bands or less were secondarily typed with pTBN12 (6, 23). Isolates were defined as clustered versus unique by use of the definition described previously (2). Spoligotyping results were available for 646 of the 648 isolates. Isolates that had spoligotype S00034, characterized by the absence of spacers 1 to 34, were classified as being of the Beijing/W lineage (14). In addition, information on the three principal genetic groups, based on the single nucleotide polymorphisms at katG codon 463 and gyrA codon 95 (19), were available for all the study isolates.
Patient data.
Patient information was obtained from the surveillance records of the Arkansas Department of Health and Human Services. This database included demographics, social and behavior characteristics, and clinical features.
The study protocols and procedures for the protection of human subjects were approved by the Health Sciences Institutional Review Boards of the University of Michigan and the University of Arkansas for Medical Sciences.
Detection of RD deletions.
A two-step experiment, that is, microarray-based hybridization followed by PCR, was conducted to determine the presence or absence of RD105, RD142, RD150, RD181, and RD239 in our isolate collection. As described below, the microarray hybridization was conducted by using the Library on a Slide platform (29). As the first step of the screening, our microarray experiment conditions were set up so that we would minimize the chance of falsely detecting the presence of the RDs under investigation, thereby maximizing the chance for catching all the existing deletions. The identification of the true RD deletions among those found by the microarray-based hybridization was done by PCR. When the size of a PCR product was different from that of positive control strain H37Rv, automated DNA sequencing was conducted to identify insertions or deletions in the RDs.
Microarray-based hybridization.
The genomic DNA of the study isolates, at a concentration of 1 μg/μl, was printed on Vivid gene array slides (Pall Life Sciences, West Chester, PA) by using a VersArray ChipWriter Pro system (Bio-Rad Laboratories, Hercules, CA). Each sample was printed twice on the same slide. A sequence within the 16S rRNA gene was used as a quantification probe to check the quantity of genomic DNA on the slides. All the hybridization probes were made by PCR with primers flanking a unique sequence within the RDs studied (Table 1). The PCR-amplified probes were purified and labeled with fluorescein-12-dCTP (Perkin-Elmer, Wellesley, MA) by using a BioPrime labeling kit (Invitrogen, Carlsbad, CA). The genomic DNA on the slides was hybridized with the RD probes by using a Super HYB kit with 50% formamide (Molecular Research Center, Inc., Cincinnati, OH) at 45°C. The slides were subsequently washed twice with a low-stringency buffer (2× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 0.1% sodium dodecyl sulfate [SDS]) for 5 min at room temperature and then twice with a high-stringency buffer (0.2× SSC, 0.1% SDS) for 20 min at 45°C. The slides were then incubated with a blocking solution containing 0.1 M Tris buffer, 0.3 M NaCl, and 10% blocking reagent (Amersham Biosciences, Piscataway, NJ) at room temperature for 1 h and then further incubated with a 5,000-fold-diluted conjugated antibody, anti-fluorescein-alkaline phosphatase Fab fragment (Roche, Basel, Switzerland) with blocking solution. The postdetection wash was done three times with a low-pH washing solution containing 0.1 M Tris (pH 7.5), 0.3 M NaCl, and 0.1% Tween 20 for 10 min and then with a high-pH washing solution containing 0.1 M Tris (pH 9.5), 0.1 M NaCl, and 0.01 M MgCl2 three times for 5 min each time. The colors of the spots on the slides were developed with an ArrayIt alkaline phosphatase kit (TeleChem, Sunnyvale, CA). The slides were scanned with an ArrayIt Microarray SpotWare colorimetric scanner (TeleChem).
TABLE 1.
RDs evaluated and primers of probes for microarray hybridization
| RD (probe no.) | Primers of probesa | Length (bp) |
|---|---|---|
| RD105 | ||
| Probe 1 | F 5′-CAA GGT ACG GCG GCT GGG GAT TC-3′ | 408 |
| R 5′-GCG GCG GGG TTA AAC AGG GTG AT-3′ | ||
| Probe 2 | F 5′-GTG CGC TTG CCA ACG ACT AAC C-3′ | 324 |
| R 5′-CGC ATG CGC ATC CAC CAG A-3′ | ||
| RD142 | ||
| Probe 1 | F 5′-GAG CGC CCG CAG GAC ATC-3′ | 923 |
| R 5′-GTT GGC GGG GTT GAG AGC-3′ | ||
| Probe 2 | F 5′-GTC CGC CGG GGC AAC CAC TTT-3′ | 1277 |
| R 5′-ACC ATC CGG GGG CAT CAC AGG-3′ | ||
| RD150 | F 5′-CGC CGC GGC AGC AAG TA-3′ | 2373 |
| R 5′-GCG CCC CAA CGG ATT TTC-3′ | ||
| RD181 | F 5′-AAC GCT GCC GCA CAA CCA ATG A-3′ | 535 |
| R 5′-TTA GCG CGA AGT GTC CGA GAT G-3′ | ||
| RD239 | F 5′-ACG GCG TGC AGG TGT GGA GTG G-3′ | 747 |
| R 5′-CGT CGG TGG GCA GTC GCA GAG C-3′ |
F, forward primer; R, reverse primer.
Microarray data analysis.
The images of the spots on the slides were read with IconoClust (Clondiag Chip Technologies GmbH, Jena, Germany). Spots were considered negative and eliminated from further analysis with the microarray if the signal values of the spots on the slide that hybridized with the 16S rRNA gene probe (4) were less than a threshold value, defined as 15% of the mean of the top 10% signal value on the slide; otherwise, the ratio of a sample was calculated as the signal value of the spot on the slide that hybridized with the probe divided by the signal value of the corresponding spot on the slide that hybridized with the 16S rRNA gene probe. When the ratio of a sample was zero, it was classified as having a deletion in the region studied, and a confirmatory PCR was conducted with primers flanking the region studied. When the ratio of the sample was greater than zero, it was classified as not having a deletion in the region studied. All calculations were done with SAS, version 9.0 (SAS Institute, Cary, NC).
PCR and sequencing.
PCR was conducted for all isolates showing RD deletions identified by microarray analysis. PCR was also done to investigate the deletion of the five RDs for isolates that did not have a sufficient amount of DNA for microarray-based hybridization (n = 102). In addition, to evaluate the sensitivity and the specificity of the microarray-based hybridization, we conducted PCR of RD105 for all 648 study isolates. The primers used for PCRs of different RDs were the same as those described previously (20), except for the primers for PCR of RD142 (5′-TCC GCG ACG ACG AAC AAC GAC GAC-3′ and 5′-GGC GGC GGA GAC GAC AGC AGG ATT-3′). The BD Advantage 2 PCR kit (BD-Biosciences Clontech, Palo Alto, CA) was used for the PCRs for RD105 and RD239. The BD Advantage GC-2 kit (BD-Biosciences Clontech) was used for the PCRs for RD142, RD150, and RD181. The thermocycling parameters for PCR assays were as follows: for RD105, 1 cycle at 94°C for 1 min, followed by 26 cycles of 94°C for 30 s, 68°C for 30 s, and 72°C for 4 min 45 s, with completion with a final cycle at 72°C for 10 min; for RD142, 1 cycle at 94°C for 2.5 min, followed by 26 cycles of 94°C for 45 s, 68°C for 45 s, and 72°C for 4.5 min, with completion with a final cycle at 72°C for 10 min; for RD150, 1 cycle at 94°C for 2.5 min, followed by 26 cycles of 94°C for 45 s, 68°C for 45 s, and 72°C for 3 min 45 s, with completion with a final cycle at 72°C for 10 min; and for RD181 and RD239, 1 cycle at 94°C for 1 min, followed by 26 cycles of 94°C for 30 s, 68°C for 30 s, and 72°C for 2 min 13 s, with completion with a final cycle at 72°C for 10 min.
When a PCR failed to generate the expected product, PCR of the 16S rRNA gene was performed to confirm the quantity and the quality of the DNA templates (4). For isolates generating a PCR product with a size different from that of positive control strain H37Rv, as visualized on a 1.0% agarose gel in 1× TBE (Tris-borate-EDTA) buffer, automated DNA sequencing was conducted to detect insertions or deletions in the RDs by using the corresponding PCR forward and reverse primers. By consideration of cost-efficiency, a two-step strategy was applied for DNA sequencing. First, we randomly selected one-third of the PCR products from each size group for DNA sequencing. Second, if the sequencing results showed differences among the PCR products within the same group, all the remaining PCR products in this group were sequenced; if the sampled PCR products showed identical sequences, the remaining products from the same group were not sequenced. Sequence comparison was performed by using the software Edit Seq 5.02 and MegAlign 5.01 (DNAStar Inc., Madison, WI).
Statistical analysis.
The distributions of epidemiological and clinical characteristics among the different RD genotypes were compared by the χ2 test or Fisher's exact test, as appropriate. When the association between clustering and the regions studied was analyzed, one isolate was randomly selected from each cluster. Considering the likelihood that cases in a cluster would not be independent, we used generalized estimating equations (GEEs) to control for potential intracluster dependence when we assessed the associations between the different RD genotypes and the clinical characteristics of the disease (15, 28). The magnitude of the associations was estimated by using the odds ratio (OR) and 95% confidence intervals (CIs). The disease sites were classified into thoracic and extrathoracic by using the definitions described previously (26). Briefly, thoracic TB was defined as disease sites confined to the lung, pleura, and intrathoracic lymph nodes, while extrathoracic TB was defined as cases of extrathoracic disease with or without concurrent disease within the thoracic cavity. Adjustment for the potential confounding by the four previously identified risk factors for extrathoracic TB was performed when the association between the RD genotype and the site of disease was analyzed by the use of logistic regression models. These factors included human immunodeficiency virus (HIV) serostatus, gender, race/ethnicity (26), and the genotype of the plcD gene (13, 27). To assess the individual effect of each of these four potential confounding factors, we first fit a base model that included only the RD genotype as an essential variable; we then added the four previously identified risk factors into the base model, one at a time, to fit an additional four models, designated models 2, 3, 4, and 5, respectively. A final model, designated model 6, was fit by adding all four risk factors for extrathoracic TB into the base model to adjust for the potential confounding of all four previously known risk factors for extrapulmonary TB. All statistical analyses were done with SAS, version 9.0 (SAS Institute).
Nucleotide sequence accession numbers.
The GenBank accession numbers for RD105 accompanied by a 9-bp insertion, an 18-bp insertion, and a deletion at the 5′ end of Rv0071 are DQ872637, DQ872638, and DQ872636, respectively. The sequence of the isolate that showed a partial deletion of RD105 can be found under GenBank accession number DQ872639. The sequences of isolates confirmed to have deletions of RD181, RD142, RD150, and RD239 can be found in GenBank under accession numbers DQ872640, DQ872641, DQ872642, and DQ872643, respectively.
RESULTS
Evaluation of the microarray-based hybridization.
Among the 648 isolates studied, 546 had DNA of sufficient quantity for the microarray hybridization experiment. All 546 isolates were subjected to PCR of RD105 in order to evaluate the sensitivity and the specificity of the microarray hybridization. On the basis of the results of microarray hybridization, 36 of the 546 isolates had the deletion of RD105. Of the 36 isolates showing the RD105 deletion on the microarray, 28 (77.8%) were confirmed to have the RD105 deletion by PCR. The sequencing results showed that the deletions of RD105 were accompanied by a 9-bp insertion (CCG GTG GAC), an 18-bp insertion (CCG GTG GAC CCG GTG GAC), or a deletion at the 5′ end of Rv0071. All except 1 of the 510 isolates that did not show the RD105 deletion in the microarray hybridization were confirmed to have RD105 by PCR; the 1 isolate that was the exception showed a partial deletion of RD105. For the other RDs investigated, the PCR assays were conducted only with isolates that showing deletions of these RDs by microarray hybridization. The proportions of isolates confirmed to have deletions of RD181, RD142, RD150, and RD239 by PCR were 91.7% (22/24), 88.9% (8/9), 18.8% (3/16), and 42.1% (16/38), respectively.
Frequency distribution of genotypes of RDs.
The results of microarray hybridization, PCR, and DNA sequencing were combined to determine the genotypes of the five RDs. Of the 648 isolates analyzed, 39 (6.0%) showed the deletion of RD105 and 1 (0.1%) had a partial deletion of RD105 (2.5 kb). The deletions of other RDs were found to be at a lower frequency compared with the frequency of the RD105 deletion. Thirteen (2.0%), 5 (0.8%), 31 (4.8%), and 18 (2.8%) isolates were found to have deletions of RD142, RD150, RD181, and RD239, respectively.
Relationship between the three principal genetic groups and RD genotypes.
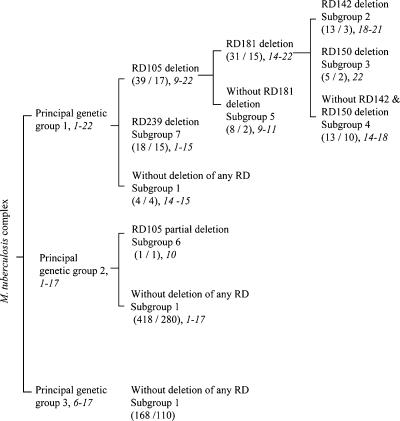
The relationship between the three principal genetic groups and the RD genotypes and the range of IS6110 copy numbers in each subgroup defined by RD genotypes were analyzed. Both principal genetic groups 1 and 2 were divided into subgroups by different combinations of deletions of the five RDs. Principal genetic group 3 remained undivided, as no deletion of the five RDs was found in this group (Fig. 1). Principal genetic group 1 was divided into three subgroups on the basis of the deletion of RD105 and RD239. None of the isolates with the RD105 deletion had the RD239 deletion, and all isolates with the RD105 or the RD239 deletion belonged to principal genetic group 1. The RD181 deletion divided the group with the RD105 deletion into two groups: one with the RD181 deletion and one without the RD181 deletion. The deletions of RD142 and RD150 further divided the group with the RD181 deletion into three groups: a group with the RD142 deletion, a group with the RD150 deletion, and a group without the RD142 and RD150 deletions. Principal genetic group 2 was divided into two groups on the basis of the RD105 deletion. In contrast to principal genetic group 1, the majority of the principal genetic group 2 isolates had no deletions in the five RDs investigated. None of the isolates in principal genetic group 3 showed any deletion in these RDs. Distinct IS6110 copy number ranges were observed for several subgroups defined by RD genotypes (Fig. 1).
FIG. 1.
Subgrouping of the three principal genetic groups of M. tuberculosis clinical isolates by deletions of RD105, RD142, RD150, RD181, and RD239. In the parentheses, the number before the slash is the number of isolates; the number after the slash is the number of strains. A strain is defined as a unique combination of IS6110 RFLP pattern and pTBN12 genotype. The range of IS6110-hybridizing band numbers for each subgroup of M. tuberculosis isolates is indicated by italicized numbers.
Relationship between Beijing/W lineage and deletion of RD105.
The RD105 deletion was previously reported to occur exclusively in all Beijing/W lineage strains (20). To explore the usefulness of RD105 as a marker for the Beijing/W lineage, we analyzed the distribution of isolates with the RD105 deletion among the Beijing/W lineage isolates and the non-Beijing/W lineage isolates. Among the 646 isolates for which spoligotyping results were available, 38 belonged to the Beijing/W linage, according to the definition of spoligotype S00034 (14). All 38 of these isolates had a deletion of RD105 and accounted for all the isolates with the RD105 deletion. These 38 isolates included 12 isolates with concurrent deletions of RD105, RD181, and RD142; 5 isolates with concurrent deletions of RD105, RD181, and RD150; 13 isolates with concurrent deletions of RD105 and RD181; and 8 isolates with the RD105 deletion alone. None of the Beijing/W lineage isolates had the RD239 deletion. The association between the Beijing/W lineage and the RD105 deletion was statistically significant (χ2 test, P < 0.0001).
Association between RD genotypes and clinical and epidemiological characteristics.
On the basis of all the combinations of RD deletions found in this study, the 648 study isolates were classified into one of seven subgroups: subgroup 1, no deletion detected; subgroup 2, the RD239 deletion; subgroup 3, concurrent deletions of RD105, RD181, and RD142; subgroup 4, concurrent deletions of RD105, RD181, and RD150; subgroup 5, concurrents deletion of RD105 and RD181; subgroup 6, deletion only of RD105; and subgroup 7, a partial deletion of RD105. The distribution of the clinical and epidemiological characteristics of the patients together with the genotypes of the plcD gene among these seven groups was analyzed. Statistically significant differences (P < 0.05) in the distributions of these deletions were found by race/ethnicity, age, plcD genotypes, and site of disease (Table 2).
TABLE 2.
Frequency distribution of clinical, epidemiological, and genotypes of plcD gene among seven RD genotypes by Fisher's exact testa
| Characteristics | No. (%) of isolates in each RD genotype-defined subgroupb
|
P | ||||||
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | ||
| Site of disease | 0.0454 | |||||||
| Thoracic | 526 (89.2) | 16 (88.9) | 10 (76.9) | 2 (40.0) | 11 (84.6) | 8 (100.0) | 1 (100.0) | |
| Extrathoracic | 64 (10.8) | 2 (11.1) | 3 (23.1) | 3 (60.0) | 2 (15.4) | 0 (0.0) | 0 (0.0) | |
| Race/ethnicity | <0.0001 | |||||||
| Non-Hispanic white | 336 (57.0) | 3 (16.7) | 7 (53.8) | 0 (0.0) | 8 (61.5) | 6 (75.0) | 0 (0.0) | |
| Non-Hispanic black | 216 (36.6) | 0 (0.0) | 4 (30.8) | 1 (20.0) | 3 (23.1) | 1 (12.5) | 0 (0.0) | |
| Hispanic | 25 (4.2) | 0 (0.0) | 1 (7.7) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (100.0) | |
| Asian/Pacific Islander | 10 (1.7) | 15 (83.3) | 1 (7.7) | 4 (80.0) | 2 (15.4) | 1 (12.5) | 0 (0.0) | |
| American Indian | 3 (0.5) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | |
| Age (yr) | 0.0004 | |||||||
| 0-14 | 6 (1.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | |
| 15-24 | 15 (2.5) | 2 (11.1) | 1 (7.7) | 1 (20.0) | 1 (7.7) | 0 (0.0) | 0 (0.0) | |
| 25-44 | 120 (20.3) | 5 (27.8) | 5 (38.5) | 3 (60.0) | 4 (30.8) | 2 (25.0) | 0 (0.0) | |
| 45-64 | 138 (23.4) | 8 (44.4) | 6 (46.2) | 1 (20.0) | 4 (30.8) | 4 (50.0) | 0 (0.0) | |
| 65+ | 311 (52.7) | 3 (16.7) | 1 (7.7) | 0 (0.0) | 4 (30.8) | 2 (25.0) | 1 (100.0) | |
| plcD genotype | <0.0001 | |||||||
| Wild type | 392 (66.4) | 17 (94.4) | 7 (53.8) | 1 (20.0) | 4 (30.8) | 1 (12.5) | 1 (100.0) | |
| plcD mutation | 35 (5.9) | 1 (5.6) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | |
| plcD and adjacent genes mutation | 163 (27.6) | 0 (0.0) | 6 (46.2) | 4 (80.0) | 9 (69.2) | 7 (87.5) | 0 (0.0) | |
Data are for 648 isolates. The variables without a statistically significant difference in the frequency distribution among seven genetic groups are not listed in this table. They include gender, city of residence, residence in a long-term-care facility, residence in a correctional facility, injection drug use, non-injection-drug use, excessive alcohol use, cavitary lung disease, and sputum smear positivity.
Subgroup 1, no deletion of RDs; subgroup 2, RD239 deletion; subgroup 3, concurrent deletion of RD105, RD181, and RD142; subgroup 4, concurrent deletion of RD105, RD181, and RD150; subgroup 5, concurrent deletion of RD105 and RD181; subgroup 6, RD105 deletion only; and subgroup 7, partial deletion of RD105.
DNA fingerprint clustering, as defined by a combination of IS6110 RFLP and pTBN12 secondary genotyping, has been used as a proxy for the transmissibility and virulence of M. tuberculosis. To assess the effect of the RD deletions on the transmissibility and virulence of the isolates, we examined the distributions of strains causing clusters of cases and strains causing only a single case among the seven subgroups by Fisher's exact test. The sample for the Fisher's exact test was composed of 337 single case isolates and 87 isolates randomly selected from each of the 87 clusters. No statistically significant association between the subgroup on the basis of the RD genotypes and clustering was found (P = 0.5534).
Multivariate logistic regression analysis of association between RD genotypes and site of disease.
The association between the site of disease and RD genotypes found by Fisher's exact test held true in all the logistic models described above. All the models showed that patients infected by an isolate with concurrent deletions of RD105, RD181, and RD142 and by an isolate with concurrent deletions of RD105, RD181, and RD150 were more likely to develop extrathoracic TB than patients infected by an isolate without any deletion of the five RDs investigated. The ORs of the RD genotypes in models 2 through 5 were similar to those obtained in the base model (data not shown). The adjustment for the potential confounding of the four previously known risk factors for extrapulmonary TB by model 6 strengthened the association between the concurrent deletions of RD105, RD181, and RD142 and the site of disease, while the association between the concurrent deletions of RD105, RD181, and RD150 and the site of disease was slightly reduced by this adjustment (Table 3).
TABLE 3.
Association between RD genotype and extrathoracic TB assessed by logistic regression models by use of the GEE method without and with adjustment for HIV serostatus, gender, race/ethnicity, and plcD genotype
| Subgroup | RD genotype | Crude OR (95% CI)a | Adjusted OR (95% CI)b |
|---|---|---|---|
| 1 | Without deletion | 1.00 | 1.00 |
| 2 | RD105, RD181, and RD142 | 2.46 (1.42, 4.26) | 3.05 (1.58, 5.90) |
| 3 | RD105, RD181, and RD150 | 12.33 (4.72, 32.23) | 11.09 (4.27, 28.80) |
| 4 | RD105 and RD181 | 1.49 (0.34, 6.54) | 2.21 (0.58, 8.44) |
| 5 | RD105c | ||
| 6 | Partial RD105c | ||
| 7 | RD239 | 1.03 (0.22, 4.76) | 1.18 (0.22, 6.36) |
Results from model 1, including only the RD genotype.
Results from model 6, including the RD genotype as well as HIV serostatus, gender, race/ethnicity, and plcD genotype.
The ORs of RD105 deletion alone and a partial RD105 deletion were not calculable because all patients infected by an isolate with an RD105 deletion alone or by an isolate with a partial RD105 deletion had thoracic TB.
DISCUSSION
This is the first population-based study to explore the epidemiological and clinical relevance of RD deletions among M. tuberculosis isolates. The major findings of this study are as follows: (i) patients infected by isolates with concurrent deletions of RD105, RD181, and RD142 and isolates with concurrent deletions of RD105, RD181 and RD150 are more likely to develop extrathoracic TB than patients infected by isolates without a deletion in any of the studied RDs; (ii) the RD105 deletion is significantly associated with the Beijing/W lineage; and (iii) two of the three principal genetic groups can be further classified into eight groups on the basis of different combinations of the deletions in the five RDs investigated.
To our knowledge, this is the first report that concurrent deletions of RD105, RD181, and RD142 and concurrent deletions of RD105, RD181, and RD150 are associated with extrathoracic TB. RD105 includes Rv0071 through Rv0074, RD181 includes Rv2262c and Rv2263c, RD142 includes Rv1189 through Rv1192, and RD150 includes Rv1671 through Rv1674c. Among these genes, Rv1189 is known to encode sigma factor I, which has been reported to be present only in M. tuberculosis and M. bovis (17) and to be involved in the survival of M. tuberculosis during the host-free aerosol particle stage (17). As bacterial sigma factors combine with RNA polymerase to regulate the transcription of other genes, the truncation of sigI might inhibit the expression of other genes. Further exploration of the genes regulated by sigI will help provide an understanding of these associations. The majority of the genes (8/14) in these RDs have no known function. Our findings might provide a clue for future functional studies of these genes.
It should be noted that the observed associations of the two combinations of the RD deletions with extrathoracic TB do not imply causality, especially given the possibility that there are likely to be a significant number of mutations in other genes that coexist in the genomes of the study isolates and that were not detected by the present investigation; any of these genes could be causing the observed phenotype. Furthermore, the sample sizes of isolates with concurrent deletions of RD105, RD181, and RD142 and isolates with concurrent deletions of RD105, RD181, and RD150 were relatively small; their associations with extrathoracic TB remain to be confirmed by using a sample with a larger number of the isolates carrying these two combinations of RD deletions. If the association is confirmed, these RD deletions would be useful for predicting the clinical outcome of an M. tuberculosis infection in the human host.
Beijing/W lineage strains have been found to be associated with nosocomial and community outbreaks, global transmission, and drug resistance (3). Consistent with previous reports (11, 20), our study also found that all the Beijing/W lineage strains had the RD105 deletion and that the RD105 deletion is exclusively found in non-Beijing/W lineage strains. This suggests that the RD105 deletion can be a useful marker for the Beijing/W lineage strain. In contrast to the previous study, which used a selected sample of isolates (11, 20), our study used a 5-year population-based collection of isolates from Arkansas, providing convincing confirmation of the relationship between the RD105 deletion and the Beijing/W lineage.
The three principal genetic groups have been considered markers for the divergence of M. tuberculosis (19), and a recent phylogenetic study further divided these three principal genetic groups into seven single-nucleotide-polymorphism cluster groups (8). We found that two of the three principal genetic groups (groups 1 and 2) can be further subgrouped on the basis of the deletions of the five RDs investigated. Further investigation of the relationship between these RD deletions and the reported single-nucleotide-polymorphism cluster groups would broaden our knowledge of the evolution of M. tuberculosis. Interestingly, most of the subgroups of the principal group 1 isolates in our study have a distinct range of IS6110 copy numbers. The IS6110 copy number ranges in the subgroups based on the deletions of RD181, RD142, and RD150 are almost nonoverlapping (Fig. 1). We previously found that IS6110 insertions are often associated with deletion events in the plcD gene region of the genome (13, 27); however, we did not observe any IS6110 element adjacent to the RD deletions, suggesting that these deletions are not due to the insertion of IS6110. The causes for the observed distribution of the IS6110 copy number range among the RD genotype-defined subgroups remain to be investigated.
In this study, we observed that the isolates in RD genotype-defined subgroups 3 and 4 had the highest IS6110 copy numbers among all the subgroups with different RD genotypes and that these two RD genotypes were associated with extrathoracic TB. We therefore hypothesize that the isolates with higher numbers of IS6110 copies might have experienced more frequent gene interruptions by the insertion of IS6110 in their genomes compared with the number of interruptions in those with lower IS6110 copy numbers. RD genotypes 3 and 4 may be markers for a high frequency of IS6110 insertion, and genes interrupted by the IS6110 insertion in these isolates may play an important role in the pathogenesis of TB.
This study also found that a larger proportion of patients infected by the isolates with the RD105 deletion are of Asian origin and younger than age 65 years compared with the proportion of patients infected by isolates without the RD105 deletion. This finding may be explained by the fact that all the isolates with RD105 deletion are in the Beijing/W lineage, and these isolates are prevalent in Asia (1, 7, 24). The latter finding might suggest that the cases infected by isolates with the RD105 deletion are more likely to have resulted from recent transmissions, because it was previously found that the proportions of cases of TB disease attributable to recent transmission are higher for young individuals than for elderly individuals (25). The other observation from the study supporting this explanation is that the majority of the isolates with the RD105 deletion belong to the Beijing/W lineage, and isolates of this lineage are known to have caused TB transmission worldwide (3).
However, we did not find a significant association between the RD genotypes and clustering. This could be due to the relatively small sample size of the Beijing/W lineage strains in our study (16 of 424 strains). In addition, as Arkansas is a state with a stable and dispersed population (5), it is possible that the clusters defined by the genotyping results in our study do not always reflect recent transmission; instead, they may represent some transmissions that occurred in the remote past.
The comparison of the results of PCR with those obtained by the microarray-based hybridization showed that the microarray technique can accurately detect those isolates that do not have large deletions of the RDs studied. However, it overestimates the number of isolates with deletions. This detection error might have been caused by printing errors that caused missing spots on the microarray slides or by the printing of a suboptimal amount of DNA on some of the spots due to an overestimation of the DNA concentration for some of the samples. Therefore, it is necessary to perform PCR and DNA sequencing to confirm any deletions found by the microarray-based method. In addition, microarray analysis can detect only the deletion of the sequence complementary to the probe sequences.
In conclusion, this study has advanced our knowledge of the potential clinical and epidemiological relevance of the RD deletions. Functional studies of the genes within these RDs will allow us to gain a better understanding of the observed associations.
Acknowledgments
This study was supported by a grant from the National Institutes of Health (grant NIH-R01-AI151975).
We acknowledge Kashef Ijaz's contribution to the establishment of the Arkansas Department of Health's surveillance database that was used for the study. We thank Dong Yang for excellent technical assistance in the culture of M. tuberculosis isolates and the preparation of the genomic DNA used for this study. We are grateful to the Tuberculosis Genotyping Laboratory of the Michigan Department of Community Health for their assistance in verifying the spoligotype of one study isolate.
Footnotes
Published ahead of print on 6 September 2006.
REFERENCES
- 1.Anh, D. D., M. W. Borgdorff, L. N. Van, N. T. Lan, T. van Gorkom, K. Kremer, and D. van Soolingen. 2000. Mycobacterium tuberculosis Beijing genotype emerging in Vietnam. Emerg. Infect. Dis. 6:302-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barnes, P. F., Z. Yang, S. Preston-Martin, J. M. Pogoda, B. E. Jones, M. Otaya, K. D. Eisenach, L. Knowles, S. Harvey, and M. D. Cave. 1997. Patterns of tuberculosis transmission in central Los Angeles. JAMA 278:1159-1163. [PubMed] [Google Scholar]
- 3.Bifani, P. J., B. Mathema, N. E. Kurepina, and B. N. Kreiswirth. 2002. Global dissemination of the Mycobacterium tuberculosis W-Beijing family strains. Trends Microbiol. 10:45-52. [DOI] [PubMed] [Google Scholar]
- 4.Boddinghaus, B., T. Rogall, T. Flohr, H. Blocker, and E. C. Bottger. 1990. Detection and identification of mycobacteria by amplification of rRNA. J. Clin. Microbiol. 28:1751-1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braden, C. R., G. L. Templeton, M. D. Cave, S. Valway, I. M. Onorato, K. G. Castro, D. Moers, Z. Yang, W. W. Stead, and J. H. Bates. 1997. Interpretation of restriction fragment length polymorphism analysis of Mycobacterium tuberculosis isolates from a state with a large rural population. J. Infect. Dis. 175:1446-1452. [DOI] [PubMed] [Google Scholar]
- 6.Chaves, F., Z. Yang, H. el Hajj, M. Alonso, W. J. Burman, K. D. Eisenach, F. Dronda, J. H. Bates, and M. D. Cave. 1996. Usefulness of the secondary probe pTBN12 in DNA fingerprinting of Mycobacterium tuberculosis. J. Clin. Microbiol. 34:1118-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dale, J. W., R. M. Nor, S. Ramayah, T. H. Tang, and Z. F. Zainuddin. 1999. Molecular epidemiology of tuberculosis in Malaysia. J. Clin. Microbiol. 37:1265-1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Filliol, I., A. S. Motiwala, M. Cavatore, W. Qi, M. H. Hazbon, M. Bobadilla del Valle, J. Fyfe, L. Garcia-Garcia, N. Rastogi, C. Sola, T. Zozio, M. I. Guerrero, C. I. Leon, J. Crabtree, S. Angiuoli, K. D. Eisenach, R. Durmaz, M. L. Joloba, A. Rendon, J. Sifuentes-Osornio, A. Ponce de Leon, M. D. Cave, R. Fleischmann, T. S. Whittam, and D. Alland. 2006. Global phylogeny of Mycobacterium tuberculosis based on single nucleotide polymorphism (SNP) analysis: insights into tuberculosis evolution, phylogenetic accuracy of other DNA fingerprinting systems, and recommendations for a minimal standard SNP set. J. Bacteriol. 188:759-772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fleischmann, R. D., D. Alland, J. A. Eisen, L. Carpenter, O. White, J. Peterson, R. DeBoy, R. Dodson, M. Gwinn, D. Haft, E. Hickey, J. F. Kolonay, W. C. Nelson, L. A. Umayam, M. Ermolaeva, S. L. Salzberg, A. Delcher, T. Utterback, J. Weidman, H. Khouri, J. Gill, A. Mikula, W. Bishai, W. R. Jacobs, Jr., J. C. Venter, and C. M. Fraser. 2002. Whole-genome comparison of Mycobacterium tuberculosis clinical and laboratory strains. J. Bacteriol. 184:5479-5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gutacker, M. M., J. C. Smoot, C. A. Migliaccio, S. M. Ricklefs, S. Hua, D. V. Cousins, E. A. Graviss, E. Shashkina, B. N. Kreiswirth, and J. M. Musser. 2002. Genome-wide analysis of synonymous single nucleotide polymorphisms in Mycobacterium tuberculosis complex organisms: resolution of genetic relationships among closely related microbial strains. Genetics 162:1533-1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hirsh, A. E., A. G. Tsolaki, K. DeRiemer, M. W. Feldman, and P. M. Small. 2004. From the cover: stable association between strains of Mycobacterium tuberculosis and their human host populations. Proc. Natl. Acad. Sci. USA 101:4871-4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hughes, A. L., R. Friedman, and M. Murray. 2002. Genomewide pattern of synonymous nucleotide substitution in two complete genomes of Mycobacterium tuberculosis. Emerg. Infect. Dis. 8:1342-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kong, Y., M. D. Cave, D. Yang, L. Zhang, C. F. Marrs, B. Foxman, J. H. Bates, F. Wilson, L. N. Mukasa, and Z. H. Yang. 2005. Distribution of insertion- and deletion-associated genetic polymorphisms among four Mycobacterium tuberculosis phospholipase C genes and associations with extrathoracic tuberculosis: a population-based study. J. Clin. Microbiol. 43:6048-6053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kremer, K., J. R. Glynn, T. Lillebaek, S. Niemann, N. E. Kurepina, B. N. Kreiswirth, P. J. Bifani, and D. van Soolingen. 2004. Definition of the Beijing/W lineage of Mycobacterium tuberculosis on the basis of genetic markers. J. Clin. Microbiol. 42:4040-4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liang, K., and S. L. Zeger. 1986. Longitudinal data analysis using generalized linear models. Biometrika 73:13-22. [Google Scholar]
- 16.Manca, C., L. Tsenova, C. E. Barry III, A. Bergtold, S. Freeman, P. A. Haslett, J. M. Musser, V. H. Freedman, and G. Kaplan. 1999. Mycobacterium tuberculosis CDC1551 induces a more vigorous host response in vivo and in vitro, but is not more virulent than other clinical isolates. J. Immunol. 162:6740-6746. [PubMed] [Google Scholar]
- 17.Manganelli, R., R. Proveddi, S. Rodrigue, J. Beaucher, L. Gaudreau, and I. Smith. 2004. Sigma factors and global gene regulation in Mycobacterium tuberculosis. J. Bacteriol. 186:895-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murray, M. G., and W. F. Thompson. 1980. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8:4321-4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sreevatsan, S., X. Pan, K. E. Stockbauer, N. D. Connell, B. N. Kreiswirth, T. S. Whittam, and J. M. Musser. 1997. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc. Natl. Acad. Sci. USA 94:9869-9874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsolaki, A. G., S. Gagneux, A. S. Pym, Y. O. Goguet de la Salmoniere, B. N. Kreiswirth, D. Van Soolingen, and P. M. Small. 2005. Genomic deletions classify the Beijing/W strains as a distinct genetic lineage of Mycobacterium tuberculosis. J. Clin. Microbiol. 43:3185-3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsolaki, A. G., A. E. Hirsh, K. DeRiemer, J. A. Enciso, M. Z. Wong, M. Hannan, Y. O. Goguet de la Salmoniere, K. Aman, M. Kato-Maeda, and P. M. Small. 2004. Functional and evolutionary genomics of Mycobacterium tuberculosis: insights from genomic deletions in 100 strains. Proc. Natl. Acad. Sci. USA 101:4865-4870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Valway, S. E., M. P. Sanchez, T. F. Shinnick, I. Orme, T. Agerton, D. Hoy, J. S. Jones, H. Westmoreland, and I. M. Onorato. 1998. An outbreak involving extensive transmission of a virulent strain of Mycobacterium tuberculosis. N. Engl. J. Med. 338:633-639. [DOI] [PubMed] [Google Scholar]
- 23.van Embden, J. D., M. D. Cave, J. T. Crawford, J. W. Dale, K. D. Eisenach, B. Gicquel, P. Hermans, C. Martin, R. McAdam, T. M. Shinnick, et al. 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 31:406-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Soolingen, D., L. Qian, P. E. de Haas, J. T. Douglas, H. Traore, F. Portaels, H. Z. Qing, D. Enkhsaikan, P. Nymadawa, and J. D. van Embden. 1995. Predominance of a single genotype of Mycobacterium tuberculosis in countries of East Asia. J. Clin. Microbiol. 33:3234-3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vynnycky, E., N. Nagelkerke, M. W. Borgdorff, D. van Soolingen, J. D. van Embden, and P. E. Fine. 2001. The effect of age and study duration on the relationship between ‘clustering’ of DNA fingerprint patterns and the proportion of tuberculosis disease attributable to recent transmission. Epidemiol. Infect. 126:43-62. [PMC free article] [PubMed] [Google Scholar]
- 26.Yang, Z., Y. Kong, F. Wilson, B. Foxman, A. H. Fowler, C. F. Marrs, M. D. Cave, and J. H. Bates. 2004. Identification of risk factors for extrapulmonary tuberculosis. Clin. Infect. Dis. 38:199-205. [DOI] [PubMed] [Google Scholar]
- 27.Yang, Z., D. Yang, Y. Kong, L. Zhang, C. F. Marrs, B. Foxman, J. H. Bates, F. Wilson, and M. D. Cave. 2005. Clinical relevance of Mycobacterium tuberculosis plcD gene mutations. Am. J. Respir. Crit. Care Med. 171:1436-1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zeger, S. L., and K. Liang. 1986. Longitudinal data analysis for discrete and continuous outcomes. Biometrics 42:121-130. [PubMed] [Google Scholar]
- 29.Zhang, L., U. Srinivasan, C. F. Marrs, D. Ghosh, J. R. Gilsdorf, and B. Foxman. 2004. Library on a slide for bacterial comparative genomics. BMC Microbiol. 4:12. [DOI] [PMC free article] [PubMed] [Google Scholar]