Abstract
Shigella sonnei contains numerous IS1 elements. The existence of polymorphisms in the length of the inter-IS1 spacer is a basis for the development of a PCR-based method for the subtyping of S. sonnei strains. The usefulness of inter-IS1 spacer typing (IST) was evaluated and compared with that of pulsed-field gel electrophoresis (PFGE) by characterization of S. sonnei isolates from epidemiologically nonrelated cases and outbreaks and of isolates that were indistinguishable by PFGE and that were collected from independent infection events. IST was less discriminatory than PFGE, with discriminatory indices of 0.96 and 0.63, respectively, but was able to compensate for the drawbacks of PFGE. PFGE exhibited a high level of discriminatory power for S. sonnei isolates; however, PFGE was also, at times, too discriminatory, which was a disadvantage in constructing the clonal relationships among strains circulating over a period of months or years. Furthermore, IST provided greater subtyping information for isolates indistinguishable by PFGE. The present study indicates that IST is more useful than PFGE for investigating the genetic relationships among S. sonnei strains circulating over a longer time span and also for discriminating certain strains which are indistinguishable by PFGE.
Shigella sonnei is the most commonly isolated species among the four Shigella species in industrialized countries (8, 10) and also the predominant species responsible for travel-associated shigellosis (7). Shigellosis is relatively infrequent and is a designated notifiable disease in Taiwan, meaning that all shigellosis cases must be reported and that the Shigella isolates must be sent to the Centers for Disease Control of Taiwan (Taiwan CDC). During the period from 1993 to 2004, 61 to 1,357 (average, 388) cases of shigellosis cases were confirmed annually in Taiwan, a country with 23 million people (3). In Taiwan, S. sonnei was responsible for most of the large shigellosis outbreaks in the industrialized western area (13) and was frequently associated with imported infections (11). In contrast, S. flexneri was mainly circulating among the aboriginal tribes in the mountainous areas (5).
Analyses of Shigella isolates by molecular subtyping methods can provide useful information on the outbreak and can help to trace the transmission and spread of the pathogen (6, 12). To date, several DNA-based methods have been developed for the subtyping of S. sonnei (14-16, 18). Among the subytping methods, pulsed-field gel electrophoresis (PFGE) has been standardized and used in the laboratories of “PulseNet,” a national molecular subtyping network for food-borne disease surveillance in the United States (19). The standardized PulseNet PFGE protocol (4) has been adopted by the Taiwan CDC and since 2003 has been used for the routine subtyping of Shigella species. PFGE has proven to be a powerful tool in our laboratory and others (1, 6) for discriminating Shigella strains for investigation of the outbreak. However, in our experience, PFGE is, in fact, at times too discriminatory for Shigella isolates. We frequently identified several PFGE genotypes among isolates collected from a single shigellosis outbreak. On the other hand, we also frequently identified isolates indistinguishable by PFGE from patients who had no apparent epidemiological link.
It has been reported that S. sonnei contains numerous insertion sequence (IS) elements (2). A recently released genomic sequence of S. sonnei reveals that the bacterial strain contains approximately 700 copies of IS elements and that 170 copies of these elements are IS1 (20). Using the Southern hybridization method, Soldati and Piffaretti (18) have demonstrated that the existence of copious IS elements in Shigella species is potentially useful for the molecular subtyping of the Shigella strains. In principle, the high frequency of IS1 elements in S. sonnei can be applied to the development of a rapid PCR-based subtyping method by amplifying the polymorphic length DNA of the inter-IS spacer. In the present study, we report an inter- IS1 spacer typing (IST) method for S. sonnei isolates. The usefulness of this novel method for the molecular subtyping of S. sonnei isolates was compared to that of PFGE. The results show the IST method is more useful than PFGE for investigating the genetic relationships among S. sonnei strains disseminated over a longer time span and for discriminating certain strains indistinguishable by PFGE.
MATERIALS AND METHODS
Bacterial strains.
S. sonnei isolates were collected in central and eastern Taiwan between 1996 and 2004. A collection of 194 isolates with no apparent epidemiological link was used to evaluate the discriminatory powers of the PFGE and the IST methods. The discriminatory powers of the two subtyping methods were evaluated by use of a discriminatory index, as calculated and reported by Hunter (9). Another collection of 150 isolates derived from 10 shigellosis outbreaks was used to evaluate the usefulness of PFGE and IST for the subtyping of isolates for the investigation of outbreaks (Table 1). The outbreaks occurred at schools, among tour groups, and in family neighborhoods over a period of weeks or months; and the epidemiological relationships among infected cases were confirmed by investigation. Another set of 20 isolates with identical PFGE patterns (pattern J16N09.0015) collected from eight epidemiologically unrelated events (Table 2) was subjected to antimicrobial susceptibility testing and IST analysis. The PFGE size reference strain, Salmonella enterica subsp. enterica serotype Braenderup H9812, was kindly provided by B. Swaminathan of the Centers for Disease Control and Prevention, Atlanta, Ga.
TABLE 1.
Characteristics of 10 shigellosis outbreaks and the genotypes of 150 Shigella sonnei isolates from the outbreaks
| Outbreak | Date of isolation (yr/mo/day) | Location (county) | No. of isolates | PFGE type (no. of isolates) | IST type (no. of isolates) |
|---|---|---|---|---|---|
| O1 | 1996/2/5-3/10 | Hwalien | 7 | J16N09.0002 (5) | IST11 (5) |
| J16N09.0003 (2) | IST11 (2) | ||||
| O2 | 1998/10/19-11/12 | Nantou | 8 | J16N09.0014 (8) | IST21 (8) |
| O3 | 1998/10/31 | Taoyuan | 8 | J16N09.0015 (4) | IST18 (4) |
| J16N09.0016 (1) | IST18 (1) | ||||
| J16N09.0129 (1) | IST18 (1) | ||||
| J16N09.0140 (1) | IST18 (1) | ||||
| J16N09.0141 (1) | IST18 (1) | ||||
| O4 | 1998/11/6 | Taoyuan | 6 | J16N09.0014 (5) | IST21 (5) |
| J16N09.0096 (1) | IST21 (1) | ||||
| O5 | 2000/10/20-11/4 | Hwalien | 49 | J16N09.0019 (17) | IST1 (17) |
| J16N09.0020 (1) | IST1 (1) | ||||
| J16N09.0023 (9) | IST1 (9) | ||||
| J16N09.0024 (1) | IST1 (1) | ||||
| J16N09.0025 (10) | IST1 (10) | ||||
| J16N09.0026 (6) | IST1 (6) | ||||
| J16N09.0027 (1) | IST1 (1) | ||||
| J16N09.0028 (1) | IST1 (1) | ||||
| J16N09.0029 (1) | IST1 (1) | ||||
| J16N09.0131 (1) | IST1 (1) | ||||
| J16N09.0127 (1) | IST23 (1) | ||||
| O6 | 2001/8/2-8/11 | Nantou | 17 | J16N09.0018 (9) | IST3 (8) |
| IST26 (1) | |||||
| J16N09.0036 (8) | IST3 (8) | ||||
| O7 | 2001/8/9-8/24 | Nantou | 27 | J16N09.0019 (18) | IST1 (18) |
| J16N09.0023 (3) | IST1 (3) | ||||
| J16N09.0050 (1) | IST1 (1) | ||||
| J16N09.0125 (1) | IST1 (1) | ||||
| J16N09.0147 (4) | IST1 (4) | ||||
| O8 | 2001/10/4-10/5 | Taoyuan | 6 | J16N09.0072 (4) | IST1 (4) |
| J16N09.0086 (1) | IST1 (1) | ||||
| J16N09.0126 (1) | IST1 (1) | ||||
| O9 | 2002/4/21-/6/5 | Nantou | 14 | J16N09.0019 (14) | IST1 (14) |
| O10 | 2003/12/18-2004/1/9 | Taitung | 8 | J16N09.0118 (8) | IST7 (8) |
TABLE 2.
Characteristics of 20 Shigella sonnei isolates with identical PFGE (J16N09.0015) genotype from eight epidemiologically unrelated events
| Event | Strain code | Date of isolation (yr.mo.day) | Country of infection | IST type | Antibiograma |
|---|---|---|---|---|---|
| E1 | Sh12569 | 1998.10.31 | Taiwan | IST18 | SSSSSSSSSSSRSSRSS |
| Sh12342 | 1998.10.31 | Taiwan | IST18 | SSSSSSSSSSSRSSRSS | |
| Sh12435 | 1998.10.31 | Taiwan | IST18 | SSSSSSSSSSSRSSRSS | |
| Sh12324 | 1998.10.31 | Taiwan | IST18 | SSSSSSSSSSSRSSRSS | |
| E2 | Sh15299 | 1999.2.4 | Taiwan | IST11 | SSSSSSSSSSSRSSRSS |
| E3 | C04.2952 | 2004.10.12 | Taiwan | IST11 | SSSSSSSSSSSRSSRRS |
| E4 | Sh33741 | 2003.11.21 | Indonesia | IST23 | SSSSSSSSSSSSSSRRS |
| Sh33680 | 2003.11.22 | Indonesia | IST23 | SSSSSSSSSSSSSSRRS | |
| Sh33592 | 2003.11.23 | Indonesia | IST23 | SSSSSSSSSSSSSSRRS | |
| Sh33597 | 2003.11.24 | Indonesia | IST23 | SSSSSSSSSSSSSSRRS | |
| Sh33651 | 2003.11.25 | Indonesia | IST23 | SSSSSSSSSSSSSSRRS | |
| Sh33683 | 2003.11.26 | Indonesia | IST23 | SSSSSSSSSSSSSSRRS | |
| Sh33766 | 2003.11.29 | Indonesia | IST23 | SSSSSSSSSSSSSSRRS | |
| E5 | S04.1133 | 2004.4.5 | Indonesia | IST1 | SSSSSSSSSSSSSSRRS |
| E6 | N04.0450 | 2004.2.1 | Vietnam | IST17 | SSSSSSSSSSSSSSRRS |
| E7 | N04.0453 | 2004.2.9 | Vietnam | IST17 | SSSSSSSSSSSSSSRRS |
| E8 | N04.1872 | 2004.5.26 | Cambodia | IST17 | SSSSSSSSSSSSSSRRS |
| N04.1873 | 2004.5.26 | Cambodia | IST17 | SSSSSSSSSSSSSSRRS | |
| N04.1876 | 2004.5.26 | Cambodia | IST17 | SSSSSSSSSSSSSSRRS | |
| N04.1870 | 2004.5.26 | Cambodia | IST17 | SSSSSSSSSSSSSSRRS |
The susceptibility (S) and resistance (R) results are presented for the antimicrobial agents in the following sequence: amikacin, ampicillin, cefazolin, cefixime, cefotaxime, cephalothin, chloramphenicol, ciprofloxacin, enrofloxacin, gentamicin, kanamycin, nalidixic acid, norfloxacin, ofloxacin, streptomycin, tetracycline, and tobramycin.. The results for nalidixic acid and streptomycin are underlined.
PCR amplification of inter-IS1 spacer.
Bacterial isolates were streaked on a Trypticase soy agar plate (Becton Dickinson and Company) and incubated at 37°C for 16 h. A single colony was transferred into 1 ml H2O to make a bacterial suspension (concentration, ca. 108 cells/ml). The suspension was then boiled for 10 min. One microliter of the heat-killed bacterial suspension (ca. 105 cells) was added into a PCR mixture as the DNA template. Each of the PCR mixtures contained 0.5 μM primers (primer IS1a [CTGAACAGGAGGGACAGCTGATAGAAACAG]; primer IS1b [CGGTGGAGCTGCATGACAAAGTCATCGGGC]), 200 μM deoxynucleoside triphosphates, 1× ExTaq buffer, and 1 U of ExTaq polymerase (Takara Shuzo Co. Ltd., Kyoto, Japan). PCR was performed at 94°C for 5 min, which was then followed by 25 cycles at 94°C for 30 s and 68°C for 8 min. The PCR amplicons were separated in a 1.2% agarose gel (15 cm by 20 cm; 20 combs) under the following conditions: 0.5× Tris-borate-EDTA buffer (5× TBE is 0.445 M Tris, 0.445 M boric acid, 0.01 M EDTA, pH 8.3) and 6 V/cm (200 V) for 80 min. A 100-bp DNA ladder (Gene Teks, Taipei, Taiwan) was used as a size standard by placing it in lanes 1, 6, 11, 16, and 20. After electrophoresis, the gel was stained with 1 mg/liter ethidium bromide, and the DNA profiles were recorded with a digital camera system (Electrophoresis Documentation and Analysis System 290; Kodak; Rochester, NY) with 1,792 by 1,200 pixels. The gels were analyzed with the fingerprint analysis software BioNumerics, version 4.5 (Applied Maths, Kortrijk, Belgium).
PFGE.
The PulseNet PFGE protocol for S. sonnei and other enterobacteria was used for PFGE analysis (4), except that 10 U of NotI was used instead of XbaI for the restriction digestion. After electrophoresis, the gel was stained with 1 mg/liter ethidium bromide, and the PFGE profiles were recorded with a digital camera system (Kodak). The PFGE gels were analyzed with the fingerprint analysis software BioNumerics (Applied Maths). A PFGE pattern with one or more DNA bands different from the others was taken to be a unique PFGE pattern.
Dendrogram.
Dendrograms were constructed with the NotI-digested PFGE patterns and the IST patterns by the unweighted pair group method with arithmetic mean algorithm by using the Dice value-predicted similarity of the two patterns and settings of 3% optimization and 1% position tolerance.
Antimicrobial susceptibility testing.
The S. sonnei isolates with the J16N09.0015 pattern were subjected to antimicrobial susceptibility testing with 17 antimicrobials agents by the disk diffusion method, which was performed as recommended by the CLSI (formerly NCCLS) (17). Disks were supplied by Becton Dickinson and Company (BD BBL Sensi-Disc antimicrobial susceptibility test disks) and contained the following antimicrobials agents (the numbers in the product designations in parentheses are in micrograms per disk): amikacin (AN30), ampicillin (AM10), cefalothin (CZ30), cefixime (CFM5), cefotaxime (CTX30), cephalothin (CF30), chloramphenicol (C30), ciprofloxacin (CIP5), enrofloxacin (ENO5), gentamicin (GM10), kanamycin (K30), nalidixic acid (NA30), norfloxacin (NOR10), ofloxacin (OFX5), streptomycin (S10), tetracycline (TE30), and tobramycin (NN10).
RESULTS
Amplification of inter-IS1 spacer.
The inter-IS1 spacers of the S. sonnei isolates were amplified by PCR, which generated six to nine DNA fragments with sizes within the range of the size standards (100 to 3,000 bp). The typeability was 100% for the isolates tested in this study.
Discriminatory powers of PFGE and IST.
A total of 77 PFGE patterns and 30 IST patterns were identified among the 194 isolates collected from patients with no apparent epidemiological links. The discriminatory indexes for PFGE and IST were 0.96 and 0.63, respectively.
Genotyping of S. sonnei isolates from 10 outbreaks.
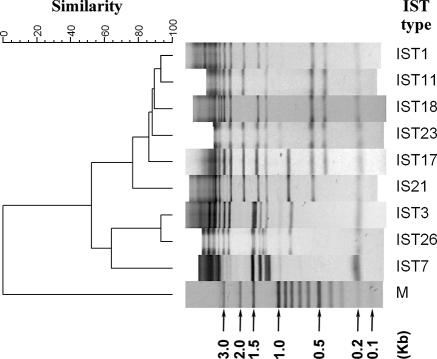
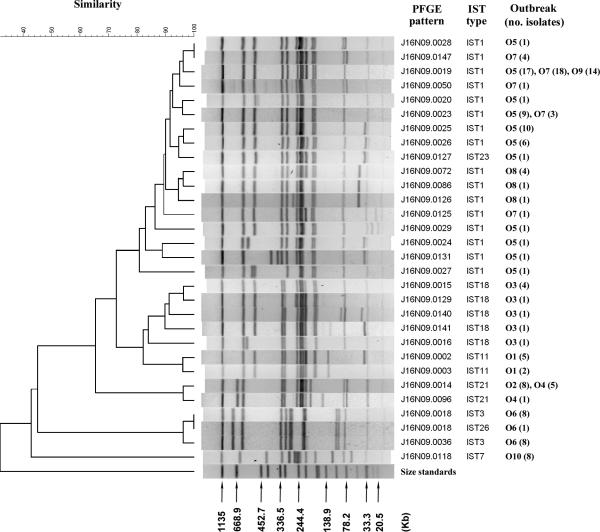
A collection of 150 S. sonnei isolates from 10 shigellosis outbreaks (Table 1) were analyzed by the IST and PFGE methods. IST identified eight IST patterns among the isolates. IST1 was detected in the isolates from four outbreaks (outbreaks O5, O7, O8, and O9) (Table 1). IST21 was detected among the isolates from two outbreaks (outbreaks O2 and O4); IST18, IST11, IST3, and IST7 were each detected among the isolates from a single outbreak. IST23 was a variant of IST1, and it was found for an isolate from outbreak O5; IST23 differed from IST1 by a 3-kb DNA fragment (Fig. 1). Similarly, IST26 was a variant of IST3, and they differed by a single DNA fragment (Fig. 1). In contrast to IST, PFGE was more sensitive at detecting genetic variations among the isolates. The results, shown in Fig. 2, indicate that isolates from a single outbreak can exhibit different PFGE patterns. Two or more PFGE patterns were identified among the isolates from seven outbreaks (Fig. 2). For the isolates from outbreak O5, which occurred at a high school, 11 PFGE patterns were identified among the 49 isolates collected from patients and their family contacts during an outbreak period of 16 days (Table 1). The most genetically divergent strains shared less than 80% pattern similarity (Fig. 2). Isolates from each of the outbreaks O7, O8, and O9 shared higher levels of PFGE pattern similarity with those from outbreak O5. The isolates from outbreaks O5, O7, and O9 had a major common genotype, J16N09.0019; in contrast, the isolates from outbreak O8 had no genotype in common with those found among isolates from other outbreaks. The clustering feature of PFGE patterns showed that the IST18 (outbreak O3) strains had a closer genetic relatedness with the IST11 (outbreak O1) and IST1 strains but had a more distant genetic relatedness with the IST21, IST3, and IST7 strains (Fig. 2). The clustering feature constructed with the IST patterns also demonstrated a similar genetic relationship among the strains (Fig. 1).
FIG. 1.
Dendrogram of nine IST patterns identified among the Shigella sonnei isolates from 10 outbreaks and the 20 isolates with an indistinguishable PFGE genotype (genotype J16N09.0015). Lane M, size standards.
FIG. 2.
Dendrogram generated with NotI-digested PFGE patterns of Shigella sonnei strains from 10 shigellosis outbreaks with their corresponding IST types and the numbers of isolates of each PFGE genotype. Size standards were XbaI-digested genomic fragments of Salmonella enterica subsp. enterica serotype Braenderup H9812.
Characterization of S. sonnei strains indistinguishable by PFGE.
Twenty S. sonnei isolates with identical PFGE patterns (pattern J16N09.0015) collected between 1996 and 2004 from eight epidemiologically unrelated events were further characterized by IST and antibiogram analysis. IST identified five IST genotypes among the isolates (Table 2). The five ISTs shared a high level of pattern similarity; IST11, IST17, IST18, and IST23 had only one DNA fragment different from the DNA fragments in IST1 (Fig. 1). The results of IST typing had higher levels of agreement with the epidemiological observations of the infection events. The four IST18 isolates were collected from an outbreak at a primary school in 1998. The seven IST23 isolates were collected from patients who had traveled to Bali Island in Indonesia in 2003 (11). The IST1 isolate (isolate S04.1133) was also collected from a patient infected in Bali Island in 2004. The six IST17 isolates were of different origins; two originated from Vietnam and four originated from Cambodia. The two IST11 isolates were collected over a 5-year interval; neither patient had a history of travel to a foreign country before the onset of the infection. Antimicrobial susceptibility tests indicated that all the isolates were resistant to streptomycin. The indigenous isolates (the first six isolates listed in Table 2) were also resistant to nalidixic acid, while all the imported isolates were susceptible to nalidixic acid. All the isolates collected after 2003 were resistant to tetracycline.
DISCUSSION
The IS1 element is abundant in the genome of S. sonnei (2). From a search of the invasive plasmid and chromosomal sequences of S. sonnei strain Ss046 that have been released (GenBank accession number CP000038), we identified 170 intact IS1 elements and 165 primer IS1a-annealing sites and 169 primer IS1b-annealing sites. The high frequency of the IS1 sequence in S. sonnei is the basis for the development of a PCR-based method to examine the inter-IS1 spacer polymorphisms. This novel method is fast, easy to handle, and cost-effective and exhibits a 100% typeability rate for the S. sonnei isolates tested in this study. IS1 has been used as a probe in the Southern hybridization method for the subtyping of Shigella species (18). The hybridization method is much more time-consuming and labor-intensive than the IST method. The hybridization method generates more DNA fragments than the IST method, and theoretically, it should thus be more discriminatory than IST. However, only small fragments can be effectively resolved in agarose gels, which decreases the discriminatory powers of the two methods.
PFGE has been adopted as a standard method for the routine subtyping of Shigella isolates by the Taiwan CDC since 2003. PFGE analysis with NotI has exhibited a high level of discriminatory power in distinguishing S. sonnei strains. In this study, for example, PFGE identified 11 genotypes among the 49 isolates collected from patients and their family contacts from outbreak O5 over a period of 16 days (Fig. 2). PFGE is a powerful tool for shigellosis outbreak investigation and has a high resolution; on the other hand, it is disadvantageous for the investigation of clonal relationships among S. sonnei strains circulating over a period of months or years. The IST method, as shown in this study, exhibits a lower level of discriminatory power than PFGE; however, the lower level of resolution was an advantage in compensating for the drawback of PFGE. In this study, outbreaks O5, O7, O8, and O9 occurred over a period of 18 months (Table 1); and all except one of the isolates from the outbreaks belonged to the IST1 genotype. The strains causing outbreaks O5, O7, and O9 were clonally related because they shared a common PFGE genotype (genotype J16N09.0019); however, the strains from outbreak O8 did not share a PFGE genotype with the strains from outbreaks O5, O7, and O9 (Fig. 2). The results of IST typing linked the clonal lineages of the strains for the four outbreaks.
IST typing identified five IST types among the 20 isolates with identical NotI-digested PFGE patterns (pattern J16N09.0015) collected from eight epidemiologically unrelated events (Table 2). The isolates were also indistinguishable by PFGE analysis with XbaI. The antibiogram analysis with 17 antimicrobial agents provided no decisive results for the discrimination of the strains collected from the events. In contrast, the results of IST typing were more informative for the independent events. However, this finding appears to be contradictory because IST typing was less discriminatory than PFGE. In a plasmid profile analysis study conducted in our laboratory, we found that most of the new PFGE patterns were generated from the acquisition or the loss of a plasmid(s) rather than from the rearrangement of chromosomal DNA (unpublished data). For example, genotype J16N09.0019 could be the primordial genotype of the IST1 clone. Except for genotypes J16N09.0024, J16N09.0131, and J16N09.0027, the other IST1 genotypes differed from J16N09.0019 by one to three plasmid DNA bands (Fig. 2). Genotype J16N09.0019 remained relatively stable, and it was the predominant type among the strains from outbreak O5 in October 2000 and outbreak O9 in April 2002 (Table 1). Genotype J16N09.0019 became genotype J16N09.0023 after it lost a 210-kb invasive plasmid (Fig. 2). IST could generate many DNA fragments, but only the small ones (those less than 3 kb) could be resolved effectively in an agarose gel under the electrophoresis conditions used in the present study. This is one of the reasons that IST exhibited a lower level of discriminatory power than PFGE. PFGE detects genetic variations that result in differences in large DNA fragments, while IST can detect variations occurring within a small region. The genotype J16N09.0015 strains could have been circulating in Taiwan and Southeast Asia for years. Although the isolates showed no apparent differences in their PFGE patterns, they already had variations within small DNA regions that could be detected by the IST method. In our study, IST3 and IST26 were detected among isolates of the J16N09.0018 type collected from outbreak O6 (Fig. 2), demonstrating that the mobility of IS1 did not result in an apparent change in the PFGE pattern, but the genetic variation was detected by IST typing.
The main advantage of the IST method for the subtyping of S. sonnei strains is that the result is more useful for investigating the spread of a clone over a period of years. In our records, shigellosis in central and eastern Taiwan was mainly caused by S. flenxeri from 1996 to mid-2000, but S. sonnei became the predominant cause from late 2000 to 2003. A total of 924 S. sonnei isolates collected between 1996 and 2004 were subjected to IST typing, and 717 isolates with the IST1 genotype were identified (data not shown). The IST1 clone, which originated from a South Asian country, emerged for the first time in 2000, continued circulating until 2003, and caused many outbreaks, including outbreaks O5, O7, O8, and O9, in central and eastern Taiwan. In contrast to IST, PFGE generated too many genotypes. Among the 717 IST1 isolates, 95 PFGE patterns were identified, and the PFGE patterns with the greatest difference shared only 55% pattern similarity. Therefore, IST typing generated more useful genotype data than PFGE for delineating the shigellosis epidemic which had been circulating in areas of outbreaks for more than 3 years.
In conclusion, IST is less discriminatory than PFGE for the subtyping of S. sonnei isolates, but it can compensate for the weakness of PFGE for evolutionary investigations. The data from IST typing are useful for the construction of clonality and for tracing of the spread of S. sonnei strains over periods of months or years. Furthermore, IST can detect genetic variations occurring within a small DNA region in S. sonnei strains that cannot be detected by PFGE analysis.
Acknowledgments
The research was supported by grants DOH90-DC-2018 and DOH91-DC-2011 from the Centers for Disease Control, DOH, Taiwan.
Pacific Edit reviewed the manuscript prior to submission.
Footnotes
Published ahead of print on 20 September 2006.
REFERENCES
- 1.Brian, M. J., R. Van, I. Townsend, B. E. Murray, T. G. Cleary, and L. K. Pickering. 1993. Evaluation of the molecular epidemiology of an outbreak of multiply resistant Shigella sonnei in a day-care center by using pulsed-field gel electrophoresis and plasmid DNA analysis. J. Clin. Microbiol. 31:2152-2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bustos-Martinez, J. A., and M. C. Gomez-Eichelmann. 1987. Frequency of IS1-mediated molecular events in different members of the family Enterobacteriaceae. J. Bacteriol. 169:4946-4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Center for Disease Control Taiwan. Center for Disease Control Taiwan. [Online.] http://www.cdc.gov.tw/en/index.asp (accessed September 1, 2006).
- 4.Centers for Disease Control and Prevention. 2004. One-day (24-28 h) standardized laboratory protocol for molecular subtyping of Escherichia coli O157:H7, non-typhoidal Salmonella serotypes, and Shigella sonnei by pulsed field gel electrophoresis. [Online.] http://www.cdc.gov/pulsenet/protocols.htm.
- 5.Chen, J. H., C. S. Chiou, P. C. Chen, T. L. Liao, T. L. Liao, J. M. Li, and W. B. Hsu. 2003. Molecular epidemiology of Shigella in a Taiwan township during 1996 to 2000. J. Clin. Microbiol. 41:3078-3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chiou, C. S., W. B. Hsu, H. L. Wei, and J. H. Chen. 2001. Molecular epidemiology of a Shigella flexneri outbreak in a mountainous township in Taiwan, Republic of China. J. Clin. Microbiol. 39:1048-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ekdahl, K., and Y. Andersson. 2005. The epidemiology of travel-associated shigellosis—regional risks, seasonality and serogroups. J. Infect. 51:222-229. [DOI] [PubMed] [Google Scholar]
- 8.Gupta, A., C. S. Polyak, R. D. Bishop, J. Sobel, and E. D. Mintz. 2004. Laboratory-confirmed shigellosis in the United States, 1989-2002: epidemiologic trends and patterns. Clin. Infect. Dis. 38:1372-1377. [DOI] [PubMed] [Google Scholar]
- 9.Hunter, P. R. 1990. Reproducibility and indices of discriminatory power of microbial typing methods. J. Clin. Microbiol. 28:1903-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kotloff, K. L., J. P. Winickoff, B. Ivanoff, J. D. Clemens, D. L. Swerdlow, P. J. Sansonetti, G. K. Adak, and M. M. Levine. 1999. Global burden of Shigella infections: implications for vaccine development and implementation of control strategies. Bull. W. H. O. 77:651-666. [PMC free article] [PubMed] [Google Scholar]
- 11.Lee, H. C., K. L. Chen, C. L. Tsai, C. H. Chen, T. N. Yeh, C. R. Yang, Y. L. Wang, H. Y. Chiu, C. L. Lee, H. P. Su, and T. H. Lin. 2004. Imported infection of Shigella sonnei molecular epidemiological investigation of cases of the Bali tours. Epidemiol. Bull. 20:23-42. [Google Scholar]
- 12.Lee, Y. S., M. C. Liu, C. F. Ko, C. H. Lu, and Y. H. Tseng. 2005. Molecular epidemiology of Shigella flexneri in a long-stay psychiatric nursing center during 2001 to 2003. J. Clin. Microbiol. 43:1353-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin, C. S., T. K. Wang, C. L. Tsai, S. Y. Ho, C. L. Lee, H. Y. Chen, and T. M. Pan. 2001. Analysis of relationships between several Shigella sonnei outbreaks in the Taoyuan area of Taiwan. Epidemiol. Bull. 17:83-97. [Google Scholar]
- 14.Liu, P. Y., Y. J. Lau, B. S. Hu, J. M. Shyr, Z. Y. Shi, W. S. Tsai, Y. H. Lin, and C. Y. Tseng. 1995. Analysis of clonal relationships among isolates of Shigella sonnei by different molecular typing methods. J. Clin. Microbiol. 33:1779-1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mendoza, M. C., A. J. Gonzalez, F. J. Mendez, and C. Hardisson. 1988. Plasmid typing of Shigella sonnei epidemic strains and molecular relationship of their R-plasmids. Eur. J. Epidemiol. 4:158-163. [DOI] [PubMed] [Google Scholar]
- 16.Mendoza, M. C., M. C. Martin, and M. A. Gonzalez-Hevia. 1996. Usefulness of ribotyping in a molecular epidemiology study of shigellosis. Epidemiol. Infect. 116:127-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.NCCLS. 2000. Performance standards for antimicrobial disk susceptibility tests. Approved standard, 7th ed. NCCLS document M2-A7. NCCLS, Wayne, Pa.
- 18.Soldati, L., and J. C. Piffaretti. 1991. Molecular typing of Shigella strains using pulsed field gel electrophoresis and genome hybridization with insertion sequences. Res. Microbiol. 142:489-498. [DOI] [PubMed] [Google Scholar]
- 19.Swaminathan, B., T. J. Barrett, S. B. Hunter, and R. V. Tauxe. 2001. PulseNet: the molecular subtyping network for foodborne bacterial disease surveillance, United States. Emerg. Infect. Dis. 7:382-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang, F., J. Yang, X. Zhang, L. Chen, Y. Jiang, Y. Yan, X. Tang, J. Wang, Z. Xiong, J. Dong, Y. Xue, Y. Zhu, X. Xu, L. Sun, S. Chen, H. Nie, J. Peng, J. Xu, Y. Wang, Z. Yuan, Y. Wen, Z. Yao, Y. Shen, B. Qiang, Y. Hou, J. Yu, and Q. Jin. 2005. Genome dynamics and diversity of Shigella species, the etiologic agents of bacillary dysentery. Nucleic Acids Res. 33:6445-6458. [DOI] [PMC free article] [PubMed] [Google Scholar]