Abstract
Gastric mucosal biopsy is widely used in the detection of Helicobacter pylori but is associated with a number of problems, including false-negative results due to sampling error and massive bleeding after biopsy. Given the extended period required to culture H. pylori, detection would be further improved by the use of rapid detection methods such as PCR. Here, we developed a rapid, safe, and convenient method for collecting H. pylori which combines endoscopic brushing with the loop-mediated isothermal amplification (LAMP) method. The specificity and sensitivity of LAMP were examined using nine urease-generating non-H. pylori bacterial species, Escherichia coli, Clostridium perfringens, Campylobacter jejuni, Helicobacter hepaticus, and 51 H. pylori strains. Results showed that H. pylori-specific LAMP primers amplified H. pylori DNA only and that the lowest detection limit of the LAMP reaction was 102 CFU. Brushing and biopsy samples taken from 200 patients with peptic ulcer at Nagoya University Hospital and a regional health care center were subjected to both LAMP and culturing. No adverse effects such as severe bleeding or penetration occurred during the procedure. By LAMP assay, 123 patients were confirmed as H. pylori positive when brushing technique samples were assayed, whereas only 100 were positive when biopsy samples were assayed. Culture assay detected H. pylori in 117 patients when it was combined with the brushing technique and in 96 when it was combined with biopsy. Combination of the endoscopic brushing technique with LAMP is considered a useful and safe system for identifying H. pylori infection.
Helicobacter pylori infection is recognized as a major cause of peptic ulcer disease and gastric cancer (8, 29). H. pylori occurs beneath the mucus layer, predominantly in the antrum (34). Infection is generally confirmed by histology (21), culturing (13), and the rapid urease test (RUT) (23) performed on endoscopic gastric mucosal biopsy samples or by the urea breath test (14). This latter test is a noninvasive method that detects 13CO2 generated by H. pylori urease (5); because it is indirect, however, it does not allow the measurement of antibiotic susceptibility, a disadvantage given that the recent gradual increase in the number of antibiotic-resistant strains has mandated susceptibility testing before eradication therapy (9, 25). The collection and identification of strains thus remain a valuable confirmation of infection and is necessary for the investigation of virulence (12, 25).
Nevertheless, biopsy carries the risk of causing bleeding and cannot be used for patients with a bleeding tendency, resulting, for example, from liver cirrhosis or idiopathic thrombocytopenic purpura, notwithstanding the possible association of these diseases with H. pylori infection (11). A second problem is the existence of urease-generating bacteria other than H. pylori; a number of bacteria produce urease and may colonize the stomach in an acid suppression environment (24, 25), potentially confounding the differentiation of H. pylori and urease-generating non-H. pylori bacteria (3).
Because it does not scratch the gastric mucosa, the brushing technique is a safe and useful way to collect bacteria (26, 36, 37). A large amount of bacteria can be collected by wide and broadly applied sweeping of the gastric mucosa. By itself, however, it is not sufficient because of the extended culture time required to determine the presence of H. pylori.
In contrast, the PCR method is faster and able to detect small amounts of bacterial chromosome (19, 33). Despite its simplicity and accuracy, however, PCR-based diagnosis is not widely used in smaller clinics because it is somewhat complicated and requires a thermal cycler. Various tests are utilized to improve diagnostic accuracy, but these increase costs and specimen requirements (2, 4, 15).
To overcome these difficulties, we developed loop-mediated isothermal amplification (LAMP), a novel gene amplification strategy in which all reactions are conducted under isothermal conditions using a single type of enzyme. This method has high amplification efficiency and provides faster amplification than the previous PCR methods (10, 27, 38). Amplification of target DNA can be conducted by simple incubation of the specimen, and all reagents are provided at a constant temperature for a fixed period of time. LAMP requires a conventional heating block only. The principle of LAMP involves autocycling strand displacement DNA synthesis using a DNA polymerase with high strand displacement activity. Recent reports demonstrating the usefulness of LAMP in detecting various bacteria and viruses (10, 16, 17, 21, 27, 28, 32, 35, 37) suggested it may be useful for the rapid detection of H. pylori. The LAMP procedure is simple, and its selectivity is extremely high owing to its use of four primers that recognize six distinct regions of the target DNA.
Here, we describe our development of a new diagnostic method to increase the sensitivity and safety of LAMP by combination with a brushing technique and evaluate the clinical utility of this method in the detection of H. pylori in 200 peptic ulcer patients.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
H. pylori 26695 and Helicobacter hepaticus 51449 were from ATCC (Manassas, VA), and 50 H. pylori clinical isolates were from Nagoya University Hospital.
H. pylori and H. hepaticus were cultured for 4 days on brucella broth agar plates (Difco Inc., Detroit, MI) supplemented with 7% heat-inactivated fetal calf serum (Gibco BRL, Rockville, MD) at 37°C in a microaerobic atmosphere. The identities of H. pylori and H. hepaticus were confirmed by characteristic colony morphology, Gram's stain, and positive reactions in urease, catalase, and oxidase tests.
Nine urease-generating non-H. pylori bacteria, including Klebsiella pneumoniae, Proteus mirabilis, Proteus vulgaris, Morganella morganii, Providencia rettgeri, Klebsiella oxytoca, Pseudomonas aeruginosa, Yersinia enterocolitica, and Staphylococcus epidermidis, as well as Escherichia coli, Clostridium perfringens, and Campylobacter jejuni, were clinical isolates from Nagoya University Hospital. They were cultured on Mueller-Hinton agar (Difco Inc., Detroit, MI) plates at 37°C for several days under suitable culture conditions. Urease-generating bacteria were confirmed to be urease positive by the RUT (Otsuka Pharmaceutical Co. Ltd, Tokyo, Japan).
Brushing technique.
To determine the reliability of LAMP combined with the brushing method in the detection of clinically isolated H. pylori specimens, brushing samples were taken from 200 peptic ulcer patients at Nagoya University Hospital and a regional health care center. All patients gave informed consent to participate in the study.
The brushing technique was performed according to a modification of the method of Sato et al. (31). After conventional endoscopic observation, a brush (163R; Mill-Rose Laboratories, Inc., Ohio) housed in guide tubing was introduced into the stomach through the biopsy channel of the endoscope. The brush was then pushed out of the guide tube and the mucosal surface from the greater curvature of the antrum to the body was stroked with the brush. The brush was then withdrawn into the guide tubing, and the brush/guide tubing was pulled out of the endoscope channel. The brush was then pushed out of the guide tubing again and introduced into 1 ml of sterilized phosphate-buffered saline (PBS). A 200-μl aliquot of this PBS containing sample was plated on an H. pylori-selective plate (Denka Seiken, Tokyo, Japan). The plate was incubated for 5 days at 37°C in a microaerobic atmosphere, and H. pylori was confirmed as a golden colony according to the manufacturer's instructions. Identification was also performed according to characteristic colony morphology, Gram's stain results, and positive reactions in urease, catalase, and oxidase tests. PBS containing H. pylori was used for both RUT and LAMP studies. After the brushing technique was used, three biopsy specimens each were taken from the greater curvature of the distal antrum and the upper body of the stomach and used for culturing, RUT, and LAMP. The results from these assays were compared to results from the same assays performed on samples obtained with the brushing technique. The biopsy sample was introduced into 50 μl of PBS and immediately plated on an H. pylori-selective plate for culturing.
RUT.
We also confirmed urease positivity by RUT using 50-μl aliquots of PBS containing brushing or biopsy samples. RUT was monitored for color change up to 2 h after the addition of each sample, and the test was scored as positive if the color changed from yellow to red.
Preparation of bacterial DNA.
Standard molecular biology methods were utilized (30). H. pylori 26695 genomic DNA was used as a positive control to determine the appropriate conditions for LAMP and to establish baseline sensitivity and specificity. Genomic DNAs of 50 H. pylori clinical isolates previously obtained from Nagoya University Hospital were used as a template for sensitivity and specificity tests of LAMP. Genomic DNAs from other non-H. pylori bacteria were used to determine the specificity of LAMP. All bacterial genomic DNA in this study was extracted with a Wizard genomic DNA purification kit (Promega) according to the manufacturer's instructions.
To confirm the suitability of LAMP studies for clinical use, genomic H. pylori DNAs from clinical samples were extracted by boiling. Briefly, samples attached to the brushing were transferred into 1 ml of sterilized PBS. Aliquots of PBS with sample after culture study were centrifuged at 12,000 × g for 5 min. The precipitates were resuspended in 100 μl of distilled water and boiled at 100°C for 5 min, and the supernatant was used as the DNA template.
16S rRNA PCR.
The quality of the DNA isolated from bacteria was confirmed by an initial PCR. Briefly, genomic DNA was amplified with 100 pmol of the 27F and 1492R primers (Fig. 1) in a 50-μl reaction mixture containing 0.25 μl of Ex Taq polymerase (Takara Biomedicals, Ohtsu, Japan) for 30 cycles of 1 min at 94°C, 1 min at 55°C, and 1 min at 72°C in a DNA thermal cycler (GeneAmp PCR system 9700; Applied Biosystems, California). The presence of a single 1,500-bp band was verified on a 1% agarose gel.
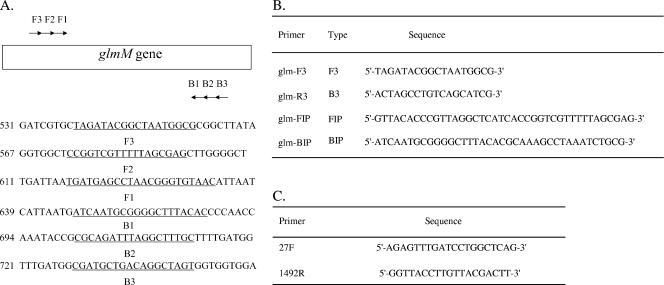
FIG. 1.
Names and locations of the target sequences used as primers for H. pylori LAMP within the H. pylori glmM gene. (A) The name and location of each target sequence as a primer in the H. pylori glmM gene are shown. LAMP primer sequences are underlined. (B) Sequences of LAMP primers used in this study. (C) Sequences of 16S rRNA primers used in this study.
LAMP assay.
LAMP primers were designed using genomic data (glmM gene; NCBI accession no. AAD07146) for the H. pylori 26695 strain. The LAMP reaction procedure was performed according to the manufacturer's instructions. LAMP requires a set of four specially designed primers (B3, F3, BIP, and FIP) which recognize a total of six distinct target DNA sequences (B1 to B3 and F1 to F3). Primers for H. pylori LAMP were designed against the glmM gene using Primer Explorer V software (Fujitsu, Tokyo, Japan). The location and sequence of each primer in the target DNA sequence are shown in Fig. 1. Primer BIP for the glmM gene consisted of the B1 direct sequence (20 nucleotides [nt]) and the B2 complementary sequence (18 nt) of the H. pylori 26695 strain. Primer FIP for the glmM gene consisted of the F1 complementary sequence (22 nt) and the F2 direct sequence (18 nt). Primers B3 and F3 for the glmM gene are complementary to sequences outside of the F2 and B2 regions. LAMP reactions were performed with a Loopamp DNA amplification kit (Eiken Chemical, Tochigi, Japan). LAMP requires the use of a 2× reaction mix consisting of 800 mM each of deoxynucleoside triphosphate, 2 M betaine, 40 mM Tris-HCl (pH 8.8), 20 mM KCl, 20 mM (NH4)2SO4, 8 mM MgSO4, and 0.2% Triton X-100 (25 μl). Total reaction mixture samples contained 1.6 μM of the FIP and BIP primers, 0.8 μM of each outer primer (F3 primer and B3 primer), 2× reaction mix (12.5 μl), Bst DNA polymerase (1 μl), and a 5-μl sample of bacterial DNA. The mixtures were incubated at 63°C for 60 min and then at 80°C for 2 min to stop the reaction. When positive, the LAMP reaction causes a level of turbidity in the reaction tube proportional to the amount of amplified DNA. Turbidity visible with the unaided eye was considered to indicate a successful LAMP procedure. After turbidity measurement, LAMP products were subjected to electrophoresis on 1% agarose gels, which were visualized under UV light after ethidium bromide staining. When amplification was successful, a ladder-like pattern containing the amplification region was seen. The results of LAMP were compared with those from plate culturing to assess the reliability of LAMP as a rapid diagnostic tool for the detection of H. pylori infection.
Great care was taken to avoid contamination between samples, and different rooms were used for DNA extraction, LAMP setup, and gel analysis. In addition, pipette tips with filters for aerosol protection were used to minimize contamination.
Case definition.
To fully evaluate and compare the diagnostic methods, no single test was used as the conclusive standard. To determine the sensitivity and specificity of each test, a patient's H. pylori infection status was established by the agreement of at least two of three methods, namely, RUT, LAMP, and culturing.
[13C]urea breath testing (Otsuka Pharmaceutical Co. Ltd, Tokyo, Japan) was done when all bacteriological testing was negative and was also used to confirm the presence of H. pylori.
Statistical analysis.
Significance in statistical analysis of the clinical study was determined using Fisher's exact test. P values of less than 0.05 were regarded as significant. The sensitivity and specificity of the LAMP assay were confirmed by repeating the procedures at least five times.
Specificity, sensitivity, and positive and negative predictive values (PPV and NPV, respectively) were calculated by the chi-square test.
RESULTS
To develop this new assay system, we first evaluated the sensitivity of the LAMP assay against H. pylori strains. We next confirmed the H. pylori detection limit. As the LAMP products contain several inverted repeat structures, positive samples demonstrate multiple bands of different sizes upon agarose gel electrophoresis. The reliability of LAMP was determined in initial validation studies with 51 H. pylori strains. All 51 isolates were successfully and rapidly detected by use of the LAMP assay (data not shown).
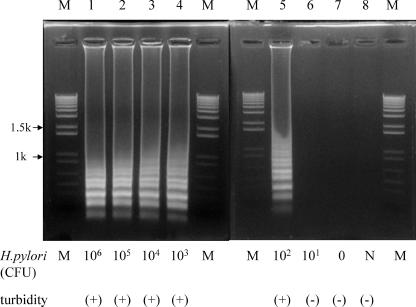
After initial validation, we checked H. pylori strain 26695, nine other urease-producing species, namely, Klebsiella pneumoniae, Proteus mirabilis, Proteus vulgaris, Morganella morganii, Providencia rettgeri, Klebsiella oxytoca, Pseudomonas aeruginosa, Yersinia enterocolitica, and Staphylococcus epidermidis, as well as Escherichia coli, Clostridium perfringens, Campylobacter jejuni, and H. hepaticus (Fig. 2). H. pylori-specific LAMP primers amplified only H. pylori DNA. To rule out the possibility that genomic DNA was absent, we confirmed the 16S rRNA PCR products of the 14 bacteria and showed that DNA extraction from the bacteria was successful (Fig. 2). The specificities of the primers were also tested by turbidity assay. Increased turbidity was limited to H. pylori DNA-containing samples only, with no increase seen in samples containing other types of bacterial DNA (Fig. 2). To determine sensitivity, serial dilutions of the H. pylori 26695 chromosomal DNA were used to determine the detection limit of LAMP and shown by agarose gel electrophoresis and turbidity assay to be 102 CFU/tube (Fig. 3). Further, no difference in sensitivity was seen between electrophoresis and turbidity assay results.
FIG. 2.
Representative results for LAMP products generated from a broad spectrum of closely related strains of urease-generating bacteria using primers for the H. pylori glmM gene. PCR conditions are described in the text. All samples were amplified by LAMP. The specificity of the method was confirmed by agarose gel electrophoresis and turbidity assay. Lanes: M, DNA size markers (kbp [k]); 1, H. pylori 26695; 2, Klebsiella pneumoniae; 3, Proteus mirabilis; 4, Proteus vulgaris; 5, Morganella morganii; 6, Providencia rettgeri; 7, Klebsiella oxytoca; 8, Pseudomonas aeruginosa; 9, Yersinia enterocolitica; 10, Staphylococcus epidermidis. 16S rRNA PCR products were used as an internal control.
FIG. 3.
Effect of the presence of H. pylori CFU on LAMP sensitivity. Various numbers of CFU of H. pylori were amplified, and the products were separated by gel electrophoresis and subjected to turbidity assay. The numbers above each lane represent 106 (lane 1), 105 (lane 2), 104 (lane 3), 103 (lane 4), 102 (lane 5), 101 (lane 6), 0 (lane 7), and negative (N; lane 8) CFU per reaction tube of template DNA of H. pylori. PCR conditions are described in the text. Lanes M, DNA size markers. k, kilobase pairs.
The results of the LAMP assay were the same as those with DNA extraction using both boiling and the extraction kit, indicating that extraction with the manufactured kit was not necessary to ensure the precision of the LAMP test (data not shown).
Finally, we compared H. pylori detection with the LAMP assay and other detection methods performed on brushing samples with detection by these same methods performed on biopsy samples in a clinical trial with 200 patients, among whom were 2 patients with liver cirrhosis and 3 with idiopathic thrombocytopenic purpura. No adverse effects (bleeding and penetration, etc.) were noted during the brushing procedure. A significantly higher prevalence of infection was demonstrated with brushing samples than with biopsy samples (P < 0.05) (Table 1). The assays combined with the brushing method showed significantly higher sensitivity than did the assays combined with the biopsy method, with a total of 123 patients confirmed as H. pylori positive versus only 100 by biopsy. Cultures from brushing-collected mucosal material were more frequently positive than those from biopsy specimens. There was a significant difference between culturing and LAMP in the detection of H. pylori. Twenty-three of 123 patients who were H. pylori positive by the brushing method combined with LAMP were not positive by the brushing method combined with culturing. The glmM gene products of H. pylori by LAMP amplified all 96 H. pylori culture-positive biopsy specimens and 117 culture-positive brushing specimens. Four H. pylori LAMP products were identified among 104 H. pylori samples which were negative by biopsy. For four patients, however, RUT results were positive, whereas biopsy and the brushing method were not. No H. pylori was detected in the 77 patients negative by the urea breath test. In the LAMP assay, no differences were seen between the electrophoresis and turbidity methods, even in the clinical trial.
TABLE 1.
Clinical detection of H. pylori in 200 patients
| No. of specimens | Result for sample obtained by:
|
|||||
|---|---|---|---|---|---|---|
| Biopsy
|
Brushing
|
|||||
| RUT | Culturing | LAMP | RUT | Culturing | LAMP | |
| 96 | − | + | + | − | + | + |
| 4 | − | − | + | − | + | + |
| 17 | − | − | − | − | + | + |
| 6 | − | − | − | − | − | + |
| 4 | + | − | − | + | − | − |
| 73 | − | − | − | − | − | − |
Even with our conservative definition of infection, the sensitivity, specificity, and positive and negative predictive values of our brushing-plus-LAMP assay were excellent and higher than those of other assays (Table 2).
TABLE 2.
Comparison of sensitivities, specificities, and PPV and NPV of H. pylori detection methods
| Method | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) |
|---|---|---|---|---|
| Brushing + LAMP | 100 | 100 | 100 | 100 |
| Brushing + culturing | 93.6 | 100 | 100 | 92.7 |
| Biopsy + LAMP | 81.3 | 100 | 100 | 77 |
| Biopsy + culturing | 78 | 100 | 100 | 74 |
DISCUSSION
To our knowledge, this is the first report of a safe and conventional method for the detection of H. pylori by the combination of LAMP and the brushing technique. Moreover, this LAMP assay is superior to all of the currently available DNA hybridization and PCR methods, owing to its lesser time requirements and simple equipment needs.
The reason for the increase in detection by this method over that by culturing may be due to the natural distribution of this bacterium. H. pylori is found mostly in the gastric surface mucus gel layer and epithelial intercellular junction of the stomach (34). Colonization is as frequent in the antrum as in the body (12), but is more dense in the antrum (1), possibly related to the fact that inflammatory responses are more prevalent and more intense in the antrum than in the body (18).
Moreover, owing to the patchy distribution of H. pylori, bacteriological culturing is the least sensitive diagnostic biopsy specimen test. Detection is dependent on the number and location of biopsy specimens, opening the possibility of sampling error (18). Thus, false-negative results in some groups of patients may be due to a relatively low density of bacteria. In the present study, 21 patients were negative by culturing from biopsy materials but positive by culturing from brushing samples. One possible reason for this is that number of bacteria in 21 cases was too low for detection by use of biopsy materials.
Several previous studies have shown the usefulness of imprint cytology in gastric biopsy and brushing cytology for the detection of H. pylori (6, 26, 36, 37). This method has not achieved broad use, however, owing to its complicated sample preparation. Antral gastric brushing has the potential to allow the sampling of a larger area of the gastric surface. The present study shows that H. pylori can be identified by the brushing method at a frequency higher than that obtained with existing methods. Antral brushing has also been used to obtain material for rapid urease testing. This procedure is inexpensive and simple, but the risk of false-positive results by urease-generating non-H. pylori bacteria has been emphasized (3, 24). Further, a false-positive rate of approximately 7% for the urea breath test has been reported (20) and is reportedly due to Klebsiella pneumoniae, Proteus mirabilis, Proteus vulgaris, Morganella morganii, Providencia rettgeri, Klebsiella oxytoca, Pseudomonas aeruginosa, Yersinia enterocolitica, and Staphylococcus epidermidis (25). In the present study, we successfully distinguished H. pylori from these nine urease-generating species with the LAMP assay. Our clinical study did not encounter any false-positive instances of the urea breath test. However, four patients who were negative by both culture and the LAMP method were positive in the RUT. The brushing method collects bacteria in the stomach widely, and the risk of sampling error through insufficient sample collection is likely to be low. We speculate that the false-positive results in these four patients may have been due to urease-generating non-H. pylori bacteria. In any case, these false positives were successfully excluded by the glmM-based LAMP assay.
H. pylori-specific genes that target the 16S rRNA (4), urease (ureA) (15), and phosphoglucosamine mutase (glmM) genes (2, 18, 32) have been proposed. The glmM gene encodes phosphoglucosamine mutase, an enzyme catalyzing the interconversion of glucosamine-6-phosphate into glucosamine-1-phosphate, which is subsequently transformed into N-acetylglucosamine (7), and PCR amplification of glmM is detected for H. pylori only. In one study which tested a wide spectrum of bacteria, primers for glmM were selective for H. pylori only and did not amplify products from non-Helicobacter spp. (19). Although the glmM gene is present in other bacteria, we constructed the LAMP primer from an H. pylori-specific glmM gene locus whose homology is low compared with those of other glmM genes. This specificity may be reflected in the high selectivity of the LAMP assay and its exclusion of false-positive urease in associated tests.
LAMP amplifies a very large amount of target DNA and produces magnesium pyrophosphate as a by-product. It is therefore possible to detect the amplification products without gel electrophoresis by using the white precipitate of magnesium pyrophosphate in the reaction mixture. This can be achieved due to the high specificity and amplification efficiency of LAMP. In all cases, we could distinguish LAMP-positive from LAMP-negative samples simply by the turbidity of the reaction mixture (Fig. 2 and 3). There was no difference between turbidity and electrophoresis results in terms of either LAMP sensitivity or LAMP specificity in our study (Fig. 2 and 3). Although this is a relatively simple approach, the macroscopic detection of a small amount of white precipitate is not always easy (27), and for strict confirmation we therefore recommend the use of electrophoresis rather than turbidity.
The LAMP-based assay identifies H. pylori from a brushing sample within about 90 min, a period comprising 20 min for DNA extraction, 60 min for the LAMP reaction, and 10 min for detection. Sensitivity decreased when the incubation time was shortened to 30 min (data not shown) but may be still high enough to identify the species of the brushing samples.
Loop primers are two primers designed outside the F3-B3 region of the target gene (Fig. 1) which are effective in amplifying PCR product by the LAMP method. However, we were unable to design these primers due to the inability to identify adequate glmM gene target lesions. Nevertheless, the LAMP assay is usually performed without loop primers, and our present LAMP assay was also successful despite the lack of loop primers.
LAMP assays provide clinicians with results quickly. Although the detection time with the rapid urease test is as the same as that by the LAMP assay, it is difficult for clinicians to rule out urease-positive non-H. pylori bacteria with the rapid urease test, potentially exacerbating the problem of inadequate eradication treatment. The LAMP assay will assist clinicians in the accurate detection of H. pylori.
The simple operation of the LAMP assay offers advantages over currently available DNA probe and PCR methods. Future development is aimed at enhancing clinical utility by increasing speed and extending coverage to all of the clinically critical H. pylori strains.
Acknowledgments
We thank the members of the Department of Bacteriology of the Nagoya University Graduate School of Medicine for their technical advice.
REFERENCES
- 1.Bayerdorffer, E., N. Lehn, R. Hatz, G. A. Mannes, H. Oertel, T. Sauerbruch, and M. Stolte. 1992. Difference in expression of Helicobacter pylori gastritis in antrum and body. Gastroenterology 102:1575-1582. [DOI] [PubMed] [Google Scholar]
- 2.Bickley, J., R. J. Owen, A. G. Fraser, and R. E. Pounder. 1993. Evaluation of the polymerase chain reaction for detecting the urease C gene of Helicobacter pylori in gastric biopsy samples and dental plaque. J. Med. Microbiol. 39:338-344. [DOI] [PubMed] [Google Scholar]
- 3.Brandi, G., B. Biavati, C. Calabrese, M. Granata, A. Nannetti, P. Mattarelli, G. Di Febo, G. Saccoccio, and G. Biasco. 2006. Urease-positive bacteria other than Helicobacter pylori in human gastric juice and mucosa. Am. J. Gastroenterol. 101:1756-1761. (First published 16 June 2006; doi: 10.1111/j.1572-0241.2006.00698.x.) [DOI] [PubMed] [Google Scholar]
- 4.Clayton, C. L., H. Kleanthous, P. J. Coates, D. D. Morgan, and S. Tabaqchali. 1992. Sensitive detection of Helicobacter pylori by using polymerase chain reaction. J. Clin. Microbiol. 30:192-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cutler, A. F., S. Havstad, C. K. Ma, M. J. Blaser, G. I. Perez-Perez, and T. T. Schubert. 1995. Accuracy of invasive and non-invasive tests to diagnose Helicobacter pylori infection. Gastroenterology 109:136-141. [DOI] [PubMed] [Google Scholar]
- 6.Dalla Libera, M., P. Pazzi, G. Carli, E. Contato, I. Piva, R. Scagliarini, A. Merighi, N. Ricci, and S. Gullini. 1996. Brushing cytology: a reliable method to detect Helicobacter pylori. J. Clin. Gastroenterol. 22:317-321. [DOI] [PubMed] [Google Scholar]
- 7.De Reuse, H., A. Labigne, and D. Mengin-Lecreulx. 1997. The Helicobacter pylori ureC gene codes for a phosphoglucosamine mutase. J. Bacteriol. 179:3488-3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dunn, B. E., H. Cohen, and M. J. Blaser. 1997. Helicobacter pylori. Clin. Microbiol. Rev. 10:720-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elviss, N. C., R. J. Owen, J. Xerry, A. M. Walker, and K. Davies. 2004. Helicobacter pylori antibiotic resistance patterns and genotypes in adult dyspeptic patients from a regional population in North Wales. J. Antimicrob. Chemother. 54:435-440. [DOI] [PubMed] [Google Scholar]
- 10.Enosawa, M., S. Kageyama, K. Sawai, K. Watanabe, T. Notomi, S. Onoe, Y. Mori, and Y. Yokomizo. 2003. Use of loop-mediated isothermal amplification of the IS900 sequence for rapid detection of cultured Mycobacterium avium subsp. paratuberculosis. J. Clin. Microbiol. 41:4359-4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gasbarrini, A., F. Franceschi, R. Tartaglione, R. Landolfi, P. Pola, and G. Gasbarrini. 1998. Regression of autoimmune thrombocytopenia after eradication of Helicobacter pylori. Lancet 352:878. [DOI] [PubMed] [Google Scholar]
- 12.Genta, R. M., R. M. Huberman, and D. Y. Graham. 1994. The gastric cardia in Helicobacter pylori infection. Hum. Pathol. 25:915-919. [DOI] [PubMed] [Google Scholar]
- 13.Goodwin, C. S., E. D. Blincow, J. R. Warren, T. E. Waters, C. R. Sanderson, and L. Easton. 1985. Evaluation of cultural techniques for isolating Campylobacter pyloridis from endoscopic biopsies of gastric mucosa. J. Clin. Pathol. 38:1127-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Graham, D. Y., P. D. Klein, D. J. Evans, D. G. Evans, L. C. Alpert, A. R. Opekun, and T. W. Boutton. 1987. Campylobacter pyloridis detected by the 13C-urea test. Lancet i:1174-1177. [DOI] [PubMed] [Google Scholar]
- 15.Ho, S. A., J. A. Hoyle, F. A. Lewis, A. D. Seker, D. Cross, N. P. Mapstone, M. F. Dixon, J. I. Wyatt, D. S. Tompkins, G. R. Taylor, and P. Quirke. 1991. Direct polymerase chain reaction test for detection of Helicobacter pylori in humans and animals. J. Clin. Microbiol. 29:2543-2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horisaka, T., K. Fujita, T. Iwata, A. Nakadai, A. T. Okatani, T. Horikita, T. Taniguchi, E. Honda, Y. Yokomizo, and H. Hayashidani. 2004. Sensitive and specific detection of Yersinia pseudotuberculosis by loop-mediated isothermal amplification. J. Clin. Microbiol. 42:5349-5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iwamoto, T., T. Sonobe, and K. Hayashi. 2003. Loop-mediated isothermal amplification for direct detection of Mycobacterium tuberculosis complex, M. avium, and M. intracellulare in sputum samples. J. Clin. Microbiol. 41:2616-2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Louw, J. A., V. Falck, C. van Rensburg, J. Zak, G. Adams, and I. N. Marks. 1993. Distribution of Helicobacter pylori colonization and associated gastric inflammatory changes: difference between patients with duodenal and gastric ulcers. J. Clin. Pathol. 46:754-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu, J.-J., C. L. Perng, R. Y. Shyu, C. H. Chen, Q. Lou, S. K. F. Chong, and C. H. Lee. 1999. Comparison of five PCR methods for detection of Helicobacter pylori DNA in gastric tissues. J. Clin. Microbiol. 37:772-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mabe, K., K. Fukase, T. Kitazawa, K. Kato, T. Matsuda, S. Kawata. 2004. The false-positive result of urea breath test. Helicobacter Res. 8:50-54.
- 21.Maeda, H., S. Kokeguchi, C. Fujimoto, I. Tanimoto, W. Yoshizumi, F. Nishimura, and S. Takashiba. 2005. Detection of periodontal pathogen Porphyromonas gingivalis by loop-mediated isothermal amplification method. FEMS Immunol. Med. Microbiol. 43:233-239. [DOI] [PubMed] [Google Scholar]
- 22.Marshall, B. J., and J. R. Warren. 1984. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet 8390:1311-1315. [DOI] [PubMed] [Google Scholar]
- 23.McNutly, C. A. M., and R. Wise. 1985. Rapid diagnosis of Campylobacter-associated gastritis. Lancet i(8443):1443-1444. [DOI] [PubMed] [Google Scholar]
- 24.Michaud, L., F. Gotirand, P. S. Ganga-Zandzou, N. Wizla-Derambure, D. Turck, and P. Vincent. 1998. Gastric bacterial overgrowth is a cause of false positive diagnosis of H. pylori infection using 13C urea breath test. Gut 42:594-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murray, P. R., E. J. Barton, and J. H. Jorgensen. 2003. Manual of clinical microbiology, 8th ed., vol 1. ASM Press, Washington, D.C.
- 26.Narváez Rodriguez, I. S., J. S. de Santamaria, M. D. M. A. Rubio, J. M. P. Acevedo, M. P. Jaen, A. M. C. de Orellana, and A. S. Monge. 1995. Cytologic brushing as a simple and rapid method in the diagnosis of Helicobacter pylori infection. Acta Cytol. 39:916-919. [PubMed] [Google Scholar]
- 27.Notomi, T., H. Okayama, H. Masubuchi, T. Yonekawa, K. Watanabe, N. Amino, and T. Hase. 2000. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 28:E63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Okamoto, S., T. Yoshikawa, M. Ihira, K. Suzuki, K. Shimokata, Y. Nishiyama, and Y. Asano. 2004. Rapid detection of varicella-zoster virus infection by a loop-mediated isothermal amplification method. J. Med. Virol. 74:677-682. [DOI] [PubMed] [Google Scholar]
- 29.Parsonnet, J., G. D. Friedman, D. P. Vandersteen, Y. Chang, J. H. Vogelman, N. Orentreich, and R. K. Sibley. 1991. Helicobacter pylori infection and the risk of gastric carcinoma. N. Engl. J. Med. 325:1127-1131. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 31.Sato, T., M. A. Fujino, Y. Kojima, F. Kithara, T. Nakamura, A. Morozumi, K. Nagata, T. Segiguchi, M. Nakamura, and H. Hosaka. 1999. Immunological rapid urease test for detecting Helicobacter pylori: comparative study of biopsy specimens and gastric mucus. Dig. Endoscopy 11:42-46. [Google Scholar]
- 32.Seki, M., Y. Yamashita, H. Torigoe, H. Tsuda, S. Sato, and M. Maeno. 2005. Loop-mediated isothermal amplification method targeting the lytA gene for detection of Streptococcus pneumoniae. J. Clin. Microbiol. 43:1581-1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shahamat, M., M. Alavi, J. E. M. Watts, M. Gonzalez, K. R. Sowers, D. W. Maeder, and F. T. Robb. 2004. Development of two PCR-based techniques for detection helical and coccoid forms of Helicobacter pylori. J. Clin. Microbiol. 42:3613-3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shimizu, T., T. Akamatsu, H. Ota, and T. Katsuyama. 1996. Immunohistochemical detection of Helicobacter pylori in the surface mucous gel layer and its clinicopathological significance. Helicobacter 1:197-206. [DOI] [PubMed] [Google Scholar]
- 35.Song, T., C. Toma, N. Nakasone, and M. Iwanaga. 2005. Sensitive and rapid detection of Shigella and enteroinvasive Escherichia coli by a loop-mediated isothermal amplification method. FEMS Microbiol. Lett. 243:259-263. [DOI] [PubMed] [Google Scholar]
- 36.Trevisani, L., S. Sartori, M. Ruina, M. Caselli, V. Abbasciano, E. Grandi, and E. Forini. 1997. Touch cytology. A reliable and cost-effective method for diagnosis of Helicobacter pylori infection. Dig. Dis. Sci. 42:2299-2303. [DOI] [PubMed] [Google Scholar]
- 37.Trevisani, L., S. Sartori, F. Galvani, M. Caselli, M. Ruina, V. Abbasciano, and E. Grandi. 1998. Usefulness of brushing urease test for diagnosis of Helicobacter pylori infection. Ital. J. Gastroenterol. Hepatol. 30:599-601. [PubMed] [Google Scholar]
- 38.Yoshida, A., S. Nagashima, T. Ansai, M. Tachibana, H. Kato, H. Watari, T. Notomi, and T. Takehara. 2005. Loop-mediated isothermal amplification method for rapid detection of the periodontopathic bacteria Porphyromonas gingivalis, Tannerella forsythia, and Treponema denticola. J. Clin. Microbiol. 43:2418-2424. [DOI] [PMC free article] [PubMed] [Google Scholar]