Abstract
Burkholderia thailandensis is closely related to Burkholderia pseudomallei, the causative agent of melioidosis. It is generally considered avirulent and previously has been reported to occur only in Southeast Asia. We report the first case of pneumonia and septicemia caused by B. thailandensis in the United States.
CASE REPORT
A healthy 2-year-old male was a restrained passenger in a single-car motor vehicle accident in northeast Texas in 2003. The overturned car came to rest in a drainage ditch beside the road, and the child was submerged for approximately 2 min. He was pulseless and apneic upon extraction from the vehicle. He was resuscitated, transported, and admitted to an intensive care unit, where he developed aspiration pneumonitis. He was afebrile for the first week of hospitalization, with a predominance of respiratory flora isolated initially, including abundant growth of nontypeable Haemophilus influenzae from endotracheal cultures. He was treated with cefuroxime, and 3 days later Stenotrophomonas maltophilia, Serratia marcescens, and an unidentified gram-negative rod (GNR) were isolated from respiratory secretions. Tobramycin was added to the antibiotic therapy, but the chest radiographs remained unchanged, with bilateral interstitial opacities. On the 12th day of hospitalization, the patient developed fever, and blood cultures were repeated. At that time, another isolate, which appeared to be the same as the unidentified GNR isolated from respiratory secretions, was recovered from blood culture. The patient was initially treated with a regimen that included ticarcillin-clavulanate and gentamicin. Treatment was changed to ceftazidime and trimethoprim-sulfamethoxazole after the unidentified GNR isolated from both the blood and respiratory secretions was presumptively identified as Burkholderia pseudomallei. Antimicrobial susceptibility testing was performed on the S. maltophilia, S. marcescens, and presumptive B. pseudomallei isolates; all three organisms were multidrug resistant. The patient improved and was discharged after a 1-month hospitalization. Ciprofloxacin and trimethoprim-sulfamethoxazole were continued for 18 weeks after discharge. Vaguely nodular bilateral pulmonary infiltrates present on chest radiographs at the time of discharge eventually resolved, and the patient is clinically well.
The blood isolate from this patient was submitted to the Centers for Disease Control and Prevention (CDC) for confirmatory identification. Standard biochemical tests were presumptive for B. pseudomallei (12); however, the isolate (CDC3015869) assimilated arabinose by use of minimal salt solution with 10% l-arabinose and therefore was identified as Burkholderia thailandensis (13).
We performed molecular characterization of this isolate using real-time PCR, 16S rRNA gene sequencing, multilocus sequence typing (MLST), and DNA-DNA hybridization. We tested the isolate by using a recently developed real-time PCR assay which amplifies a region of orf2 of the B. pseudomallei type III secretion system gene cluster (10). This assay is specific for B. pseudomallei and does not amplify DNA from other Burkholderia spp., including B. thailandensis. DNA from CDC3015869 was not amplified using the type III secretion system PCR.
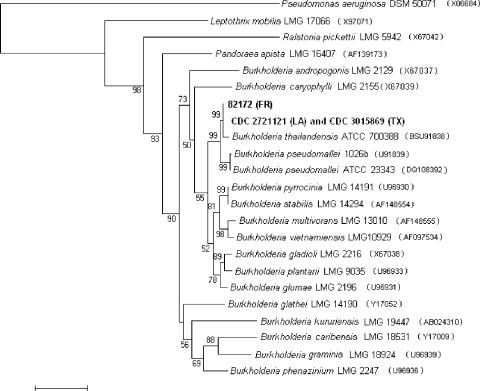
16S rRNA gene sequencing was performed as previously described (6) and yielded a sequence of approximately 1.5 kbp for CDC3015869. BESTFIT analysis (Wisconsin package, v. 10.3; Accelrys, San Diego, CA) indicated 99.4% identity with the B. thailandensis type strain (GenBank accession no. BSU91838), with a 2-bp insertion and 9-bp differences. An analysis comparing the 16S rRNA gene sequence of CDC3015869 with those of related Burkholderia strains in the CDC collection indicated that it was identical to strain CDC2721121, which was isolated from a pleural wound from a 76-year-old Louisiana man in 1997. Additionally, there was only a 1-bp difference (99.9% identity) between the 16S rRNA gene sequences of CDC3015869/CDC2721121 and that of 82172, another isolate recovered in 1982 from the intestines of a foal in France. Analysis comparing all three 16S rRNA gene sequences indicated they clustered together and most closely with B. thailandensis (Fig. 1).
FIG. 1.
16S rRNA gene sequence analysis. The neighbor-joining phylogenetic tree of Burkholderia thailandensis strains was based on comparisons with 16S rRNA gene sequences of related bacteria. Pseudomonas aeruginosa DSM 50071 (GenBank accession no. X06684) was used as an outgroup for this analysis. Sequences were aligned and trimmed and gaps removed using the Wisconsin package, v. 10.3. Analysis was then done with MEGA 3.1 (Kimura 2-parameter, 1,000-step bootstrap) (http://www.megasoftware.net/). The bar indicates 2% sequence dissimilarity.
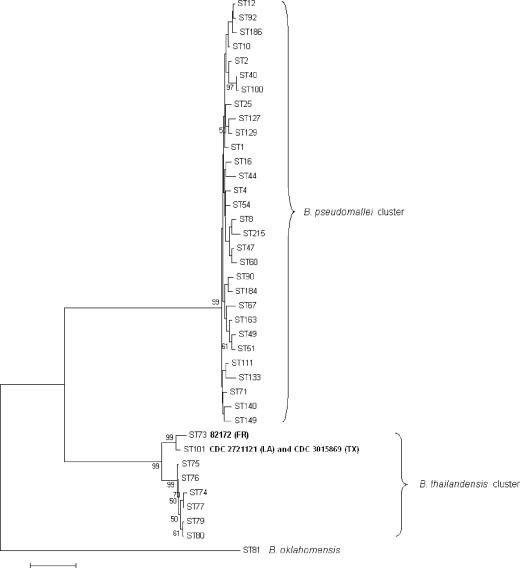
Although sequencing of the 16S rRNA gene can reveal differences among Burkholderia species, MLST is based upon differences in the sequences of seven housekeeping genes and has been shown to be not only useful for subtyping strains of B. pseudomallei but also capable of resolving genetic differences among close relatives of B. pseudomallei (6, 7, 8). CDC3015869 and CDC2721121 were identical by MLST and were designated sequence type (ST) 101 (7, 8). Strain 82172 was previously designated ST 73, and although it did not share any alleles with the six STs of B. thailandensis previously identified, it clustered most closely with B. thailandensis based on analysis of the concatenated sequences of the MLST loci (8). ST 101 shares two alleles, gltB and gmhD, with ST 73, and both STs cluster with other B. thailandensis isolates (Fig. 2). The allele designations and sequences are available on the MLST website (http://bpseudomallei.mlst.net).
FIG. 2.
Multilocus sequence typing. The neighbor-joining phylogenetic tree was derived from concatenating all seven allele sequences for each MLST locus, using a representative set of STs from the MLST website (www.mlst.net). Analysis of concatenated sequences was done per Godoy et al. (8) using MEGA 3.1 (Kimura 2-parameter, 1,000-step bootstrap) (http://www.megasoftware.net/). The bar indicates 0.5% sequence dissimilarity.
DNA-DNA hybridization was performed as previously described (2). The relative binding ratios (RBR) of CDC2721121 were 100% to both CDC3015869 and 82172 and 96% to the type strain of B. thailandensis but only 76% to the type strain of B. pseudomallei under the optimum reassociation conditions (65°C) (Table 1) . Divergence for strains CDC3015869 and 82172 was <1, and the RBR remained high (91% to 95%) under stringent reassociation (80°C) conditions. The divergence for B. pseudomallei was 4.5, and RBR dropped to 59% under the stringent reassociation conditions.
TABLE 1.
DNA-DNA hybridization of Burkholderia spp. and Burkholderia-like strains
| Source of unlabeled DNA | Result with labeled DNA from strain CDC2721121 (LA)
|
||
|---|---|---|---|
| RBRa (%) (65°C) | Db | RBRa (%) (80°C) | |
| CDC2721121 (LA) | 100 | 0.0 | 100 |
| CDC3015869 (TX) | 100 | 1.0 | 91 |
| 82172 (France) | 100 | 0.5 | 95 |
| Burkholderia thailandensis ATCC 700388 | 96 | 1.0 | 91 |
| Burkholderia pseudomallei ATCC 23343 | 76 | 4.5 | 59 |
| Burkholderia oklahomensis CCUG 51349 | 73 | 5.5 | 52 |
| Burkholderia cepacia ATCC 25416 | 35 | 10.5 | 20 |
The RBR is the amount of double-stranded DNA formed between labeled and unlabeled DNAs from different strains divided by the amount of double-stranded DNA formed between labeled and unlabeled DNA from the same strain.
Divergence (D) within related sequences was calculated on the assumption that each 1°C decrease in the thermal stability of a DNA duplex is caused by 1% of unpaired bases within that duplex. Divergence was calculated to the nearest 0.5%.
On the basis of our phenotypic and molecular data, including DNA-DNA hybridization, CDC3015869 is conclusively identified as B. thailandensis. Based on MLST, all three strains described above were closely related to B. thailandensis isolates from Southeast Asia, although they form a distinct subcluster.
Burkholderia pseudomallei is the causative agent of melioidosis, a human and animal disease endemic in Southeast Asia and northern Australia. It has been reported to occur in tropical latitudes between 20°N and 20°S, including Central and South America (4). Melioidosis can present with a variety of clinical manifestations, including acute pneumonia with septicemia. Septicemic disease can have high mortality even after aggressive antimicrobial therapy is initiated (4). Infection can result from cutaneous inoculation, inhalation, or ingestion; circumstances involving heavy inoculations, such as near drowning, may shorten onset of infection (4).
B. pseudomallei is a gram-negative rod and soil saprophyte. Environmental investigations to collect and characterize soilborne B. pseudomallei in Thailand resulted in isolation of a closely related yet distinct B. pseudomallei-like organism, now recognized as B. thailandensis (3). The most reliable biochemical test for differentiation of these two species is assimilation of arabinose by B. thailandensis (11). Numerous Burkholderia-specific PCR assays can also differentiate the two organisms (4). Genetically, both 16S rRNA gene sequencing and DNA-DNA hybridization studies have confirmed the designation of B. thailandensis as a unique species (3, 14). B. thailandensis is much less virulent in animal models than B. pseudomallei, with a mean 50% lethal dose of 109 CFU/mouse, versus 182 CFU/mouse for B. pseudomallei (11). With B. thailandensis generally considered avirulent, disease due to B. thailandensis is extremely rare. No B. thailandensis isolates were identified among 1,200 patients with melioidosis (11). There has been at least one reported case of melioidosis where B. thailandensis was cultured from the purulent material of an amputated knee sustained after a motorcycle accident in Thailand (5, 9).
Our data demonstrate that infection with B. thailandensis can occur in humans and corroborate two other similar reports from Thailand (5, 9). It is likely that the Texas case of pneumonia and septicemia resulted from aspiration of drainage ditch water at the accident site. Similar routes of infection, including vehicular accidents (9) and near drowning (1, 4), are described for melioidosis. We were unable to isolate B. thailandensis from soil or water samples collected 1 year after the accident at the Texas site; therefore, we could not establish evidence of environmental persistence in the United States. No information documenting exposure risks, such as military service or reported motor vehicle accident, is available for the LA case, so the source of infection remains unknown.
On the basis of our phenotypic and molecular data, including DNA-DNA hybridization, the isolates described here are definitively identified as B. thailandensis. Further studies are needed to identify the virulence factors associated with these and other pathogenic B. thailandensis isolates and to determine their molecular and evolutionary relationships to other B. thailandensis and B. pseudomallei strains.
Melioidosis was diagnosed when this case was presumed to be caused by B. pseudomallei. Emergency response preparedness procedures emphasize the importance of definitively identifying the causative organism of disease. In this case, arabinose assimilation proved to be a simple, straightforward, and useful method for differentiating B. pseudomallei and B. thailandensis. When B. pseudomallei is presumptively identified, the use of tests such as arabinose assimilation in clinical laboratories, as well as B. pseudomallei-specific PCR assays, 16S rRNA gene sequencing, and MLST in reference laboratories, can differentiate B. pseudomallei from close relatives such as B. thailandensis. Our findings emphasize the importance of obtaining definitive identification of the causative organism in order to monitor for emergence of novel pathogens and to rule out possible infection by a bioterror agent.
Nucleotide sequence accession numbers.
The 16S rRNA gene sequences of CDC3015869, CDC2721121, and 82172 were deposited in GenBank with accession no. DQ388535, DQ388536, and DQ388537, respectively.
Acknowledgments
We thank the Texas Department of Health for their assistance in providing the Texas isolate and environmental samples and Tanja Popovic for her critical review of the manuscript.
Footnotes
Published ahead of print on 18 October 2006.
REFERENCES
- 1.Achana, A., K. Silpapojakul, W. Thininta, and S. Kalnaowakul. 1985. Acute Pseudomonas pseudomallei pneumonia and septicemia following aspiration of contaminated water: a case report. Southeast Asian J. Trop. Med. Public Health 16:500-504. [PubMed] [Google Scholar]
- 2.Brenner, D. J., A. C. McWhorter, J. K. Knutson, and A. G. Steigerwalt. 1982. Escherichia vulneris: a new species of Enterobacteriaceae associated with human wounds. J. Clin. Microbiol. 15:1133-1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brett, P. J., D. DeShazer, and D. E. Woods. 1998. Burkholderia thailandensis sp. nov., a Burkholderia pseudomallei-like species. Int. J. Syst. Bacteriol. 48:317-320. [DOI] [PubMed] [Google Scholar]
- 4.Cheng, A. C., and B. J. Currie. 2005. Melioidosis: epidemiology, pathophysiology, and management. Clin. Microbiol. Rev. 18:383-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dharakul, T., B. Tassaneetrithep, S. Trakulsomboon, and S. Songsivilai. 1999. Phylogenetic analysis of Ara+ and Ara− Burkholderia pseudomallei isolates and development of a multiplex PCR procedure for rapid discrimination between the two biotypes. J. Clin. Microbiol. 37:1906-1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gee, J. E., C. T. Sacchi, M. B. Glass, B. K. De, R. S. Weyant, P. N. Levett, A. M. Whitney, A. R. Hoffmaster, and T. Popovic. 2003. Use of 16S rRNA gene sequencing for rapid identification and differentiation of Burkholderia pseudomallei and B. mallei. J. Clin. Microbiol. 41:4647-4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Glass, M. B., A. G. Steigerwalt, J. G. Jordon, P. P. Wilkins, and J. E. Gee. 2006. Burkholderia oklahomensis sp. nov., a Burkholderia pseudomallei-like species formerly known as the Oklahoma strain of Pseudomonas pseudomallei. Int. J. Syst. Evol. Microbiol. 56:2171-2176. [DOI] [PubMed]
- 8.Godoy, D., G. Randle, A. J. Simpson, D. M. Aanensen, T. L. Pitt, R. Kinoshita, and B. G. Spratt. 2003. Multilocus sequence typing and evolutionary relationships among the causative agents of melioidosis and glanders, Burkholderia pseudomallei and Burkholderia mallei. J. Clin. Microbiol. 41:2068-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lertpatanasuwan, N., K. Sermsri, A. Petkaseam, S. Trakulsomboon, V. Thamlikitkul, and Y. Suputtamongkol. 1999. Arabinose-positive Burkholderia pseudomallei infection in humans: case report. Clin. Infect. Dis. 28:927-928. [DOI] [PubMed] [Google Scholar]
- 10.Novak, R. T., M. B. Glass, J. E. Gee, D. Gal, M. J. Mayo, B. J. Currie, and P. P. Wilkins. 2006. Development and evaluation of a real-time PCR assay targeting the type III secretion system of Burkholderia pseudomallei. J. Clin. Microbiol. 44:85-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith, M. D., B. J. Angus, V. Wuthiekanun, and N. J. White. 1997. Arabinose assimilation defines a nonvirulent biotype of Burkholderia pseudomallei. Infect. Immun. 65:4319-4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weyant, R. S., C. W. Moss, R. E. Weaver, D. G. Hollis, J. G. Jordan, E. C. Cook, and M. I. Daneshvar. 1996. Identification of unusual pathogenic gram-negative aerobic and facultatively anaerobic bacteria, 2nd ed. Williams & Wilkins, Baltimore, Md.
- 13.Wuthiekanun, V., M. D. Smith, D. A. Dance, A. L. Walsh, T. L. Pitt, and N. J. White. 1996. Biochemical characteristics of clinical and environmental isolates of Burkholderia pseudomallei. J. Med. Microbiol. 45:408-412. [DOI] [PubMed] [Google Scholar]
- 14.Yabuuchi, E., Y. Kawamura, T. Ezaki, M. Ikedo, S. Dejsirilert, N. Fujiwara, T. Naka, and K. Kobayashi. 2000. Burkholderia uboniae sp. nov., l-arabinose-assimilating but different from Burkholderia thailandensis and Burkholderia vietnamiensis. Microbiol. Immunol. 44:307-317. [DOI] [PubMed] [Google Scholar]