Abstract
Laser capture microdissection in combination with fluorescent in situ hybridization was used to identify an unknown species of spirochetes from the pig colonic mucosa. The 16S rRNA gene was PCR amplified, and the closest related type strain was Treponema bryantiiT (90.1%). The spirochete, here named “Candidatus Treponema suis,” was associated with colitis, including invasion of the surface epithelium as well as superficial parts of the mucosa.
Helical-shaped bacteria are commonly present in the gastrointestinal tract of animals and humans. Intestinal spirochetes of pigs include several species of the genus Brachyspira, of which Brachyspira hyodysenteriae and Brachyspira pilosicoli are important pathogens causing swine dysentery, a severe mucohemorrhagic colitis, and spirochetal colitis, respectively (14, 15). Culturing of Brachyspira as well as Treponema species is fastidious and not always successful. Although ultrastructural differentiation is possible, the various spirochetes are quite similar (typically 0.2 to 0.4 μm in width and 4 to 12 μm long) (11), and the size is not a useful criterion for distinction in histopathological specimens. Alternative methods for species identification include fluorescent in situ hybridization (FISH) with specific oligonucleotide probes targeting 16S or 23S rRNA of the porcine pathogens B. hyodysenteriae and B. pilosicoli (2). Here, a novel technique, laser capture microdissection (LCM)-FISH (7), was used to identify an unknown spirochete and to establish its spatial distribution in the pig colonic tissue on archival tissue samples originating from a study by Fossi et al. (4). The bacterial cells were visualized by FISH with a 16S rRNA-targeting oligonucleotide probe, followed by laser capture microdissection (LCM) of the targeted microcolony. The PCR was done directly on the captured cell material, and the dissected bacterial cells were subsequently identified by analyses of the amplified 16S rRNA gene sequence.
Twenty-three formalin-fixed paraffin-embedded biopsy specimens of pig colon were obtained from the National Veterinary and Food Research Institute, Seinäjoki Unit, PB 60101, Seinäjoki, Finland. The samples originated from the previously mentioned study included two sets of eight piglets challenged with one of two B. pilosicoli strains (Br1622 and P43/6/8T, respectively) and eight uninfected control pigs (4). However, upon both histopathological and in situ hybridization examination of all 23 pigs, an unknown species of spirochetes rather than the challenged bacteria was found to have invaded the colonic mucosa of several of the pigs, including the control pigs (4). For laser capture microdissection (LCM), 3-μm-thick sections of the samples were mounted on 0.17-mm PALM POL-Membrane Slides (P.A.L.M. Microlaser Technologies AG, Bernried, Germany) and kept at 4°C until use. For in situ hybridization, sections were mounted on Superfrost Slides (Erie Scientific Company, Portsmouth, NH). The tissue sections were first hybridized with an oligonucleotide probe S-G-Leptospira-1414-a-A-18, originally designed to target the Leptospira interrogans group (4), and subsequently with S-S-Treponema-0833-a-A-18, targeting the novel “Candidatus Treponema suis” (this study). Probes were selected using the software ARB (http://www.arb-home.de). Both oligonucleotide probes were 5′ labeled with Alexa Fluor 488 (MWG-BIOTECH AG, Ebersberg, Germany). The hybridization temperature for both probes was 45°C for 16 h. Hybridization, laser capture microdissection, and PCR were done according to Klitgaard et al. (7). The primer sets (MWG-BIOTECH AG, Ebersberg, Germany) used for 16S rRNA gene amplification are listed in Table 1. The Treponema-specific primers were designed in ARB based on primer walking of the sequenced 16S rRNA gene. The amplicons were purified by using the QIAquick spin PCR purification kit (QIAGEN, Hilden, Germany) and were sequenced at Macrogen, Seoul, Korea (www.macrogen.com). The sequence was assembled in Bionumerics version 4.0 (Applied Math, Sint-Martens-Latem, Belgium) and checked for chimeras both by blasting the six individual sequences in GenBank (http://www.ncbi.nlm.nih.gov) and by the software Pintail version 1.1 (http://www.cardiff.ac.uk/biosi/research/biosoft/). The phylogenetic analysis was done by downloading 16S rRNA gene sequences longer than 1,200 bp from the RDP database of the Treponema type strains plus the most similar clones of the new spirochete sequence (http://rdp.cme.msu.edu). Before alignment, the sequences were trimmed at the ends so the sequences started at Escherichia coli position 47 and ended at position 1418. The sequences were first aligned pairwise (WARD) followed by a global sequence alignment. A phylogenetic tree was constructed by using the maximum parsimony method with 500 bootstrap simulations using the software Bionumerics. B. hyodysenteriae (GenBank accession no. M57743) was used as an outgroup.
TABLE 1.
Names and sequences of 16S rRNA oligonucleotide probes used for in situ hybridization and 16S rRNA gene primers used for PCR and sequencing
| Primer and probea | Sequenceb (5′-3′) | Reference or source |
|---|---|---|
| Oligonucleotide probes | ||
| S-S-Treponema-0833-a-A-18 | CCCAGTCCTCATGACCAG | This study |
| S-G-Leptospira-1414-a-A-18 | CGGGTGCTCCCCACTCAG | 4 |
| Primer sets | ||
| S-D-Bact-0010-a-S-20 | AGAGTTTGATCCTGGCTNAG | Modified from reference 17 |
| S-P-Group-0246-a-S-18 | AGCTAGTTGGYGRGGTAAC | This study |
| S-P-Group-0246-a-A-18 | GTTACCYCRCCAACTAGCT | This study |
| S-*-Univ-0519-a-S-19 | CCAGCAGCCGCGGTAATAC | Modified from reference 17 |
| S-*-Trep-0391-a-S-18 | GAGCGACGCCGCGTGGAT | This study |
| S-D-Bact-0804-a-A-21 | GACTACCNGGGTATCTAATCC | Modified from reference 17 |
| S-*-Trep-0683-a-S-18 | TGTAGGGGTGAAATCTGT | This study |
| S-D-Bact-1054-a-A-20 | ACGAGCTGACGACRRCCATG | Modified from reference 17 |
| S-*-Trep-0974-a-S-19 | GGAACCTTACCTGGGTTTG | This study |
| S-D-Bact-1392-a-A-19 | TGACGGGCGGTGTGTACAA | Modified from reference 17 |
| S-*-Trep-1271-a-S-18 | TGGAGCAAAACGCAGAAA | This study |
| S-*-Univ-1492-a-A-21 | GTTACCTTGTTACGACTTCAC | Modified from reference 17 |
Names are according to Alm et al. (1).
N signifies A, T, G, or C; R signifies A or G; Y signifies C or T.
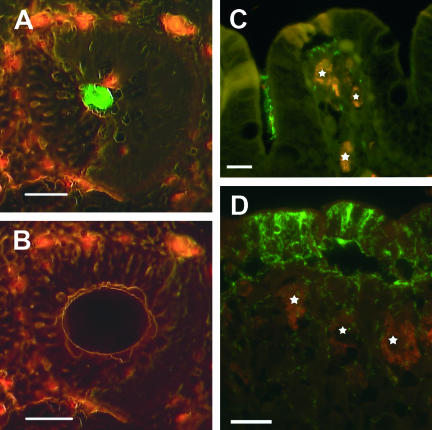
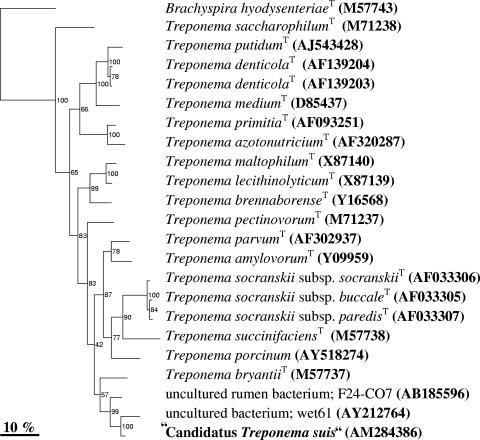
The bacterial microcolonies were sampled by LCM (Fig. 1A and B). Due to the formalin fixation and storage of the tissue samples, the bacterial DNA had been fragmented. Consequently, the fragmented 16S rRNA gene was PCR amplified by six primer sets. The resulting PCR products were sequenced and assembled in a 1,462-bp sequence, from now on named “Candidatus Treponema suis” (“Ca. Treponema suis”) (GenBank accession number AM284386). A phylogenetic analysis clustered “Ca. Treponema suis” in the Treponema genus group, hence the name (Fig. 2). The closest matches of the 16S rRNA gene sequence in the phylogenetic alignment were two clones, wet61 (94.4% pair wise homology) and F24-CO7 (95.3% pairwise homology), originating from horse feces and a cow rumen, respectively (9, 12). The three sequences clustered together with a bootstrap value of 99%. The closest related type strain was Treponema bryantiiT (90.1% pairwise homology), but the polygenetic affiliation of the strain with “Ca. Treponema suis” was not as obvious, given the relative low bootstrap values of 57%. Because of the relative low similarity to the closest isolate, it is very likely that “Ca. Treponema suis” is a new phylogenetic species, even though the “phylophasic” approach concept of species for prokaryotes, currently in use by taxonomists, is based on a combined genomic, phenotypic, and phylogenetic characterization of bacterial isolates (16). Empirically, it is unlikely that bacteria sharing less than 97% 16S rRNA gene sequence similarity will be identified as the same species in the polyphasic species concept (10, 13). Consequently, in this study, a potential new invasive species of Treponema (“Candidatus Treponema suis”) was identified.
FIG. 1.
In situ visualization of the novel “Ca. Treponema suis” colonizing colonic tissue samples of pigs. The bacteria were visualized with green fluorescent 16S rRNA oligonucleotide probes, and the intestinal cell material exhibited red autofluorescence. (A and B) Cross-section of a colonic crypt before and after microdissection, respectively. The bacterial cells were visualized with a probe targeting spirochete species (S-G-Leptospira-1414-a-A-18). Scale bar, 50 μm. (C) Fluorescent in situ hybridization with a probe specific for “Ca. Treponema suis” (S-S-Treponema-0833-a-A-18) demonstrating a cluster of “Ca. Treponema suis” in a crypt and single spirochetes within lamina propria surrounded by large macrophages (stars). Bar, 15 μm. (D) Porcine colonic mucosa with severe focal spirochetal infiltration by “Ca. Treponema suis” (S-S-Treponema-0833-a-A-18). High numbers of the spirochete are demonstrated within the surface epithelium and adjacent lamina propria, while colonization of the surface is absent. Large macrophages are indicated by stars. Bar, 20 μm.
FIG. 2.
Neighbor-joining tree of 16S rRNA gene sequence similarity, showing the phylogenetic position of “Ca. Treponema suis” and representatives of closely related clones and Treponema type strains. Bootstrap percentages are indicated at branches and are calculated from 500 trees. Brachyspira hyodysenteriae (M57743) was used as an outgroup. Sequence accession numbers are shown in parentheses.
From the 16S rRNA gene sequence of “Ca. Treponema suis,” a new specific hybridization probe was designed (S-G-L-1414-a-A-18). Applying the species-specific probe on the colonic tissue samples, the results were in accordance with the ones obtained with the S-G-Leptospira-1414-a-A-18 probe, where the spirochete was demonstrated in the colonic contents in small clusters close to the epithelium and as single organisms within lamina propria (Fig. 1C). In total, the new species-specific “Ca. Treponema suis” probe revealed spirochetes in 60% of the B. pilosicoli experimentally infected pigs and in 43% of the control pigs from the previously mentioned infection study (4). The fact that B. pilosicoli wasn't observed in any of the pigs, challenged as well as unchallenged, makes an influence of the B. pilosicoli strain with respect to the “Ca. Treponema suis”-associated colitis unlikely. Still, the importance of this bacterium as an emergent pathogen is at present not known. In five of the “Ca. Treponema suis”-positive pigs the spirochete was revealed to infiltrate massively the surface epithelium and underlying superficial parts of the mucosa in multiple foci, while the adjacent crypts appeared normal and were only colonized sporadically (Fig. 1D).
Compared to in situ hybridization studies of B. hyodysenteriae and B. pilosicoli infections, in which spirochetal colonization of the epithelium is associated with severe to moderate loss of enterocytes (5, 6), the “Ca. Treponema suis”-infiltrated epithelium was characterized by only a small amount of necrotic cells and only a sparse surface colonization. The inflammatory response in the subepithelial part of the mucosa infiltrated by spirochetes was characterized by large macrophages, while the cellular infiltration in swine dysentery and spirochetal colitis predominantly consists of monocytes (5, 6).
By transmission electron microscopy, cells of “Ca. Treponema suis” were found to be longer (6 to 11 μm) than those of other Treponema species (4 to 8 μm) previously found in pigs (3, 4, 8). Another difference was the number of periplasmatic flagella; T. succinifaciens, T. berlinense, and T. porcinum have two inserted subterminally, whereas the intestinal cross-sections of “Ca. Treponema suis” revealed 10 to 14 flagella (4). Thus, in size and number of flagella, “Ca. Treponema suis” more resembles the porcine species of the genus Brachyspira (11).
In conclusion, the LCM-FISH method was successfully used to access the genomic information and thus the phylogenetic affiliation of an unknown, invasive spirochete (“Ca. Treponema suis”) by combining histological recognition of the bacterium with molecular analysis of 16S rRNA genes. The technique turned out to be very useful, as tissue samples from an old experiment could be used in a retrospective way to identify a potentially unknown pathogen.
Nucleotide sequence accession number.
The sequence for “Candidatus Treponema suis” has been deposited in GenBank under accession number AM284386.
Acknowledgments
We thank Katja Kristensen, Annie Ravn Pedersen, and Ulla Andreasen for excellent technical assistance.
The work was supported by a grant from the Danish Agricultural and Veterinary Research Council (project no. 23-02-0137) to Kirstine Klitgaard and a grant from The Ministry of Food, Agriculture, and Fisheries (project no. 3401-65-03-745) to Lars Mølbak.
Footnotes
Published ahead of print on 27 September 2006.
REFERENCES
- 1.Alm, E. W., D. B. Oerther, N. Larsen, D. A. Stahl, and L. Raskin. 1996. The oligonucleotide probe database. Appl. Environ. Microbiol. 62:3557-3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boye, M., T. K. Jensen, K. Moller, T. D. Leser, and S. E. Jorsal. 1998. Specific detection of the genus Serpulina, S. hyodysenteriae and S. pilosicoli in porcine intestines by fluorescent rRNA in situ hybridization. Mol. Cell Probes 12:323-330. [DOI] [PubMed] [Google Scholar]
- 3.Cwyk, W. M., and E. Canale-Parola. 1979. Treponema succinifaciens sp. nov., an anaerobic spirochete from the swine intestine. Arch. Microbiol. 122:231-239. [DOI] [PubMed] [Google Scholar]
- 4.Fossi, M., K. Ahlsten, T. Pohjanvirta, M. Anttila, T. Kokkonen, T. K. Jensen, M. Boye, A. Sukura, K. Pelkola, and S. Pelkonen. 2005. Neither hippurate-negative Brachyspira pilosicoli nor Brachyspira pilosicoli type strain caused diarrhoea in early-weaned pigs by experimental infection. Acta Vet. Scand. 46:257-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jensen, T. K., M. Boye, K. Moller, T. D. Leser, and S. E. Jorsal. 1998. Association of Serpulina hyodysenteriae with the colonic mucosa in experimental swine dysentery studied by fluorescent in situ hybridization. APMIS 106:1061-1068. [PubMed] [Google Scholar]
- 6.Jensen, T. K., K. Moller, M. Boye, T. D. Leser, and S. E. Jorsal. 2000. Scanning electron microscopy and fluorescent in situ hybridization of experimental Brachyspira (Serpulina) pilosicoli infection in growing pigs. Vet. Pathol. 37:22-32. [DOI] [PubMed] [Google Scholar]
- 7.Klitgaard, K., L. Mølbak, T. K. Jensen, C. F. Lindboe, and M. Boye. 2005. Laser capture microdissection of bacterial cells targeted by fluorescence in situ hybridization. BioTechniques 39:864-868. [DOI] [PubMed] [Google Scholar]
- 8.Nordhoff, M., D. Taras, M. Macha, K. Tedin, H. J. Busse, and L. H. Wieler. 2005. Treponema berlinense sp. nov. and Treponema porcinum sp. nov., novel spirochaetes isolated from porcine faeces. Int. J. Syst. Evol. Microbiol. 55:1675-1680. [DOI] [PubMed] [Google Scholar]
- 9.Ozutsumi, Y., K. Tajima, A. Takenaka, and H. Itabashi. 2005. The effect of protozoa on the composition of rumen bacteria in cattle using 16S rRNA gene clone libraries. Biosci. Biotechnol. Biochem. 69:499-506. [DOI] [PubMed] [Google Scholar]
- 10.Rossello-Mora, R., and R. Amann. 2001. The species concept for prokaryotes. FEMS Microbiol. Rev. 25:39-67. [DOI] [PubMed] [Google Scholar]
- 11.Sellwood, R., and A. P. Bland. 1997. Ultrastructure of intestinal spirochaetes, p. 109-150. In D. J. Hampson and T. B. Stanton (ed.), Intestinal spirochaetes in domestic animals and humans. Cab International, Wallingford, United Kingdom.
- 12.Simpson, J. M., J. W. Santo Domingo, and D. J. Reasoner. 2003. Assessment of equine fecal contamination: the search for alternative bacterial source-tracking targets. FEMS Microbiol. Ecol. 47:65-75. [DOI] [PubMed] [Google Scholar]
- 13.Stackebrandt, E., and B. M. Goebel. 1994. A place for DNA-DNA reassociation and 16S ribosomal-RNA sequence-analysis in the present species definition in bacteriology. Int. J. Syst. Evol. Microbiol. 44:846-849. [Google Scholar]
- 14.Taylor, D. J., and T. J. Alexander. 1971. The production of dysentery in swine by feeding cultures containing a spirochaete. Br. Vet. J. 127:58-61. [DOI] [PubMed] [Google Scholar]
- 15.Trott, D. J., T. B. Stanton, N. S. Jensen, G. E. Duhamel, J. L. Johnson, and D. J. Hampson. 1996. Serpulina pilosicoli sp. nov., the agent of porcine intestinal spirochetosis. Int. J. Syst. Evol. Microbiol. 46:206-215. [DOI] [PubMed] [Google Scholar]
- 16.Wayne, L. G., D. J. Brenner, R. R. Colwell, P. A. D. Grimont, O. Kandler, M. I. Krichevsky, L. H. Moore, W. E. C. Moore, R. G. E. Murray, E. Stackebrandt, M. P. Starr, and H. G. Truper. 1987. Report of the Ad-Hoc-Committee on Reconciliation of Approaches to Bacterial Systematics. Int. J. Syst. Evol. Microbiol. 37:463-464. [Google Scholar]
- 17.Weisburg, W. G., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]