Streptococcus pneumoniae is a frequent cause of potentially life-threatening infections, such as pneumonia, meningitis, and bacteremia (14). However, the global emergence of antimicrobial resistance in S. pneumoniae is a serious concern (4). Recent data from 12 Asian countries showed high resistance rates (17, 18). We studied prospective resistance to a large number of antimicrobial agents in pneumococcal isolates. We also analyzed macrolide resistance, with an emphasis on resistance genes, molecular epidemiology, and serotype patterns.
From 1999 to 2002, S. pneumoniae isolates (n = 136) from blood and cerebrospinal fluid (CSF) (n = 60) and nasopharynx (n = 76) of children (<5 years) with pneumonia and meningitis from three hospitals in Dhaka, Bangladesh, were studied. MICs were determined by CLSI broth microdilution method (1) and by Etest (AB Biodisk, Solna, Sweden). Macrolide resistance phenotypes were determined by triple-disk test (11, 12) and macrolide resistance genotypes by a light cycler protocol (15). Isolates were serotyped by antisera (Statens Seruminstitut, Copenhagen, Denmark). Multilocus sequence typing (MLST) was carried out (2), and two predominant sequence types (ST) were analyzed by use of the eBURST2 program (3).
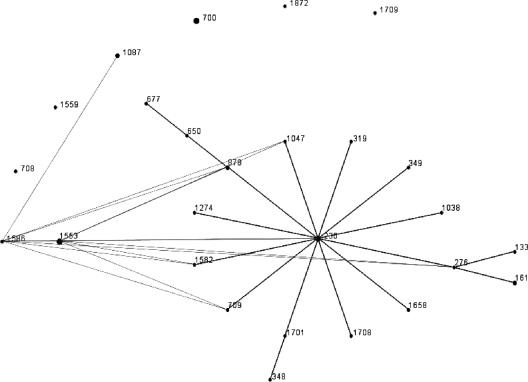
MIC results for S. pneumoniae (Table 1) showed high rates of resistance to sulfamethoxazole-trimethoprim (77.9%) and tetracycline (46.3%). The resistance rates of other drugs were low. The rates of multiply-resistant isolates were 27.9% and 11.7% against 2 and ≥3 classes of antibiotics, respectively. Six (4.4%) isolates (Table 2) were resistant to erythromycin A; five of them exhibited the partially inducible iMcLSB phenotype (susceptible or intermediate to rokitamycin but developing no induction resistance to rokitamycin in the presence of erythromycin) and one strain the M phenotype (resistant to erythromycin, azithromycin, and clarithromycin but susceptible to clindamycin and streptogramin B). Isolates with the iMcLSB phenotype were positive for the erm(B) gene, and the isolate displaying the M phenotype was positive for mef(A). Macrolide-resistant isolates were serotypes 7B (n = 4), 9V (n = 1), and 18C (n = 1). MLST results of serotype 7B macrolide-resistant strains established two sequence types: ST 1553 (strains 14, 39, and 94) and ST 1586 (strain 61). ST 1586 is a single-locus variant (SLV) of ST 1553, and both appear to belong to a single clonal complex. In addition, one isolate was a serotype 9V variant of ST 1553 (strain 68), indicating serotype switching. The mef(A)-positive strain was not closely genetically related to this clonal complex (strain 28, ST 113). eBURST analysis (Fig. 1) showed ST 1553 and ST 1586 to form a pair of SLVs. No other SLVs were found in the MLST database (www.mlst.net). An analysis for double-locus variants determined these two ST to be members of a group of 26 ST represented by 37 isolates with ST 230 as the predicted group founder. ST 230 is represented in the MLST database by two erythromycin-sensitive serotype 14 isolates from Denmark and Italy and an erythromycin-resistant serotype 24F isolate, also from Italy. Six ST in the group are double-locus variants of either ST 1586 or ST 1553 or both.
TABLE 1.
MIC ranges, MIC50s, and MIC90s of 136 S. pneumoniae strains isolated in Bangladesh
| Antibiotica | MIC (μg/ml)
|
% Resistant | ||
|---|---|---|---|---|
| Range | MIC50 | MIC90 | ||
| Penicillin G | 0.016-0.5 | 0.016 | 0.03 | 8.8 |
| Amoxicillin | 0.016-0.5 | 0.016 | 0.06 | 0 |
| Cefotaxime | 0.016-0.25 | 0.016 | 0.06 | 0 |
| Gatifloxacin | 0.016-1 | 0.25 | 0.25 | 0 |
| Ciprofloxacin | 0.06->32 | 1 | 1 | 2.9 |
| Teicoplanin | 0.016-0.125 | 0.06 | 0.06 | 0 |
| Vancomycin | 0.03-0.25 | 0.25 | 0.25 | 0 |
| Telithromycin | 0.016-0.06 | 0.016 | 0.016 | 0 |
| Tetracycline | 0.125->32 | 0.5 | >32 | 46.3 |
| Clindamycin | 0.06->32 | 0.06 | 0.06 | 3.7 |
| Erythromycin A | 0.06->32 | 0.06 | 0.06 | 4.4 |
| SXT* | 0.03->32 | 4 | 6 | 77.9 |
| Chloramphenicol* | 1-6 | 2 | 2 | 8.1 |
SXT, sulfamethoxazole-trimethoprim. *, MICs were determined by Etest (see text). For ciprofloxacin, a breakpoint of >4 μg/ml was used.
TABLE 2.
Characteristics of macrolide-resistant S. pneumoniae strains isolated from children in Bangladesha
| Strain | Source | MIC (μg/ml)
|
Macrolide resistance
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ERY | CLI | PEN | SXT | TET | CHL | Genotype | Phenotype | Serotype | STb | ||
| 14 | CSF | ≥256 | >32 | 0.25 | 16 | 16 | 2 | erm(B) | iMcLSB | 7B | 1553 |
| 39 | Blood | ≥256 | >32 | 0.5 | 16 | >32 | 2 | erm(B) | iMcLSB | 7B | 1553 |
| 61 | NP | ≥256 | >32 | 0.5 | 16 | >32 | 2 | erm(B) | iMcLSB | 7B | 1586 |
| 68 | NP | ≥256 | >32 | 0.25 | >32 | >32 | 2 | erm(B) | iMcLSB | 9V | 1553 |
| 94 | NP | ≥256 | 16 | 0.5 | 8 | 1 | 2 | erm(B) | iMcLSB | 7B | 1553 |
| 28 | CSF | 1 | 0.06 | 0.03 | 0.5 | 8 | 2 | mef(A) | M | 18C | 113 |
Abbreviations: NP, nasopharynx; ERY, erythromycin A; CLI, clindamycin; PEN, penicillin G; SXT, sulfamethoxazole-trimethoprim; TET, tetracycline; CHL, chloramphenicol.
ST 1553 showed alleles 6, 5, 2, 17, 6, 22, and 14; ST 1586 showed alleles 43, 5, 2, 17, 6, 22, and 14; and ST 133 showed alleles 7, 2, 1, 1, 10, 1, and 21.
FIG. 1.
eBURST analysis of ST of two multiply-resistant serotype 7B strains in this study (ST 1586 and ST 1553) and 2,681 different ST of S. pneumoniae available in the MLST database (www.mlst.net). Single-locus variants are connected by black lines. Double-locus variants are not connected by lines, with the exception of the double-locus variants of ST 1553 and ST 1586, which are connected by gray lines.
Our study highlights a high level of tetracycline and sulfamethoxazole-trimethoprim resistance in Bangladesh. There is increasing concern over resistance in pneumococci to sulfamethoxazole-trimethoprim, which is recommended by the WHO as a first-line drug for treating nonsevere pneumonia. Our study supports the view that this recommendation may not be optimal for Bangladesh; however, changing to alternative agents, such as amoxicillin, is costly (http://www.who.int/child-adolescent-health/publications/referral_care/chap3/chap31.htm). Moreover, the widespread use of sulfamethoxazole-trimethoprim may further drive the spread of multiply-resistant pneumococcal clones and may also select resistance to penicillin G, chloramphenicol, and macrolides. Our observation of a high level of resistance to sulfamethoxazole-trimethoprim in Bangladesh is consistent with findings from the mid-1990s (16). In contrast, the rates of resistance to penicillin G, macrolides, and other drugs are relatively low compared to those described in recent reports from other Asian countries, except India (8, 17, 18). Multiply-resistant S. pneumoniae is a problem in Bangladesh. Although an increasing trend of fluoroquinolone resistance has been reported in Hong Kong, Spain, and Canada (7, 9), a low rate (2.9%) of ciprofloxacin resistance was observed in our study. However, gatifloxacin and moxifloxacin remained active in vitro, indicating their potential utility in Bangladesh.
The predominance of the MLSB phenotype (resistance to all macrolides, lincosamides, and streptogramin B)/erm(B) genotype in our study is consistent with recent findings from Sri Lanka, Korea, China, Taiwan, Japan, Spain, Italy, France, and South Africa (6, 9, 15, 17). All strains with the MLSB phenotype exhibited a partially macrolide-inducible iMcLSB phenotype, i.e., they were susceptible or had intermediate resistance to rokitamycin and had no resistance induction to rokitamycin in the presence of erythromycin (13). This phenotype is the most common mechanism of macrolide resistance in Bangladesh, although the sample size is small. Of note, the majority of macrolide-resistant strains belong to a single serotype, 7B. Serotype 7B infections were recorded only in India in the late 1990s (5). Three 7B isolates and the 9V variant have the same sequence type, ST 1553, and form a clonal complex. In addition, one 7B isolate is a single-locus (aroE) variant (ST 1586) of ST 1553. Of note, ST 1553 and ST 1586 are not closely related to any other clone in the MLST database, as shown by eBURST analysis. The strain exhibiting a mef genotype belongs to ST 113. Strains of this clone (global clone 36 of the Pneumococcal Molecular Epidemiology Network [http://www.mlst.net; http://www.sph.emory.edu/PMEN]) (10) are serotype 18C and were isolated from meningitis in The Netherlands, the United Kingdom, and Spain in the 1980s and 1990s. Interestingly, the present report is the first to document macrolide resistance in an isolate of this particular clone.
In summary, our report shows that resistance to beta-lactams, macrolides, and fluoroquinolones in pneumococci is not as yet a serious problem in Bangladesh, unlike in many other Asian countries. However, the emergence of a unique multiply-resistant serotype 7B clone reiterates the need for continual surveillance of antimicrobial resistance in pneumococci.
Acknowledgments
This research protocol was funded by USAID (Washington, D.C.) grant number 00097 and by a visiting scientist grant from the German National Reference Center for Streptococci. ICDDR,B acknowledges with gratitude the commitment of USAID and the German National Reference Center for Streptococci to the Center's research efforts.
Footnotes
Published ahead of print on 27 September 2006.
REFERENCES
- 1.Clinical and Laboratory Standards Institute. 2005. Performance standards for antimicrobial susceptibility testing; fifteenth informational supplement. Clinical and Laboratory Standards Institute, Wayne, Pa.
- 2.Enright, M. C., and B. G. Spratt. 1998. A multilocus sequence typing scheme for Streptococcus pneumoniae identification of clones associated with serious invasive disease. Microbiology 144:3049-3060. [DOI] [PubMed] [Google Scholar]
- 3.Feil, E. J., B. C. Li, D. M. Aanensen, W. P. Hanage, and B. G. Spratt. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 186:1518-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Felmingham, D., D. J. Farrell, R. R. Reinert, and I. Morrissey. 2004. Antibacterial resistance among children with community-acquired respiratory tract infections (PROTEKT 1999-2000). J. Infect. 48:39-55. [DOI] [PubMed] [Google Scholar]
- 5.Lalitha, M. K., K. Thomas, A. Manoharan, J. H. Song, and M. C. Steinhoff. 1999. Changing trend in susceptibility pattern of Streptococcus pneumoniae to penicillin in India. Indian J. Med. Res. 110:164-168. [PubMed] [Google Scholar]
- 6.Leclercq, R. 2002. Mechanisms of resistance to macrolides and lincosamides: nature of the resistance elements and their clinical implications. Clin. Infect. Dis. 34:482-492. [DOI] [PubMed] [Google Scholar]
- 7.Low, D. E. 2004. Quinolone resistance among pneumococci: therapeutic and diagnostic implications. Clin. Infect. Dis. 38(Suppl. 4):S357-S362. [DOI] [PubMed] [Google Scholar]
- 8.McGee, L., L. McDougal, J. Zhou, B. G. Spratt, F. C. Tenover, R. George, R. Hakenbeck, W. Hryniewicz, J. C. Lefévre, A. Tomasz, and K. P. Klugman. 2001. Nomenclature of major antimicrobial-resistant clones of Streptococcus pneumoniae defined by the Pneumococcal Molecular Epidemiology Network. J. Clin. Microbiol. 39:2565-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Montanari, M. P., I. Cochetti, M. Mingoia, and P. E. Varaldo. 2003. Phenotypic and molecular characterization of tetracycline- and erythromycin-resistant strains of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 47:2236-2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Montanari, M. P., M. Mingoia, I. Cochetti, and P. E. Varaldo. 2003. Phenotypes and genotypes of erythromycin-resistant pneumococci in Italy. J. Clin. Microbiol. 41:428-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Montanari, M. P., M. Mingoia, E. Giovanetti, and P. E. Varaldo. 2001. Differentiation of resistance phenotypes among erythromycin-resistant pneumococci. J. Clin. Microbiol. 39:1311-1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Musher, D. M. 1992. Infections caused by Streptococcus pneumoniae: clinical spectrum, pathogenesis, immunity, and treatment. Clin. Infect. Dis. 14:801-807. [DOI] [PubMed] [Google Scholar]
- 13.Reinert, R. R., C. Franken, M. van der Linden, R. Lütticken, M. Cil, and A. Al-Lahham. 2004. Molecular characterisation of macrolide resistance mechanisms of Streptococcus pneumoniae and Streptococcus pyogenes isolated in Germany, 2002-2003. Int. J. Antimicrob. Agents 24:43-47. [DOI] [PubMed] [Google Scholar]
- 14.Reinert, R. R., S. Reinert, M. van der Linden, M. Y. Cil, A. Al-Lahham, and P. Appelbaum. 2005. Antimicrobial susceptibility of Streptococcus pneumoniae in eight European countries from 2001 to 2003. Antimicrob. Agents Chemother. 49:2903-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reinert, R. R., A. Ringelstein, M. van der Linden, M. Y. Cil, A. Al-Lahham, and F. J. Schmitz. 2005. Molecular epidemiology of macrolide-resistant Streptococcus pneumoniae isolates in Europe. J. Clin. Microbiol. 43:1294-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saha, S. K., N. Rikitomi, D. Biswas, K. Watanabe, M. Ruhulamin, K. Ahmed, M. Hanif, K. Matsumoto, R. B. Sack, and T. Nagatake. 1997. Serotypes of Streptococcus pneumoniae causing invasive childhood infections in Bangladesh, 1992 to 1995. J. Clin. Microbiol. 35:785-787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Song, J. H., H. H. Chang, J. Y. Suh, K. S. Ko, S. I. Jung, W. S. Oh, K. R. Peck, N. Y. Lee, Y. Yang, A. Chongthaleong, N. Aswapokee, C. H. Chiu, M. K. Lalitha, J. Perera, T. T. Yee, G. Kumararasinghe, F. Jamal, A. Kamarulazaman, N. Parasakthi, P. H. Van, T. So, and T. K. Ng. 2004. Macrolide resistance and genotypic characterization of Streptococcus pneumoniae in Asian countries: a study of the Asian Network for Surveillance of Resistant Pathogens (ANSORP). J. Antimicrob. Chemother. 53:457-463. [DOI] [PubMed] [Google Scholar]
- 18.Song, J.-H., S.-I. Jung, K. S. Ko, N. Y. Kim, J. S. Son, H.-H. Chang, H. K. Ki, W. S. Oh, J. Y. Suh, K. R. Peck, N. Y. Lee, Y. Yang, Q. Lu, A. Chongthaleong, C.-H. Chiu, M. K. Lalitha, J. Perera, T. T. Yee, G. Kumarasinghe, F. Jamal, A. Kamarulzaman, N. Parasakthi, P. H. Van, C. Carlos, T. So, T. K. Ng, and A. Shibl. 2004. High prevalence of antimicrobial resistance among clinical Streptococcus pneumoniae isolates in Asia (an ANSORP study). Antimicrob. Agents Chemother. 48:2101-2107. [DOI] [PMC free article] [PubMed] [Google Scholar]