Abstract
Bacteremia due to Helicobacter canis has been reported in a patient with X-linked hypogammaglobulinemia. Here we report on the first human case of H. canis bacteremia in an immunocompetent host. Identification of the organism was made by genetic and phylogenetic analyses of the complete 16S rRNA sequence.
CASE REPORT
A 44-year-old, previously healthy man presented in August 2005 with fever and a red plaque on the shin after returning from a trip to Ardèche, France. There was no history of insect bite or local trauma. His symptoms resolved with a 10-day course of oral amoxicillin-clavulanate but recurred 2 days later, with additional red plaques appearing elsewhere. The patient owned two healthy cats. In France, he had petted a domestic dog on several occasions but had had no facial contact with the dog. The patient reported no homosexual contact, arthritis, asthma, or allergies. He had no recurrent infections and no complications after receiving live vaccines (Mycobacterium bovis BCG in 1962, oral polio vaccine in 1985, yellow fever vaccine in 1991). The family history was noncontributory. His temperature was 38°C; and there were multiple erythematous plaques and small purpuric lesions on his legs, trunk, and arms. His peripheral white blood cell count was 14,000/mm3, with a normal differential count. Serum creatinine, liver, and coagulation tests were normal; and serologic tests for cytomegalovirus, Epstein-Barr virus, Borrelia burgdorferi, Brucella, Yersinia, syphilis, human immunodeficiency virus, hepatitis C virus, rheumatoid factor, and antinuclear antibodies were negative. Quantitative immunoglobulin levels, the numbers of circulating T and B lymphocytes, and antibody responses to primary vaccination with pneumococcal polysaccharide and a tetanus diphtheria toxoid booster were normal. Histological examination of a skin biopsy specimen showed cellulitis. A blood culture grew Helicobacter canis (see below). Gram stain, bacterial culture, PCR for B. burgdorferi, and broad-range eubacterial PCR of the skin biopsy specimen were negative. Treatment with intravenous ceftriaxone (2 g daily for 2 weeks) was followed by the complete resolution of the fever and skin lesions. Cultures of stool samples from the patient's cats did not yield Helicobacter spp. In April 2006, the patient was well, without any other infectious illness. In August 2006, he presented with a “flu-like” illness of 2 weeks' duration. Blood cultures remained sterile, and infectious mononucleosis due to Epstein-Barr virus was diagnosed by serology.
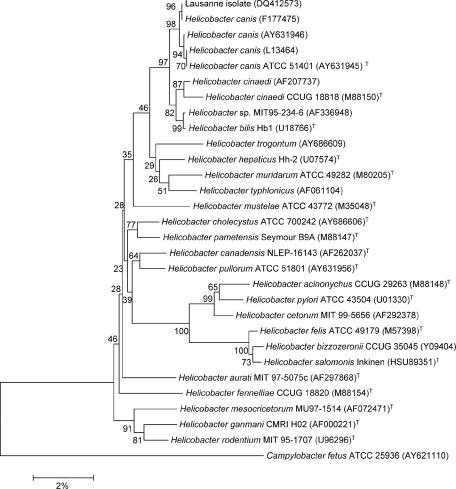
After 3 days of incubation in the BACTEC 9240 blood culture system (Becton Dickinson, Sparks, MD), one of two aerobic blood culture sets showed spiral-shaped bacteria on acridine orange staining. A single morphotype of small grayish colonies was obtained after subculture on both brucella agar supplemented with hemin, vitamin K1, cysteine, and 5% sheep blood under anaerobic conditions and blood agar under microaerophilic (5% O2) conditions at 37°C. The isolate was oxidase positive but was negative for catalase and urease. Identification was by 16S rRNA sequence analysis, based on the blood agar isolate. DNA was extracted with a MagNAPure LC DNA isolation kit (Roche, Mannheim, Germany). PCR amplification of the entire 16S rRNA gene was performed with primers fD1 and rP2 (15) and Taq polymerase (Applied Biosystems, Rotkreuz, Switzerland). PCR products were purified with a QIAquick kit (QIAGEN, Basel, Switzerland) and sequenced with a BigDye Terminator cycle sequencing kit (Applied Biosystems) with one of six different primers (1). The sequences were aligned and combined into a single 16S rRNA sequence by using Contig Express (Vector NTI; Informax, Frederick, MD). The sequence was compared with all 16S rRNA sequences available in GenBank by using the BLASTN 2.2.2 program (www.ncbi.nlm.nih.gov). The sequence of the Lausanne strain (GenBank accession number DQ412573) exhibited 100% identity with that of a feline H. canis strain (GenBank accession number AF177475) (6) and 99.8% similarity with that of type strain H. canis NCTC 12739. The next closest species were H. bilis ATCC 56130T and H. cinaedi CCUG 18818 (99.0% and 98.3% sequence similarities, respectively), whereas its sequence was <98% similar to those of all other Helicobacter species. The affiliation of the isolate to the H. canis species was confirmed by phylogenetic analysis with Mega 3.0 software (10). The most similar sequences determined by the BLASTN analysis and the sequences of the Helicobacter type strains were included. Sequences were edited by removal of the longer 5′ and 3′ ends so that their lengths matched that of the shortest sequence. The sequence with GenBank accession number DQ412573 clustered with the H. canis sequences. In phylogenetic analyses performed by using Kimura's corrected p distance and the complete deletion option of the Mega software, bootstrap values of 97%, 93%, and 80% by neighbor-joining, minimal evolution, and parsimony analyses, respectively, supported the node separating this group from their closest neighbor, the H. bilis and H. cinaedi cluster (Fig. 1).
FIG. 1.
Phylogenetic tree showing the close relationship of the Lausanne strain (GenBank accession number DQ412573) to Helicobacter canis. The tree was inferred from 1,646 bp of the 16S rRNA sequence data. Campylobacter fetus was used as the outgroup. Bootstrap analysis was done with 500 resamplings; the percentage is shown at each node. The scale bar represents a 2% nucleotide difference. GenBank accession numbers are shown in parentheses.
Antimicrobial susceptibility testing was performed by the E-test method (AB Biodisk, Solna, Sweden). MICs were as follows: amoxicillin, 0.38 μg/dl; amoxicillin-clavulanate, 0.094 μg/dl; ceftriaxone 0.75 μg/dl; piperacillin-tazobactam, 1 μg/dl; imipenem, 0.047 μg/dl; metronidazole, 0.064 μg/dl; and clindamycin, 0.094 μg/dl. No beta-lactamase production was detected by the nitrocefin disk method (Cefinase; Becton Dickinson, Cockeysville, MD).
To our knowledge, this is the second human case of H. canis bacteremia reported and the first case to be reported in an immunocompetent host. Identification of the isolate was confirmed by the congruent and robust results of genetic and phylogenetic analyses of the entire 16S rRNA sequence. The patient's illness responded only temporarily to oral amoxicillin-clavulanate, perhaps due to suboptimal compliance or serum drug levels insufficient for the treatment of bacteremia; but the illness completely resolved with ceftriaxone. Similar to other non-H. pylori Helicobacter species (5), the isolate exhibited susceptibility in vitro to both of these antibiotics; but the best method for susceptibility testing of these organisms remains unclear, and no interpretation guidelines are available. The bacteremia was associated with multifocal cellulitis, similar to that seen in patients with bacteremia due to other non-H. pylori Helicobacter species, most of whom have been immunosuppressed (2, 4, 9, 14). H. canis has been recovered from humans on two occasions: from the stool of a 5-year-old, healthy boy with uncomplicated gastroenteritis (3) and from the stool of a man with X-linked hypogammaglobulinemia who developed recurrent cellulitis and bacteremia with H. canis and “H. rappini” linked to his recent acquisition of a puppy (8). Non-H. pylori Helicobacter species are colonizers of certain animals, and human infection probably occurs as a zoonosis in most cases (7, 12). H. canis has been recovered from the stools of both symptomatic and asymptomatic cats (11), Bengal cats (6), and dogs (13). We hypothesize that our patient's bacteremia was related to pet exposure, although cultures of stools from his cats were negative and his dog exposure was not very close. Physicians should consider the possibility of non-H. pylori Helicobacter bacteremia, including H. canis bacteremia, in cases of multifocal cellulitis, particularly in immunosuppressed patients with animal exposure. Correct identification generally requires the use of molecular methods.
Acknowledgments
Written consent for publication of this case report was obtained from the patient. The patient has seen and approved the final version of the manuscript.
Footnotes
Published ahead of print on 27 September 2006.
REFERENCES
- 1.Adékambi, T., and M. Drancourt. 2004. Dissection of phylogenetic relationships among 19 rapidly growing Mycobacterium species by 16S rRNA, hsp65, sodA, recA and rpoB gene sequencing. Int. J. Syst. Evol. Microbiol. 54:2095-2105. [DOI] [PubMed] [Google Scholar]
- 2.Burman, W. J., D. L. Cohn, R. R. Reves, and M. L. Wilson. 1995. Multifocal cellulitis and monoarticular arthritis as manifestations of Helicobacter cinaedi bacteremia. Clin. Infect. Dis. 20:564-570. [DOI] [PubMed] [Google Scholar]
- 3.Burnens, A. P., J. Stanley, U. B. Schaad, and J. Nicolet. 1993. Novel Campylobacter-like organism resembling Helicobacter fennelliae isolated from a boy with gastroenteritis and from dogs. J. Clin. Microbiol. 31:1916-1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cuccherini, B., K. Chua, V. Gill, S. Weir, B. Wray, D. Stewart, D. Nelson, I. Fuss, and W. Strober. 2000. Bacteremia and skin/bone infections in two patients with X-linked agammaglobulinemia caused by an unusual organism related to Flexispira/Helicobacter species. Clin. Immunol. 97:121-129. [DOI] [PubMed] [Google Scholar]
- 5.Flores, B. M., C. L. Fennell, K. K. Holmes, and W. E. Stamm. 1985. In vitro susceptibilities of Campylobacter-like organisms to twenty antimicrobial agents. Antimicrob. Agents Chemother. 28:188-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Foley, J. E., S. L. Marks, L. Munson, A. Melli, F. E. Dewhirst, S. Yu, Z. Shen, and J. G. Fox. 1999. Isolation of Helicobacter canis from a colony of Bengal cats with endemic diarrhea. J. Clin. Microbiol. 37:3271-3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fox, J. G. 2002. The non-H. pylori helicobacters: their expanding role in gastrointestinal and systemic diseases. Gut 50:273-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gerrard, J., D. Alfredson, and I. Smith. 2001. Recurrent bacteremia and multifocal lower limb cellulitis due to Helicobacter-like organisms in a patient with X-linked hypogammagloblinemia. Clin. Infect. Dis. 33:116-118. [DOI] [PubMed] [Google Scholar]
- 9.Kiehlbauch, J. A., R. V. Tauxe, C. N. Baker, and I. K. Wachsmuth. 1994. Helicobacter cinaedi-associated bacteremia and cellulitis in immunocompromised patients. Ann. Intern. Med. 121:90-93. [DOI] [PubMed] [Google Scholar]
- 10.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignement. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 11.Shen, Z., Y. Feng, F. E. Dewhirst, and J. G. Fox. 2001. Coinfection of enteric Helicobacter spp. and Campylobacter spp. in cats. J. Clin. Microbiol. 39:2166-2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Solnick, J. V., and D. B. Schauer. 2001. Emergence of diverse Helicobacter species in the pathogenesis of gastric and enterohepatic diseases. Clin. Microbiol. Rev. 14:59-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stanley, J., D. Linton, A. P. Burnens, F. E. Dewirst, R. J. Owen, A. Porter, S. L. On, and M. Costas. 1993. Helicobacter canis sp. nov., a new species from dogs: an integrated study of phenotype and genotype. J. Gen. Microbiol. 139:2495-2504. [DOI] [PubMed] [Google Scholar]
- 14.Trivett-Moore, N. L., W. D. Rawlinson, M. Yuen, and G. L. Gilbert. 1997. Helicobacter westmeadii sp. nov., a new species isolated from blood cultures of two AIDS patients. J. Clin. Microbiol. 35:1144-1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weisburg, W. G., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]