Abstract
Ganciclovir (GCV) resistance is an emerging problem for transplant recipients. A sensitive and rapid real-time PCR approach for simultaneous and semiquantitative detection of human cytomegalovirus (HCMV) UL97 mutations in codons 460/520 was established by LightCycler and confirmed by restriction fragment length polymorphism and sequencing. Results from HCMV laboratory strains were compared with results from 11 GCV-resistant clinical isolates.
Mutations in the human cytomegalovirus (HCMV) phosphotransferase gene (UL97) and polymerase gene (UL54) are responsible for resistance to ganciclovir (GCV) (UL97/UL54), cidofovir (UL54), or foscarnet (UL54) (2). Over 90% of GCV-resistant clinical HCMV isolates have mutations in the UL97 gene (10). Most of the GCV-resistant clinical HCMV isolates feature mutations in the UL97 gene between codons 400 and 665 (9). Some of the most frequent mutations occur in codons 460, 594, and 595 (4, 7). There is a high impact for genotypic resistance screening with respect to known UL97 mutations conferring GCV resistance (4). The ratio of the wild-type and mutant strains present in HCMV-infected allogeneic hematopoietic stem cell transplant recipients may reflect clinical outcome (5, 11). Up to now, genotypic resistance screening has been performed by PCR-based restriction fragment length polymorphism (RFLP) assay and sequencing. Combinations of RFLP assays and sequencing analysis need at least 3 to 4 days for completion. RFLP assays for the detection of mutations in codons 460 and 520 have been described previously (3, 12). The relative frequencies of the UL97 mutations M460V, M460I, and H520Q were given previously, with the mutations found in 20 out of 75 (27%) unrelated ganciclovir-resistant isolates (4).
We present a new LightCycler (LC) PCR assay using specific hybridization probes with melting point analysis for simultaneous characterization of the relevant UL97 codons 460 and 520 by a dual-color format with two specific pairs of hybridization probes for both codons which are each labeled with different fluorescence dyes (LC Red dye 640 and LC Red dye 705). The binding of the hybridization probe leads to a specific melting point for the wild-type sequence. In mutant strains, however, the mismatch between the hybridization probe and the mutant target sequence results in unstable binding of the probes, decreasing the melting temperature. The evaluation for the different codons can be performed by the use of different fluorescence channels. The reaction mixture utilized a master mix containing a LightCycler Fast Start hybridization probe kit (Roche Diagnostics, Mannheim, Germany) 1×, 4.5 mM MgCl2, 0.6 μM of each primer, 0.2 μM of each hybridization probe, and an additional 2.5 U/reaction Fast Start Taq DNA polymerase (Roche Diagnostics, Mannheim, Germany) for increase of the sensitivity of the amplification reaction. An enhanced concentration of Fast Start Taq DNA polymerase (5 U/μl) in the reaction mixture resulted in a higher sensitivity of our assay (8). The following program was used for cycling: initial denaturation, enzyme activation at 95°C for 10 min; denaturation step at 95°C for 15 s; annealing step at 50°C for 20 s, elongation step at 72°C for 20 s, repeat for 50 cycles; and melting curve analysis at 95°C for 0 s, 50°C for 15 s, and 85°C for 0 s, with a slope of 0.1 (°C/s). The time was 80 min per LightCycler assay. The amplicon size of our UL97 PCR was 470 bp using primers 405F and 550R. Primer design was performed using Oligo primer analysis software, version 5.0 (NBI, Plymouth, United Kingdom). Hybridization probes were designed by TIB-MolBiol (Berlin, Germany) and purchased from this manufacturer. Sequences of primers and specific hybridization probes are shown in Table 1. Proportions of UL97 wild-type and mutant strains in mixed viral populations can be determined by finding the area under the melting curve. Laboratory strains AD169 and Towne and 11 of our own clinical isolates of stem cell transplant recipients with a specific point mutation in codon 460 were investigated by the LightCycler real-time PCR assay.
TABLE 1.
Sequences of HCMV UL97 primers and hybridization probes for the LightCycler assay for codons 460/520 and the site-directed mutagenesis of mutation H520Q
| Primer or probe designation (nucleotide position) | Function | Sequence (5′-3′)a |
|---|---|---|
| 405F (141693-141712) | Forward primer | CTGCTGCACAACGTCAAGGT |
| 550R (142131-142153) | Reverse primer | CCCAGCGCCGACAGCTCCGACAT |
| 460Sensor (141853-141874) | Hybridization probe | ATGAGCACGTTCATGGGTGTAA-Fl |
| 460Anchor (141823-141851) | Hybridization probe | LC640-GTCAAAGTGGCATACACGACACTGGTGAT-P |
| 520Sensor (142032-142053) | Hybridization probe | GAATGTTACCACCCTGCTTTCC-Fl |
| 520Anchor (142055-142076) | Hybridization probe | LC705-ACCCATGCCGCTGCAGAAGCTG-P |
| m520F (142031-142049) | Forward mismatch primer | CGAATGTTACCAGCCTGCTT |
| m520R (142031-142049) | Reverse mismatch primer | AAGCAGGCTGGTAACATTCG |
Fl, fluorescein; LC640, fluorescence dye LC Red dye 640; LC705, fluorescence dye LC Red dye 705. Underlined, boldface residues indicate sequence mismatches.
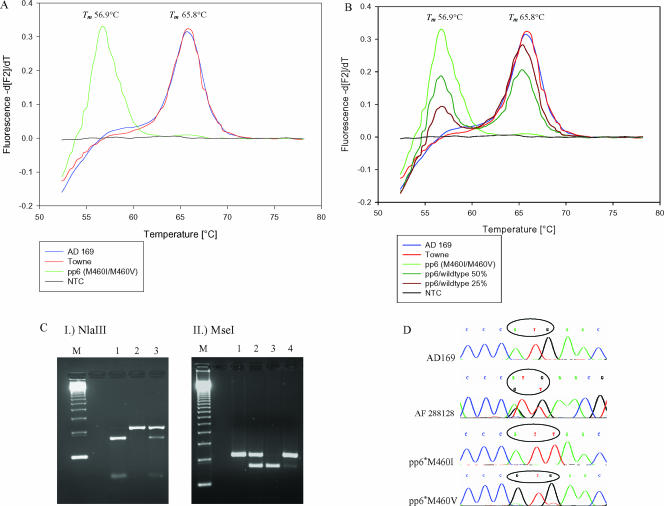
The laboratory strains AD169 and Towne had a specific melting point of 65.8°C, while the point mutations in codon 460 reduced the specific melting point to 56.9°C (Fig. 1A and B). The difference between melting points for the laboratory strains and the UL97 mutant strain pp6 (M460I, M460V) was 8.9°C. In contrast to AD169 and Towne, all other investigated clinical isolates from stem cell transplant recipients with specific mutations in codon 460 showed the same melting point as the reference strain pp6, which was derived from an AIDS patient (Dana Wolf, Jerusalem, Israel, personal communication). It was not possible to discriminate among all three different point mutations potentially present in codon 460 (M460I/V; ATT, ATA; GTG) (6), since the different nucleotide substitutions did not alter melting points. However, we were able to give proportions of coexisting wild-type and mutant strains in mixed viral populations for any mutation in codon 460 (Fig. 1B). In a mixture of defined proportions of UL97 mutant and wild-type strains, it was possible to detect 5% mutant or wild-type strain, respectively. The detection limit of the PCR was 14 copies/assay by use of a commercially available HCMV AD169 quantitated DNA control (tebu-bio, Offenbach, Germany) with defined copy number per microliter.
FIG. 1.
LightCycler assay for UL97 codon 460. (A) Detection of mutations in codon 460 for laboratory strains (AD169 and Towne) and a mutant strain (pp6). (B) Semiquantitative detection of mixed viral populations in codon 460. (C) For comparison, RFLP results of the different mutations in codon 460 (6) are given. Panel I shows an NlaIII digest to discriminate between wild-type and mutant strains (lane M, marker; lane 1, AD169; lane 2, pp6; lane 3, GCV-resistant isolate [288128]) from a hematopoietic stem cell transplant recipient with mixed viral populations. Panel II shows an MseI digest to discriminate between different mutations in codon 460 (lane M, marker; lane 1, AD169; lane 2, pp6; lane 3, M460I mutant [plaque purified from strain pp6; pp6*M460I] and lane 4, M460V mutant [plaque purified from strain pp6; pp6*M460V]). (D) Corresponding sequences of wild-type and mixed mutant strains (288128) in codon 460 are shown. Tm, melting temperature; NTC, no template control.
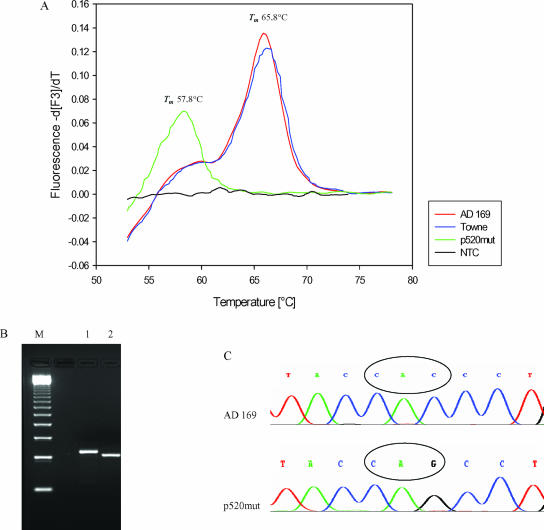
The H520Q mutation was generated by site-directed mutagenesis and overlap extension. The resulting PCR product was cloned into the pGEM-T Easy vector system (Promega, Mannheim, Germany). After cloning and linearization, the vector was investigated by LightCycler real-time PCR. The laboratory strains showed a specific melting point of 65.8°C. The H520Q mutation could be detected with a decreased specific melting point of 57.8°C (Fig. 2A). The detection limit was 14 copies/assay by use of the HCMV AD169 DNA control.
FIG. 2.
Light Cycler assay for codon 520. (A) Detection of the H520Q mutation using melting point analysis. (B) AluI digest for the detection of the H520Q mutation by RFLP analysis (12). Lane M, marker; lane 1, AD169; lane 2, p520mut plasmid. (C) Corresponding sequence analysis is shown. Tm, melting temperature; NTC, no template control.
Concluding, we established real-time PCR assays for the simultaneous and semiquantitative detection of different UL97 mutations in codons 460 (3) and 520 (12). Using melting point analysis, we can also give semiquantitative results of the wild-type/mutant ratios in mixed viral strains. In comparison to RFLP and sequencing (Fig. 1D and 2C), the real-time UL97 mutation analysis enabled a very rapid detection of specific UL97 point mutations associated with GCV resistance, allowing semiquantitative estimations of proportions of UL97 wild-type and mutant strains coexisting in vivo. With RFLP analysis and direct sequencing, it was not always possible to detect small amounts of mutant strains in mixed viral populations, but the real-time PCR described here enabled a sensitive detection of nondominant ex vivo mutant strains (1, 5, 11).
In comparison to the real-time PCR assay using molecular beacons (13), our LightCycler assay offers the possibility of simultaneous detection of the H520Q mutation. The development of new semiquantitative assays for other relevant UL97 mutations conferring GCV resistance may contribute to a better understanding of the in vivo dynamics and replication of HCMV UL97 mutant and wild-type strains.
Acknowledgments
The UL97 mutant strain pp6 was a generous gift from Dana Wolf, Jerusalem, Israel.
Footnotes
Published ahead of print on 11 October 2006.
REFERENCES
- 1.Baldanti, F., L. Simoncini, A. Sarasini, M. Zavattoni, P. Grossi, M. G. Revello, and G. Gerna. 1998. Ganciclovir resistance as a result of oral ganciclovir in a heart transplant recipient with multiple human cytomegalovirus strains in blood. Transplantation 66:324-329. [DOI] [PubMed] [Google Scholar]
- 2.Baldanti, F., N. Lurain, and G. Gerna. 2004. Clinical and biologic aspects of human cytomegalovirus resistance to antiviral drugs. Hum. Immunol. 65:403-409. [DOI] [PubMed] [Google Scholar]
- 3.Chou, S., A. Erice, and M. C. Jordan. 1995. Analysis of UL97 phosphotransferase coding sequence in clinical cytomegalovirus isolates and identification of mutations conferring ganciclovir resistance. J. Infect. Dis. 171:576-583. [DOI] [PubMed] [Google Scholar]
- 4.Chou, S. 1999. Antiviral drug resistance in human cytomegalovirus. Transplant Infect. Dis. 1:105-114. [DOI] [PubMed] [Google Scholar]
- 5.Eckle, T., G. Jahn, and K. Hamprecht. 2004. The influence of mixed HCMV UL97 wildtype and mutant strains on ganciclovir susceptibility in a cell associated plaque reduction assay. J. Clin. Virol. 30:50-56. [DOI] [PubMed] [Google Scholar]
- 6.Eckle, T., G. Jahn, and K. Hamprecht. 2003. High impact of an expanded restriction fragment length polymorphism assay on detection of ganciclovir-resistant UL97 mutants of human cytomegalovirus. Antimicrob. Agents Chemother. 47:442-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Erice, A. 1999. Resistance of human cytomegalovirus to antiviral drugs. Clin. Microbiol. Rev. 12:268-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Exner, M. M., and M. A. Lewinski. 2002. Sensitivity of multiplex real-time PCR reactions, using the LightCycler and ABI PRISM 7700 Sequence Detection System, is dependent on concentration of the DNA polymerase. Mol. Cell. Probes 16:351-357. [DOI] [PubMed] [Google Scholar]
- 9.Gilbert, C., J. Bestman-Smith, and G. Boivin. 2002. Resistance of herpesviruses to antiviral drugs: clinical impacts and molecular mechanism. Drug Res. Update 5:88-114. [DOI] [PubMed] [Google Scholar]
- 10.Gilbert, C., and G. Boivin. 2005. Human cytomegalovirus resistance to antiviral drugs. Antimicrob. Agents Chemother. 49:873-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamprecht, K., T. Eckle, L. Prix, C. Faul, H. Einsele, and G. Jahn. 2003. Ganciclovir-resistant cytomegalovirus disease after allogeneic stem cell transplantation: pitfalls of phenotypic diagnosis by in vitro selection of UL97 mutant strain. J. Infect. Dis. 187:139-143. [DOI] [PubMed] [Google Scholar]
- 12.Hanson, M. N., L. C. Preheim, S. Chou, C. L. Talarico, K. K. Biron, and A. Erice. 1995. Novel mutation in UL97 gene of a clinical cytomegalovirus strain conferring resistance to ganciclovir. Antimicrob. Agents Chemother. 39:1204-1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yeo, A. C., K. P. Chan, G. Kumarasinghe, and H. K. Yap. 2005. Rapid detection of codon 460 mutations in the UL97 gene of ganciclovir-resistant cytomegalovirus clinical isolates by real-time PCR using molecular beacons. Mol. Cell. Probes 19:389-393. [DOI] [PubMed] [Google Scholar]