Abstract
Bovine tuberculosis is a major problem in many countries; hence, new and better diagnostic tools are urgently needed. In this work, we have tested ESAT6, CFP10, PE13, PE5, MPB70, TB10.4, and TB27.4 for their potentials as diagnostic markers in field animals from Northern Ireland, Mexico, and Argentina, regions with low, medium, and high prevalences of bovine tuberculosis, respectively. At all three sites, ESAT6 and CFP10 were superior diagnostic antigens, while their combination performed even better at the two sites where the combination was tested, providing the best coverage for the detection of diseased populations. The high sensitivity in the skin test reactor groups, combined with the high specificity in the tuberculosis-free groups, indicated that a diagnosis could correctly be made for 85% of the infected animals, based on their responses to these two antigens. Furthermore, TB10.4, PE13, and PE5 have the potential to supplement ESAT6 and CFP10 in a future five-component diagnostic cocktail.
In recent decades, bovine tuberculosis (BTB) has been spreading in cattle herds in several countries around the world and is now a major problem in many countries or parts thereof. This is the case, for example, in parts of the United Kingdom, Argentina, and Mexico (2, 13, 17). Elsewhere, the disease occurs sporadically, and even in officially tuberculosis (TB)-free areas it can prove to be very difficult to eradicate biologically. BTB is a zoonotic disease; and although introduction of milk pasteurization in developed countries dramatically reduced the rates of transmission of TB from cattle to humans (20), the persistence of Mycobacterium bovis, the causative agent of BTB, still accounts for up to 10% of the human cases of tuberculosis in some parts of the world (12). Thus, the persistence or an increase in M. bovis infections poses not only a major economic problem but also an increased human health risk, particularly in areas where human immunodeficiency virus and AIDS are endemic. Strict import and export regulations and extensive movement restrictions, combined with skin test and slaughter policies, have significantly reduced the incidence of disease in certain circumstances. However, it is now clear that more efficient diagnostic tools with increased specificity for eradication, control, and surveillance programs, as well as new and better vaccination strategies, are required to control BTB in the future.
M. bovis-infected cattle primarily mount a cellular immune response (cell-mediated immunity [CMI]) (28), and therefore, skin testing with the bovine purified protein derivative (PPD) (PPDB) has been used for many years in control programs. Since it shares many antigenic components with avian PPD (PPDA), the test readout can be compromised (3). For example, calves exposed to environmental mycobacteria early after birth show responses to PPDA within 6 weeks (8). Furthermore, skin testing requires immobilization of the animals twice: for the injection of PPD and for the readout. Moreover, it lacks objectivity, and in case of uncertainty it cannot be repeated for 2 months due to desensitization. As a consequence, blood-based diagnostic assays have been developed in which an antigen's ability to induce the cellular release of cytokines, especially gamma interferon (IFN-γ), is measured (36). To avoid specificity problems and at the same time have a strong response in infected animals, the field validation of the antigens selected for this in vitro assay is extremely important. The diagnostic antigen selected should ideally accommodate the possible application of the M. bovis bacillus Calmette-Guérin (BCG) vaccine in the future (9). Genetic comparison of M. bovis and a number of BCG strains has revealed several large genomic regions in BCG strains that have been lost during the attenuation process (5, 18); and because of their theoretical lack of interference with a BCG vaccine, the proteins encoded by these regions have received growing attention as diagnostic antigens, especially antigens from region of difference (RD) RD1 (1, 10, 19, 26, 30). All studies have clearly identified the two RD1 antigens ESAT6 and CFP10 as being the most immune dominant of those tested with experimentally infected cattle. Furthermore, both can be used to distinguish between BCG-vaccinated and M. bovis-infected cattle (7, 33).
Previously, we have screened a panel of 28 carefully selected mycobacterial antigens, including antigens from selected RDs, for their diagnostic potential using experimentally infected animals (1). In the present study, we have selected the four best-performing antigens identified in the previous study and added an additional known TB antigen and two representatives of the large PE antigen family, members of which are known to induce a cellular immune response (14). These seven antigens were tested and compared as diagnostic markers by using the IFN-γ whole-blood-based assay against the PPDB-PPDA set. The strategy was to determine their diagnostic performance in the field with naturally infected cattle and thereby identify antigens with the potential for use in a future diagnostic cocktail. To take into account different epidemiological backgrounds (genetic and environmental), the study was carried out both with skin test-positive animals and with animals from BTB-free areas in Argentina, Mexico, and Northern Ireland, three countries with different incidences of BTB.
MATERIALS AND METHODS
Selection of mycobacterial antigens for evaluation.
Seven proteins among the 3,924 open reading frames of M. bovis were selected for testing (Table 1). ESAT6, CFP10, TB10.4, and TB27.4 were primarily selected because they have shown the highest sensitivities and specificities in a previous screening program with experimentally infected animals (1). The three antigens (ESAT6, CFP10, TB27.4) from RD1 have extra specificity potential since they have no close orthologues in most environmental strains. As an example, the closest orthologues in the environmental mycobacterium Mycobacterium avium subsp. avium have homologies of less than 30% (Table 1). The genes encoding these three proteins have been lost early in the attenuation process of M. bovis BCG, and none of the vaccine strains therefore express these proteins. MPB70 is a well-known BTB antigen expressed at high levels in M. bovis and has been shown to act as a potent T-cell antigen in cattle (25). It shares sequence similarity (>70%) with MPB83, another M. bovis antigen in cattle, but has no close orthologue in most environmental species. PE13 and PE5 were selected after prescreening of a subset of the 33-member M. bovis PE family (M. Govaerts, personal communication). The broad distribution and number of homologues within mycobacteria illustrate the importance of this family. PE13 has several paralogues in M. bovis, with PE19, PE18, and PE31 being the closest ones, having identities of >70%. PE5 has PE29 and PE15 as its closest paralogues in M. bovis, with identities of 80.4 and 65.7%, respectively. Both PE13 and PE5 are found in all BCG vaccine strains and have orthologues in the environmental mycobacterium M. avium subsp. avium strain (59 and 86% homologies, respectively) and should theoretically be the two least-specific antigens of the seven antigens selected.
TABLE 1.
Characteristics of proteins used in this study
| Protein | M. bovis Mb no.a | M. tuberculosis Rv no.b | Present in BCG | % Homology in M. avium subsp. aviumc | Protein size (no. of amino acids) | Comment |
|---|---|---|---|---|---|---|
| ESAT6 | Mb3905 | Rv3875 | No | 29 | 94 | 6-kDa early secretory antigenic target |
| CFP10 | Mb3904 | Rv3874 | No | 24 | 100 | Exported protein cotranscribed with Rv3875 |
| PE13 | Mb1227 | Rv1195 | Yes | 54 | 99 | PE family protein |
| MPB70 | Mb2900 | Rv2875 | Yes | 25 | 193 | Major secreted immunogenic protein MPT70 |
| TB10.4 | Mb0296 | Rv0288 | Yes | 79 | 96 | Low-molecular-wt protein antigen 7 |
| PE5 | Mb0293 | Rv0285 | Yes | 88 | 102 | PE family protein |
| TB27.4 | Mb3908 | Rv3878 | No | 24 | 280 | Conserved hypothetical alanine-rich protein |
The numbering system for Mycobacterium bovis strain AF2122/97 was used (16).
The Mycobacterium tuberculosis numbering system for laboratory strain H37Rv was used (11).
TBLASTN with a blosum 62 scoring matrix. To allow homology searches in regions of low complexity, searches were done without a filter.
Antigen preparation and purification.
ESAT6, CFP10, PE13, MPB70, TB10.4, PE5, and, TB27.4 (Table 1) were prepared by growing 6 liters of transformed Escherichia coli BL21 culture in Luria-Bertani broth to an optical density (OD) at 600 nm of ∼0.4 at 30°C, after which recombinant protein expression was induced by either 1 mM isopropyl-β-d-thiogalactopyranoside or 0.3 M NaCl for another 4 h. The bacteria were harvested, and the inclusion bodies were released by disrupting the membrane with a mild detergent (B-PER; Pierce, Rockford, IL). The inclusion bodies were washed three times in 100× diluted B-PER solution. The recombinant proteins, found primarily in inclusion bodies for all proteins, were dissolved in 50 to 100 ml 8 M urea-50 mM Na2HPO4 (pH 8) (buffer A) and metal affinity purified under denaturing conditions by exploiting the six-His tag fused to the proteins. The dissolved proteins were bound to 20 ml Talon column material (Clontech, Mountain View, CA) that had been preequilibrated in buffer A and washed by alternating between 10 mM Tris HCl (pH 8.0)-60% 2-propanol (buffer B) and buffer A. Each preparation was washed three times with 2 column volumes of buffer A and B a total of six times before elution in buffer A supplemented with 200 mM imidazole. The eluted fractions, run-through, and washes were analyzed by Coomassie blue staining of sodium dodecyl sulfate (SDS)-polyacrylamide gels. Fractions containing recombinant proteins were inspected, and the purest proteins were pooled. To allow anion-exchange chromatography, the buffer was changed by dialysis against 25 mM Tris HCl (pH 8.0)-3 M urea (buffer C) before being applied to a Mono Q column (GE Healthcare, Denmark), washed with 5 column volumes of buffer C, and eluted with a 0 to 1 M linear NaCl gradient made up in buffer C. All fractions were again analyzed for recombinant proteins, and the purest fractions were pooled. For the third, chromatography-based purification step, the buffer was changed by dialysis against 25 mM acetic acid (pH 4.5)-3 M urea (buffer D), and the sample was loaded onto an SP column (GE Healthcare), washed with 5 column volumes of buffer D, and eluted with a 0 to 0.5 M linear NaCl gradient. The fractions were analyzed as described above; and pooled fractions were concentrated 5 to 10 times, depending on their absorbance at 214 nm, to be finally refolded by dialysis against 25 mM HEPES (pH 7.5)-10% glycerol-0.15 M NaCl.
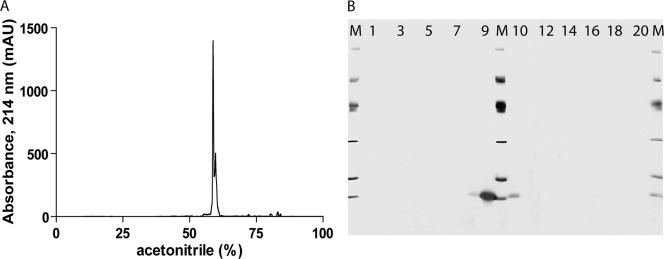
After purification and refolding, the protein concentration was measured by a bicinchoninic acid test (Pierce) by using bovine serum albumin as the standard, and the lipopolysaccharide content was determined by a kinetic turbidimetric Limulus amebocyte lysate assay (Charles River Laboratories). No toxicity or nonspecific stimulation was found for any of the antigens on naïve murine peripheral blood mononuclear cells when they were tested at concentrations up to 10 μg/ml. To detect possible impurities, 50 μg of each recombinant protein was evaluated by analytical reverse-phase high-pressure liquid chromatography on a hydrophobic C4 column (Grace Vydac, United Kingdom). After the proteins, which were in refolding buffer, bound to the column, the recombinant proteins were eluted with a linear 0 to 100% gradient of acetonitrile (Fig. 1A). Protein visualization was done by separation on SDS-polyacrylamide gels, followed by silver staining (Fig. 1B) and Coomassie blue staining and by Western blotting of the same fractions with two different polyclonal anti-E. coli antibodies. The optimal antigen concentration for the IFN-γ release assay was determined by titrating all proteins against TB-free and experimentally infected cattle. Based upon receiver operator curve (ROC) analysis, 4 μg/ml was selected as the optimal concentration for all antigens tested.
FIG. 1.
Quality control of CFP10. Fifty micrograms of recombinant protein was loaded onto reverse-phase C4 columns and eluted by using a linear acetonitrile gradient. The elution profile was monitored by continuously measuring the absorbance at 214 nm (mAU, milli-absorbance units) (A). Over the 0 to 100% elution gradient, 20 fractions were collected and applied onto SDS-polyacrylamide gels, followed by silver staining. Lanes M, SDS-polyacrylamide gel electrophoresis size marker (97, 66, 45, 31, 21, and 14 kDa); lanes 1 to 20, the 20 eluted fractions, respectively. Fraction 1 corresponds to 0% acetonitrile on the chromatogram, and fraction 20 corresponds to 100% acetonitrile (B).
Animal selection. (i) Skin test reactors.
In all three countries, animals were selected for study if they were reactors when they were skin tested with bovine tuberculin, in accordance with national guidelines, 1 to 2 months before slaughter. In Northern Ireland, animals were considered positive if the single intradermal comparative test showed a PPDB testing bias (compared to the results of PPDA testing) greater than 4 mm; in Mexico the PPDB bias had to be greater than 6 mm, and in Argentina the animals were considered positive if the cervical skin test with PPDB alone showed an increase in skin thickness greater than 5 mm. All animals were sampled for blood and nasal mucus 24 h before slaughter. Subsequently, delayed-type hypersensitivity testing results and in vitro IFN-γ assay results were collated. Following slaughter, the retropharyngeal, mediastinal, and bronchial lymph nodes from the skin test-positive animals were examined for the presence of tuberculous lesions; and samples from suspicious organs were taken for confirmation by bacterial culture, PCR, and/or histopathology. The results for all animals with lesions were confirmed in the second test. In total, 124 animals were included in the skin test reactor group: 49 from Northern Ireland, 39 from Argentina, and 36 from Mexico.
(ii) Negative controls.
The main selection criteria for the negative controls were that the herds came from BTB-free areas, with no history of tuberculosis during the previous 10 years; that there had been two clear skin tests for each herd; and that they all tested skin test negative 1 to 2 months before the blood sample was taken. Forty-seven animals were included in this group: 20 from Northern Ireland, 18 from Argentina, and 9 from Mexico.
Blood collection and IFN-γ release assay.
Blood samples were shipped to the laboratories at ambient temperature and were processed within 10 h of collection. To measure the T-cell responses in whole-blood cultures after a 20-h in vitro antigen stimulation, a commercial bovine IFN-γ microplate enzyme-linked immunosorbent assay (ELISA; Bovigam; Commonwealth Serum Laboratories, Australia) was used according to the manufacturer's instructions. Briefly, samples were collected from the jugular vein and placed into heparinized tubes, and 200 μl of blood was incubated in microplates, in duplicate for each antigen. Single batches were used throughout the study for each antigen at each of the three locations. The optimal antigen concentrations were determined by comparing the area-under-the-curve values obtained by ROC analysis at eight dilutions points for BTB-infected and BTB-free animals (1), and the final concentration was set at 4 μg/ml for all antigens tested. Negative control wells, to which only phosphate-buffered saline (PBS) was added, were included for each animal tested, as were as positive controls containing 1 μg/ml pokeweed mitogen (Sigma-Aldrich, United Kingdom), before incubation in a humidified 5% CO2 incubator at 37°C for 20 h. Culture supernatants were pooled before measuring IFN-γ release by ELISA, since the coefficient of variation between duplicate wells had previously been found to be less than 5% (1). The OD for the PBS controls, which was usually approximately 0.1, was used to normalize individual readouts and to calculate optical density indices (ODIs), where the results obtained by antigen stimulation were divided by the results for the PBS-stimulated cultures.
Collection of nasal swabs.
Nasal mucus samples were collected from cattle by using either a sterile 13-cm wooden swab (Sterilin, United Kingdom) with a compact cotton wool tip or a 50-cm homemade sterile swab made of aluminum wire and gauze. The swabs were expunged into 5 to 10 ml of sterile PBS for transport to the laboratory, where the supernatant was centrifuged at 4,000 × g for 15 min to pellet the mycobacteria, if any were present.
DNA extraction from nasal swabs.
Pellets from the nasal swabs were resuspended in 400 μl of 1× Tris-EDTA buffer and heated to 80°C for 30 min to inactivate the bacteria in the samples before 50 μl of 10 mg/ml lysozyme were added. The mixture was then incubated overnight at 37°C. On the next day, 100 μl of 10% SDS and 10 μl of 10 mg/ml proteinase K were added; and the mixture was incubated for 10 min at 65°C before 100 μl of 5 M NaCl and 100 μl of N-cetyl-N,N,N-trimethylammonium bromide-NaCl were added. The suspensions were mixed thoroughly, incubated for 10 min at 65°C, treated with chloroform-isoamyl alcohol (24:1; vol/vol), and centrifuged at 15,000 × g for 10 min. The supernatants were transferred to fresh tubes, 0.6 volume of isopropanol was added, and the DNA was precipitated by leaving the samples overnight at −20°C, followed by centrifugation at 15,000 × g for 15 min. The pellets were washed twice with 70% ethanol before finally being dried and resuspended in 20 μl of water.
Mycobacterium tuberculosis complex-specific PCR.
By using 2 μl of the extracted DNA preparations and oligonucleotides 5′-CGTGAGGGCATCGAGGTGGC-3′ and 5′-GCGTAGGCGTCGGTGACAAA-3′, which are specific for a 245-bp IS6110 amplicon (21), touchdown amplification was performed with an initial denaturation step at 96°C for 3 min, followed by eight cycles of 96°C for 1 min, annealing starting at 72°C for 1 min (decreasing by 1°C/cycle), and extension for 1 min at 72°C. This initial touchdown amplification was followed by 30 cycles of 96°C for 1 min, 65°C for 1 min, 72°C for 2 min, with a final extension at 72°C for 8 min.
Radiometric detection of Mycobacterium bovis in nasal mucus.
The supernatant derived from the cotton wool swab was decontaminated with 6% NaOH and 2.9% sodium citrate. Acetyl-l-cysteine was also added as a decongestant to solubilize the mucus. This preparation was mixed on a rotary mixer for 15 min at 15°C, and the volume was then adjusted to 10 ml with sterile PBS (pH 6.8). This mixture was centrifuged at 3,000 rpm for 15 min, the supernatant was removed, and the sediment was resuspended in 2 ml sterile PBS. If the sample was not used immediately, it was stored at −20°C. The samples were inoculated into BACTEC vials (BACTEC 460; Becton Dickinson, United Kingdom) to determine the presence of viable mycobacteria. Five hundred microliters of the sample was inoculated into each of two BACTEC vials containing 5 ml Middlebrook 7H12 with antibiotic supplement and polyethylene stearate. All cultures were incubated at 37°C with a microenvironment of 5% CO2. Growth index readings were obtained on days 7, 14, 21, 28, 42, 56, and 84. During the measurements, the vials were in a 5% CO2 microenvironment. When a growth index reading indicated the presence of organisms, Ziehl-Neelsen staining was carried out to confirm the presence of acid alcohol-resistant bacilli.
Data analysis.
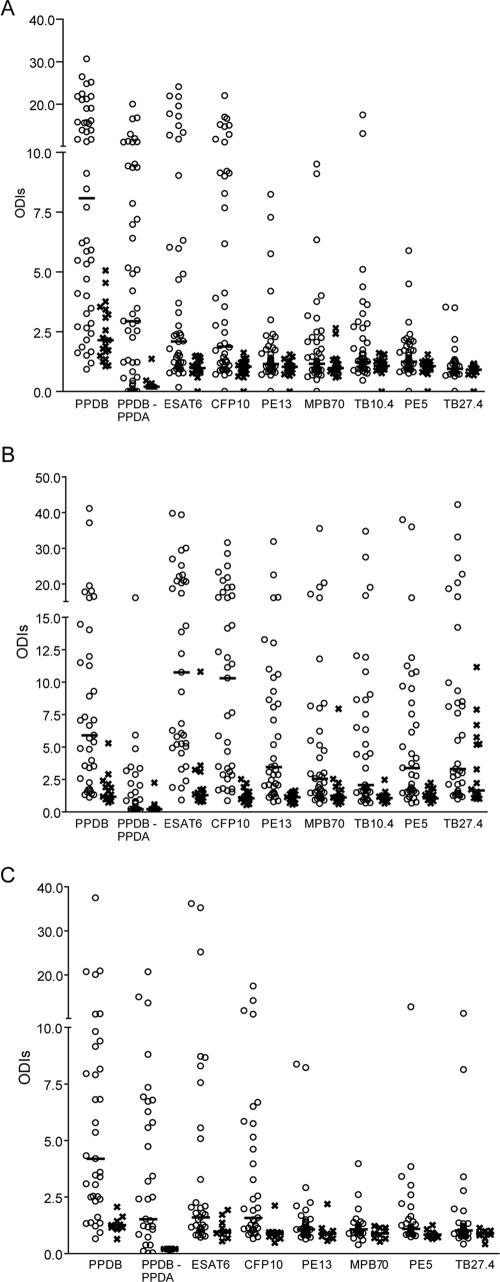
To evaluate their potential as diagnostic markers and inclusion in a diagnostic cocktail, the performance characteristics of seven selected recombinant proteins were compared with those of PPDB and PPDA in the blood-based CMI test. All animals in the skin test-positive groups were first included, regardless of the results of the confirmatory test (Fig. 2 and 3). Since these data were not normally distributed, the statistical analysis was carried out by using the nonparametric Mann-Whitney test. Next, to calculate the sensitivity and the specificity of each antigen, only skin test-positive animals that could be confirmed to have BTB by the presence of lesions, bacterial culture, or PCR were included in the infected group (Table 2). Due to the data distribution, the need for a single cutoff in a practical application, ROC analysis profiles (GraphPad), and published studies (24, 32), for all antigens tested an ODI value of 2 was selected as the cutoff value to indicate a positive result.
FIG. 2.
Whole-blood IFN-γ release assay with single antigens. Blood collected from skin test reactors (○) or TB-free animals (×) was incubated with different mycobacterial antigens, and the amount of IFN-γ released was measured by ELISA after 20 h of incubation in a humidified incubator at 37°C with 5% CO2. Samples were collected from animals from Northern Ireland (A), Argentina (B), and Mexico (C). The results are given as ODI units. Horizontal bars represent median values. The numbers of animals were 49, 39, and 36 for reactors in Northern Ireland, Argentina and México, respectively, and 20, 18, and 9 for TB-free animals from the three countries, respectively. The data for the PPDB-PPDA group are calculated by subtraction of the value for PPDA from the value for PPDB for a given animal.
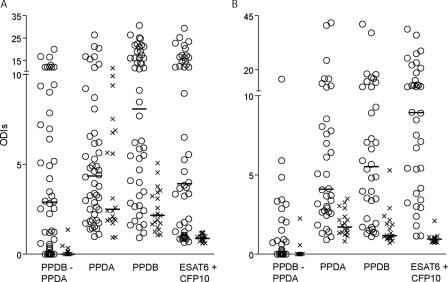
FIG. 3.
IFN-γ cytokine release in a one-well assay with ESAT6 and CFP10. ESAT6 and CFP10 were mixed in equal amounts (total, 4 μg per well), and the ability to release IFN-γ from peripheral blood mononuclear cells was compared to those for PPDB and PPDA after incubation with blood from skin test reactors (○) or TB-free animals (×) for 20 h at 37°C with 5% CO2. The amount of IFN-γ released was measured by ELISA. The animals came from Northern Ireland (A) and Argentina (B). The results are given as ODI units. Horizontal bars represent the median value. The numbers of animals from each location are as provided in the legend to Fig. 2. The data for the PPDB-PPDA group were calculated by subtraction.
TABLE 2.
Regional differences in specificities and sensitivities based on a defined cutoffa
| Antigen | Northern Ireland
|
Argentina
|
Mexico
|
Combined
|
||||
|---|---|---|---|---|---|---|---|---|
| Specificity (%) | Sensitivity (%) | Specificity (%) | Sensitivity (%) | Specificity (%) | Sensitivity (%) | Specificity (%) | Sensitivity (%) | |
| PPDA | 35 | 86 | 61 | 81 | 89 | 65 | 55 | 78 |
| PPDB | 45 | 91 | 78 | 75 | 89 | 90 | 66 | 84 |
| ESAT6 | 100 | 59 | 78 | 94 | 100 | 40 | 91 | 69 |
| CFP10 | 100 | 50 | 89 | 91 | 89 | 50 | 94 | 68 |
| PE13 | 100 | 23 | 100 | 78 | 89 | 10 | 98 | 43 |
| MPB70 | 90 | 36 | 83 | 56 | 100 | 5 | 89 | 36 |
| TB10.4 | 100 | 45 | 94 | 50 | NDd | ND | 97b | 48b |
| PE5 | 100 | 27 | 89 | 56 | 100 | 25 | 96 | 39 |
| TB27.4 | 100 | 14 | 56 | 72 | 100 | 5 | 83 | 36 |
| ESAT6 + CFP10c | 100 | 73 | 94 | 94 | ND | ND | 97b | 85b |
Specificity and sensitivity are based on cutoff values calculated as two times the ODI for each antigen and country. Only data from confirmed cases were used. Comfirmatory assays were lesion analysis, bacterial culture, or PCR analysis. n = 22 for Northern Ireland, n = 34 for Argentina, and n = 21 for Mexico.
Based only on data from Northern Ireland and Argentina.
The ESAT6 and CFP10 antigens were tested together in the same well (one-well assay).
ND, not determined.
RESULTS
Production and purification of recombinant mycobacterial proteins.
When all seven recombinant M. bovis proteins were expressed in Escherichia coli BL21-SI under the transcriptional control of the T7 promoter, all seven proteins formed insoluble inclusion bodies. This insoluble state was exploited in the first step of an extensive purification process, in which the bacteria were lysed in mild detergent and the insoluble material was pelleted and washed several times to remove the soluble E. coli proteins. At this stage, the proteins were dissolved in an urea-containing buffer and bound via the N-terminal His tag to a metal affinity column. The proteins were washed extensively before they were eluted from the column. The two final purification steps consisted of binding of the proteins first to an anion column and second to a cation column, both of which were washed and eluted with a linear salt gradient. The purity of the final product was determined and analyzed by reverse-phase high-pressure liquid chromatography (Fig. 1A), in which the percent purity was calculated by integrating the peaks in the chromatograms. To confirm and identify possible contaminants, the eluted fractions were separated on polyacrylamide gels, followed by staining for visual inspection (Fig. 1B). All seven proteins were found to have purities of at least 98% and to be practically free of lipopolysaccharide (<0.01 endotoxin unit/mg).
CMI responses to mycobacterial antigens at three test sites with different prevalences.
IFN-γ assays were performed in Northern Ireland, Argentina, and Mexico by using blood obtained from tuberculin skin test reactors and from cattle originating from known tuberculosis-free herds. In each of the three countries, the herds were selected from areas where BTB was endemic and where skin test-positive herds were previously characterized by the disclosure of tuberculous lesions at postmortem examination. Blood samples were obtained from 49, 39, and 36 skin test reactor animals from Northern Ireland, Argentinean, and Mexican farms, respectively, and from 20, 18, and 9 nonreactor cattle in tuberculosis-free herds in the same three countries, respectively. As expected, almost all the animals in the skin test-positive group at all three sites had strong responses to PPDB, with median ODIs of 8.1 (Northern Ireland), 5.9 (Argentina), and 4.2 (México); and the responses were significantly different from those of the skin test-negative groups (P < 0.019, Mann-Whitney test). In several countries, skin tests are implemented as a comparative test, in which the reading for PPBA is subtracted from the reading for PPDB. For this reason, the corresponding CMI groups were included in this study (Fig. 2A to C). Animals in the skin test-negative group from Northern Ireland in general had stronger responses against PPDA (median, 2.5 ODI units [data not shown]) than the same groups from Latin America (median, 1.7 ODI units for both groups), but for all three sites the median ODI for the skin test-negative group was higher for PPDA than for PPDB, an indication that most animals had been exposed to environmental mycobacteria.
When the skin test-positive and -negative groups were compared, all recombinant antigens tested induced their highest levels of IFN-γ release in the Argentinean population, and they were all able to discriminate the positive and negative Argentinean groups (P < 0.026, Mann-Whitney test). In Northern Ireland, the only recombinant protein that failed to induce a significant difference between the skin test-positive and -negative groups was TB27.4 (P < 0.121). Due to the relatively low number of animals (n = 9) in the skin test-negative group from Mexico, it was possible to demonstrate significant differences only for CFP10 (P < 0.029) and ESAT6 (P < 0.032). By comparing the data from the three sampling sites (Fig. 2A to C), ESAT6 and CFP10 stand out as the best-performing diagnostic antigens among those tested. Regional differences in the responses to PE13 and PE5, MPB70, TB27.4, and TB10.4 were apparent. In the Argentinean data set, there was a strong recognition of all antigens in the skin test-positive group, whereas in the Mexican data set, the animals recognized the antigens at a much lower frequency and, in general, developed weaker responses. PE13, MPB70, and TB10.4 were satisfactorily detected in Northern Ireland and Argentina, whereas PE13 and MPB70 induced relatively weak responses in Mexico, while TB10.4 was not tested there. Finally, there was clearly a specificity problem with TB27.4 in the Argentinean data set.
Sensitivity and specificity based upon confirmed cases: comparison of the different areas.
In order to estimate the sensitivities and specificities of the different antigens, a cutoff of 2 ODI units was used for all antigens assayed at the three sites (Table 2). Estimates were calculated for each site separately, as well as in combination. In the skin test-positive groups, only data for animals in which infection was confirmed by lesion analysis (Northern Ireland, Mexico) or lesion analysis and BTB-specific PCR with nasal mucus samples (Argentina) were used. This definition reduced the numbers of positive reference samples to 22 in Northern Ireland, 34 in Argentina, and 21 in Mexico. In the samples from Northern Ireland, PPDB and PPDA both had very high levels of sensitivity (greater than 85%), but as expected, there was clearly a lack of specificity. Six of the seven recombinant antigens tested also reached 100% specificity with the sample set, but with much lower sensitivities. ESAT6, CFP10, and TB10.4 were superior to the other antigens, reaching sensitivities between 45 and 59% when they were used as single antigens. With the Argentinean samples, the sensitivities of PPDB and PPDA were lower than those with the samples from Northern Ireland, but the specificities were clearly higher. PE13 was the only single antigen that reached 100% specificity; but apart from TB27.4, all of the antigens had specificities equal to or greater than the 78% specificity reached by PPDB. ESAT6 and CFP10 were both greater than 90% sensitive, and PE13 and TB27.4 reached percent sensitivities in the 70s. PPDB was the most effective with the Mexican samples, with a sensitivity of 90% and a specificity of 89%. This level of specificity was also reached with the CFP10 protein, although the sensitivity was lower (50%). ESAT6 had 100% specificity and 40% sensitivity. The PE5 protein had a relatively low sensitivity of 25% but had 100% specificity. By combining the results for the three sites, ESAT6 and CFP10 were superior, with sensitivities of 69 and 68%, respectively, while PE13, PE5, and TB10.4 appeared to be highly specific, with values greater than 95%.
Combining high-sensitivity antigens ESAT6 and CFP10 for use for diagnosis.
In an effort to increase the sensitivities of the recombinant antigens to a level above that for PPDB, the two antigens with the highest sensitivities as single antigens, ESAT6 and CFP10, were combined in a one-well assay and their sensitivities were compared with those of PPDB, PPDA, and PPDA subtracted from PPDB, with the last test being performed as a two-well assay (Fig. 3). In Northern Ireland, the median values for ESAT6 and CFP10 as single antigens were about 2 ODI units for the skin test-positive group (Fig. 2A). In other words, roughly 50% of the samples had values above the chosen cutoff and would be considered positive. Combining the two proteins increased the response strength, especially for those samples with ODIs close to 2 with single antigens, increasing the median value almost twofold to 3.9 ODI units for the skin test-positive group. Importantly, the median for the skin test-negative group did not increase but dropped from approximately 1.0 ODI unit for each antigen to 0.9 ODI unit when the antigens were combined. In Argentina, the median values for ESAT6 and CFP10 as single antigens were extremely high (10 ODI units) (Fig. 2B) and were higher than the value of 5.9 ODI units obtained for PPDB with the same sample set. By combining the two antigens, this median dropped to 8.9 ODI units, while the values for the skin test-negative group decreased from 1.2 ODI units to 0.9 ODI unit. By using 2 ODI units as the cutoff, the specificities of the cocktail were 100% for the samples from Northern Ireland and 94% for the samples from Argentina, and the sensitivities were 73% and 94%, respectively (Table 2). Combining these two data sets gave a specificity of 97% and a sensitivity of 85%. As an alternative way to calculate sensitivities and specificities, ROC analysis estimated the sensitivity to be 91% with a 100% specificity cutoff with the ESAT6-CFP10 cocktail both in Northern Ireland and Argentina and the sensitivity to be 86% with a 96% specificity cutoff for the combined data sets. In order to enhance the performance of the ESAT6-CFP10 antigen cocktail by adding multiple antigens, the sensitivity would need to be improved without impairing the high specificity.
Can use of high-specificity antigens PE13, PE5, and TB10.4 improve the sensitivities?
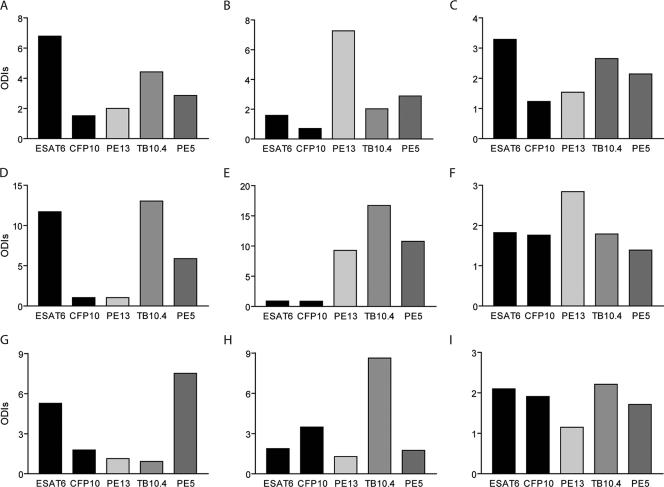
Using the data for the three high-specificity antigens PE13, PE5, and TB10.4, we examined those few samples that ESAT6-CFP10 did not identify as positive. We found that when the antigens were used in a two-well assay and when the definition of a positive test was a positive result by either assay, only PE13 was capable of complementing ESAT6-CFP10, and this occurred only with the Argentinean samples. However, in this two-well assay ESAT6-CFP10 and PE13 correctly identified all infected and noninfected animals. Given that PE13, PE5, and TB10.4 have extremely high specificities, it would be promising to combine these in a cocktail in a diagnostic one-well assay, thereby benefiting from the additive value of having more antigens in the cocktail. A broader range of epitopes should lead to a more robust assay that would benefit field situations with large test numbers and herds of doubtful infection status. In order to illustrate this, nine animals that gave a response only to ESAT6 and not to CFP10 (Fig. 4A, D, and G) or that gave a response to either one of them that was marginally above or that was below the 2-ODI-unit cutoff (Fig. 4B, C, E, F, H, and I) were selected. The ESAT6 and CFP10 responses of these nine animals were measured in combination with their responses to the three highly specific antigens to examine the potential to lead to a more robust positive response. Variations in the abilities of the animals' immune systems to recognize the given peptides made it impossible to select only one additional antigen. For animal B (Fig 4B), PE13 was clearly the most recognized of all five antigens tested. For animal H, TB10.4 would complement ESAT6 and CFP10, and for animal G, only PE5 gave a better value. For animals C and I, none of the three specific antigens had any strong additive value on its own, and it is likely that all three antigens would be required to clearly identify these as M. tuberculosis-infected animals. For animal E, all three antigens were strongly recognized, whereas neither ESAT6 nor CFP10 was recognized. It is therefore likely that the next generation of diagnostic antigen cocktails should contain ESAT6, CFP10, PE13, PE5, and TB10.4.
FIG. 4.
Amount of IFN-γ released in blood after stimulation with PE5, PE13, TB10.4, ESAT6, or CFP10 for 20 h at 37°C with 5% CO2. Blood was collected from nine skin test reactors and was incubated with single mycobacterial antigens at a concentration of 4 μg/ml, after which the release of IFN-γ was measured by ELISA. All antigens were tested singly. Note that the use of PE5, PE13, and TB10.4 can improve the assay readout. (A to I) Results for animals A to I, respectively.
DISCUSSION
In this work, we demonstrate that earlier findings from antigen screening experiments with experimentally infected cattle and the most immunodominant antigens can be extended to the diagnosis of tuberculosis in the field, regardless of the disease prevalence. In addition, a cocktail of carefully selected antigens has the potential to be a novel diagnostic reagent. On the basis of the prevalence of tuberculin skin test reactors, countries have been divided into three categories: (i) those with a prevalence of less than 0.1%, (ii) those with a prevalence of between 0.1% and 1.0%, and (iii) those with a prevalence of greater than 1% or an unknown prevalence (13, 37). Seven recombinant antigens (Table 1) were tested in Northern Ireland (low prevalence), Mexico (medium prevalence), and Argentina (high prevalence), with sensitivities ranging from 5 to 94% (Table 2). This broad range is interesting and gives insight into the immunogenicities of mycobacterial antigens. It is likely that bacterial parameters, such as protein abundance, subcellular localization, posttranslational modifications, and temporal expression/regulation, together with immune parameters, such as localization of the antigen in the phagosome, proteolytic sensitivity, and the presence of major histocompatibility complex binding motifs, play a role in the efficiency of protein epitope presentation and recognition. Given the large site-to-site variations in the sensitivities of several of the proteins tested, it is also likely that the genetic background of the host, environmental sensitizations, and the M. bovis strains involved also play significant roles.
ESAT6 is the best-documented TB antigen among those tested, and sensitivities in the range from 66% to 78% have already been reported in the field in Europe (26, 27, 30, 32). We found an overall specificity of 91% and a sensitivity of 69%, which is at the lower end of the reported interval, due to variations from site to site (Table 2). The gene encoding CFP10 (esxB) is cotranscribed with the ESAT6-encoding esxA gene (6), and the two encoded proteins have been shown to form a stable heterogeneous secreted complex in mycobacteria that acts as a signaling molecule (31). The colocalization is a likely explanation why CFP10 results in the same strong responses as ESAT6 in skin test-positive animals and with minimal background in TB-free animals (Table 2). It is also recognized at approximately the same frequency as ESAT6, but not by exactly the same animals. TB27.4, the third protein transcribed from the RD1 region used in this study, has no known function but was included since it had previously been shown to be detected in samples from experimentally infected animals that were both ESAT6 and CFP10 negative (1). In the field, TB27.4 proved poorly sensitive (5 and 14%) in Northern Ireland and Mexico, but it was more sensitive (72%) in Argentina (Table 2). Unfortunately, the specificity in Argentina was only 56%, and all skin test reactors that were TB27.4 positive were also ESAT6 and/or CFP10 positive.
The PE/PE-PGRS multigene family counts 96 members in M. bovis (16), but little is known about their function or immunogenicity. Since members of the family are present in the mycobacterial cell wall, it has been speculated that they could generate antigenic diversity and thereby help mycobacteria escaping the immune system (11). On the basis of a preliminary screening of seven proteins from the PE subfamily (M. Govaerts, personal communication), two PE proteins were selected for testing (Table 1). PE13 gave highly variable sensitivities at the three sites, ranging from 10% to 78%, whereas PE5 gave sensitivities ranging from 25 to 56%. For both proteins, the sensitivity was the highest with the Argentinean samples (Table 2). PE5 is substantially homologous to PE29 (80%) and PE15 (66%) in M. bovis, and PE13 is substantially homologous to PE19 (75%) and PE18 (71%) in M. bovis. Considering the high degree of homology to PE proteins in environmental mycobacteria (Table 1), it is surprising that no major background problem was found with the TB-free control groups (Fig. 2). Their major immunogenic epitopes are most likely found in the variable regions.
The MPB70 protein is highly homologous to the MPB83 protein (approximately 70% identity), and both can be found in many mycobacterial species, including M. bovis and M. tuberculosis. Their function is unknown; but MPB70 is one of the major antigens secreted from M. bovis, while MPB83 is a cell wall lipoprotein. In vivo, both proteins are expressed at low levels in M. tuberculosis and some BCG strains (such as BCG strains Moscow, Brazil, Sweden, and Tokyo) but at high levels in M. bovis and BCG strains lacking RD2 (e.g., BCG strains Pasteur, Copenhagen, and Glaxo). Both proteins are highly immunogenic after M. bovis infection in mice (22). MPB70 has previously been used in ELISAs to detect antibodies in cattle and was found to have a high specificity (96%) but a relatively low sensitivity (18%) (35). Using the CMI-based whole-blood assay, we found sensitivities of 36% in Northern Ireland, 5% in Mexico, and 56% in Argentina, with specificities of 90, 100, and 83%, respectively (Table 2). MPB70 together with TB27.4 was the least discriminating of the antigens tested, with both antigens having specificities and sensitivities that were quite low. Neither of them has the potential to be used in a single-antigen-based diagnostic assay or, because of a lack of specificity, to be part of a diagnostic cocktail.
The highly immunogenic nature of ESAT6 and CFP10 has previously been reported across a broad range of species, including mice, guinea pigs, humans, and cattle (4, 26, 27, 30, 32). Their potential to differentiate between BCG infection and M. tuberculosis or M. bovis infection has been recognized for several years, and a number of reports have demonstrated the high degrees of specificity and sensitivity imparted by the use of these two proteins with cattle (1, 28, 34) and humans (24, 30).
In order to increase their overall sensitivities, they were tested not only as individual antigens but also as a cocktail. The combination of the two led to an impressive synergistic effect (Fig. 2 and 3), with increased responses in the skin test-positive groups and decreased responses in the skin test-negative groups compared with the responses to the antigens tested singly. With the samples from Northern Ireland, the sensitivity increased to 73%, with a specificity of 100%, whereas with the samples from Argentina, the specificity increased to 94%, while the sensitivity stayed at 94% (Table 2). Use of the combination of the antigens in a one-well assay was clearly the best-performing system tested in this study, with an overall specificity of 97% and an overall sensitivity of 85% (Table 2). In order to further improve the sensitivity of the CMI-based whole-blood assay, more antigens will be needed in an optimal diagnostic cocktail. Among the proteins tested in this study, TB10.4, PE5, and PE13 are the obvious choices. They have reasonably high sensitivities, but more importantly, they all have very high specificities, regardless of the BTB prevalence and other environmental factors. Furthermore, in several cases they will add to the strength of the response in infected animals, especially when the responses to ESAT6 and CFP10 are weak, thereby, at the same time, making the assay much more robust, presumably by an increase in the number of epitopes detected (Fig. 4).
Of 124 animals whose infection status could not be confirmed by postmortem confirmatory tests, we found 47 skin test-positive animals (95% confidence interval [CI], 29 to 46%). More than half of these (27 of 49; 95% CI, 43 to 71%) were from the low-incidence area (Northern Ireland), 15 of 36 (95% CI, 26 to 58%) were from the medium-incidence area (Mexico), and only 5 of 39 (95% CI, 2 to 23%) were from the high-incidence area (Argentina). One explanation for the discrepancy could be that there is a direct relationship between exposure, infection, and how rapidly lesions develop, combined with the application of different rates of removal of reactor cattle among these areas.
Since the CFP10 and ESAT6 antigens are much more specific than the PPD used in the skin test, we used the CMI-based assay to examine the responses to both antigens. Twenty-five of the 47 animals were positive for a response to ESAT6 and CFP10 in a one-well assay, whereas the remaining 22 animals were negative by the CMI-based test, regardless of the antigen used. Given that CMI assays based on the IFN-γ readout have been found to be particularly useful for the detection of bovine tuberculosis during the early stages of infection (29) and that both the ESAT6 and the CFP10 antigens are useful for estimation of the degree of latent infection in humans (15, 23), animals that are skin test and CMI test positive should be considered highly suspect of being infected. Such subclinically infected animals may account for the significant number of reactor cattle appearing at slaughter without lesions. Furthermore, these subclinical reactor cattle could, over time, develop disease, contributing to a significant number of herd breakdowns which are difficult to attribute to a precise source.
This is the first reported field trial to have used several recombinant antigens in three countries across the world with different disease prevalences. Given the high specificity and sensitivity obtained with the ESAT6-CFP10 mixture and the ability of CMI-based assays to detect bovine TB infections at early times, we consider this method of great value for the routine testing of cattle, especially if the sensitivity were to be raised even further by including more antigens. On the basis of the findings from this work, we suggest that a specific cocktail consisting of ESAT6, CFP10, PE5, PE13, and TB10.4 be tested in the near future with large numbers of field animals.
Acknowledgments
We gratefully acknowledge the assistance of Vivi Andersen and Cristina Parada for excellent technical help with this study.
This work was supported by INCO-EC grant CT2000-30023.
Footnotes
Published ahead of print on 27 September 2006.
REFERENCES
- 1.Aagaard, C., M. Govaerts, L. Meng Okkels, P. Andersen, and J. M. Pollock. 2003. Genomic approach to identification of Mycobacterium bovis diagnostic antigens in cattle. J. Clin. Microbiol. 41:3719-3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abernethy, D. A., G. O. Denny, F. D. Menzies, P. McGuckian, N. Honhold, and A. R. Roberts. 2006. The Northern Ireland programme for the control and eradication of Mycobacterium bovis. Vet. Microbiol. 112:231-237. [DOI] [PubMed] [Google Scholar]
- 3.Amadori, M., S. Tagliabue, S. Lauzi, G. Finazzi, G. Lombardi, P. Telo, L. Pacciarini, and L. Bonizzi. 2002. Diagnosis of Mycobacterium bovis infection in calves sensitized by mycobacteria of the avium/intracellulare group. J. Vet. Med. B Infect. Dis. Vet. Public Health 49:89-96. [DOI] [PubMed] [Google Scholar]
- 4.Andersen, P., A. B. Andersen, A. L. Sorensen, and S. Nagai. 1995. Recall of long-lived immunity to Mycobacterium tuberculosis infection in mice. J. Immunol. 154:3359-3372. [PubMed] [Google Scholar]
- 5.Behr, M. A., M. A. Wilson, W. P. Gill, H. Salamon, G. K. Schoolnik, S. Rane, and P. M. Small. 1999. Comparative genomics of BCG vaccines by whole-genome DNA microarray. Science 284:1520-1523. [DOI] [PubMed] [Google Scholar]
- 6.Berthet, F. X., P. B. Rasmussen, I. Rosenkrands, P. Andersen, and B. Gicquel. 1998. A Mycobacterium tuberculosis operon encoding ESAT-6 and a novel low-molecular-mass culture filtrate protein (CFP-10). Microbiology 144(Pt 11):3195-3203. [DOI] [PubMed] [Google Scholar]
- 7.Buddle, B. M., N. A. Parlane, D. L. Keen, F. E. Aldwell, J. M. Pollock, K. Lightbody, and P. Andersen. 1999. Differentiation between Mycobacterium bovis BCG-vaccinated and M. bovis-infected cattle by using recombinant mycobacterial antigens. Clin. Diagn. Lab. Immunol. 6:1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buddle, B. M., B. J. Wards, F. E. Aldwell, D. M. Collins, and G. W. de Lisle. 2002. Influence of sensitisation to environmental mycobacteria on subsequent vaccination against bovine tuberculosis. Vaccine 20:1126-1133. [DOI] [PubMed] [Google Scholar]
- 9.Buddle, B. M., D. N. Wedlock, N. A. Parlane, L. A. Corner, G. W. De Lisle, and M. A. Skinner. 2003. Revaccination of neonatal calves with Mycobacterium bovis BCG reduces the level of protection against bovine tuberculosis induced by a single vaccination. Infect. Immun. 71:6411-6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cockle, P. J., S. V. Gordon, A. Lalvani, B. M. Buddle, R. G. Hewinson, and H. M. Vordermeier. 2002. Identification of novel Mycobacterium tuberculosis antigens with potential as diagnostic reagents or subunit vaccine candidates by comparative genomics. Infect. Immun. 70:6996-7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, A. Krogh, J. McLean, S. Moule, L. Murphy, K. Oliver, J. Osborne, M. A. Quail, M. A. Rajandream, J. Rogers, S. Rutter, K. Seeger, J. Skelton, R. Squares, S. Squares, J. E. Sulston, K. Taylor, S. Whitehead, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 12.Cosivi, O., J. M. Grange, C. J. Daborn, M. C. Raviglione, T. Fujikura, D. Cousins, R. A. Robinson, H. F. Huchzermeyer, I. de Kantor, and F. X. Meslin. 1998. Zoonotic tuberculosis due to Mycobacterium bovis in developing countries. Emerg. Infect. Dis. 4:59-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Kantor, I. N., and V. Ritacco. 2006. An update on bovine tuberculosis programmes in Latin American and Caribbean countries. Vet. Microbiol. 112:111-118. [DOI] [PubMed] [Google Scholar]
- 14.Delogu, G., and M. J. Brennan. 2001. Comparative immune response to PE and PE_PGRS antigens of Mycobacterium tuberculosis. Infect. Immun. 69:5606-5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doherty, T. M., A. Demissie, J. Olobo, D. Wolday, S. Britton, T. Eguale, P. Ravn, and P. Andersen. 2002. Immune responses to the Mycobacterium tuberculosis-specific antigen ESAT-6 signal subclinical infection among contacts of tuberculosis patients. J. Clin. Microbiol. 40:704-706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garnier, T., K. Eiglmeier, J. C. Camus, N. Medina, H. Mansoor, M. Pryor, S. Duthoy, S. Grondin, C. Lacroix, C. Monsempe, S. Simon, B. Harris, R. Atkin, J. Doggett, R. Mayes, L. Keating, P. R. Wheeler, J. Parkhill, B. G. Barrell, S. T. Cole, S. V. Gordon, and R. G. Hewinson. 2003. The complete genome sequence of Mycobacterium bovis. Proc. Natl. Acad. Sci. USA 100:7877-7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gilbert, M., A. Mitchell, D. Bourn, J. Mawdsley, R. Clifton-Hadley, and W. Wint. 2005. Cattle movements and bovine tuberculosis in Great Britain. Nature 435:491-496. [DOI] [PubMed] [Google Scholar]
- 18.Gordon, S. V., R. Brosch, A. Billault, T. Garnier, K. Eiglmeier, and S. T. Cole. 1999. Identification of variable regions in the genomes of tubercle bacilli using bacterial artificial chromosome arrays. Mol. Microbiol. 32:643-655. [DOI] [PubMed] [Google Scholar]
- 19.Harboe, M., T. Oettinger, H. G. Wiker, I. Rosenkrands, and P. Andersen. 1996. Evidence for occurrence of the ESAT-6 protein in Mycobacterium tuberculosis and virulent Mycobacterium bovis and for its absence in Mycobacterium bovis BCG. Infect. Immun. 64:16-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hardie, R. M., and J. M. Watson. 1992. Mycobacterium bovis in England and Wales: past, present and future. Epidemiol. Infect. 109:23-33. [PMC free article] [PubMed] [Google Scholar]
- 21.Hermans, P. W., D. van Soolingen, J. W. Dale, A. R. Schuitema, et al. 1990. Insertion element IS986 from Mycobacterium tuberculosis: a useful tool for diagnosis and epidemiology of tuberculosis. J. Clin. Microbiol. 28:2051-2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hewinson, R. G., S. L. Michell, W. P. Russell, R. A. McAdam, and W. R. Jacobs, Jr. 1996. Molecular characterization of MPT83: a seroreactive antigen of Mycobacterium tuberculosis with homology to MPT70. Scand. J. Immunol. 43:490-499. [DOI] [PubMed] [Google Scholar]
- 23.Lalvani, A., P. Nagvenkar, Z. Udwadia, A. A. Pathan, K. A. Wilkinson, J. S. Shastri, K. Ewer, A. V. Hill, A. Mehta, and C. Rodrigues. 2001. Enumeration of T cells specific for RD1-encoded antigens suggests a high prevalence of latent Mycobacterium tuberculosis infection in healthy urban Indians. J. Infect. Dis. 183:469-477. [DOI] [PubMed] [Google Scholar]
- 24.Lalvani, A., A. A. Pathan, H. McShane, R. J. Wilkinson, M. Latif, C. P. Conlon, G. Pasvol, and A. V. Hill. 2001. Rapid detection of Mycobacterium tuberculosis infection by enumeration of antigen-specific T cells. Am. J. Respir. Crit. Care Med. 163:824-828. [DOI] [PubMed] [Google Scholar]
- 25.Lightbody, K. A., R. M. Girvin, D. P. Mackie, S. D. Neill, and J. M. Pollock. 1998. T-cell recognition of mycobacterial proteins MPB70 and MPB64 in cattle immunized with antigen and infected with Mycobacterium bovis. Scand. J. Immunol. 48:44-51. [DOI] [PubMed] [Google Scholar]
- 26.Mustafa, A. S., F. Oftung, H. A. Amoudy, N. M. Madi, A. T. Abal, F. Shaban, I. Rosen Krands, and P. Andersen. 2000. Multiple epitopes from the Mycobacterium tuberculosis ESAT-6 antigen are recognized by antigen-specific human T cell lines. Clin. Infect. Dis. 30(Suppl. 3):S201-S205. [DOI] [PubMed] [Google Scholar]
- 27.Pollock, J. M., and P. Andersen. 1997. The potential of the ESAT-6 antigen secreted by virulent mycobacteria for specific diagnosis of tuberculosis. J. Infect. Dis. 175:1251-1254. [DOI] [PubMed] [Google Scholar]
- 28.Pollock, J. M., R. M. Girvin, K. A. Lightbody, R. A. Clements, S. D. Neill, B. M. Buddle, and P. Andersen. 2000. Assessment of defined antigens for the diagnosis of bovine tuberculosis in skin test-reactor cattle. Vet. Rec. 146:659-665. [DOI] [PubMed] [Google Scholar]
- 29.Pollock, J. M., and S. D. Neill. 2002. Mycobacterium bovis infection and tuberculosis in cattle. Vet. J. 163:115-127. [DOI] [PubMed] [Google Scholar]
- 30.Ravn, P., A. Demissie, T. Eguale, H. Wondwosson, D. Lein, H. A. Amoudy, A. S. Mustafa, A. K. Jensen, A. Holm, I. Rosenkrands, F. Oftung, J. Olobo, F. von Reyn, and P. Andersen. 1999. Human T cell responses to the ESAT-6 antigen from Mycobacterium tuberculosis. J. Infect. Dis. 179:637-645. [DOI] [PubMed] [Google Scholar]
- 31.Renshaw, P. S., K. L. Lightbody, V. Veverka, F. W. Muskett, G. Kelly, T. A. Frenkiel, S. V. Gordon, R. G. Hewinson, B. Burke, J. Norman, R. A. Williamson, and M. D. Carr. 2005. Structure and function of the complex formed by the tuberculosis virulence factors CFP-10 and ESAT-6. EMBO J. 24:2491-2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Pinxteren, L. A., P. Ravn, E. M. Agger, J. Pollock, and P. Andersen. 2000. Diagnosis of tuberculosis based on the two specific antigens ESAT-6 and CFP10. Clin. Diagn. Lab. Immunol. 7:155-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vordermeier, H. M., P. C. Cockle, A. Whelan, S. Rhodes, N. Palmer, D. Bakker, and R. G. Hewinson. 1999. Development of diagnostic reagents to differentiate between Mycobacterium bovis BCG vaccination and M. bovis infection in cattle. Clin. Diagn. Lab. Immunol. 6:675-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vordermeier, H. M., A. Whelan, P. J. Cockle, L. Farrant, N. Palmer, and R. G. Hewinson. 2001. Use of synthetic peptides derived from the antigens ESAT-6 and CFP-10 for differential diagnosis of bovine tuberculosis in cattle. Clin. Diagn. Lab. Immunol. 8:571-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wood, P. R., L. A. Corner, J. S. Rothel, J. L. Ripper, T. Fifis, B. S. McCormick, B. Francis, L. Melville, K. Small, K. de Witte, et al. 1992. A field evaluation of serological and cellular diagnostic tests for bovine tuberculosis. Vet. Microbiol. 31:71-79. [DOI] [PubMed] [Google Scholar]
- 36.Wood, P. R., and J. S. Rothel. 1994. In vitro immunodiagnostic assays for bovine tuberculosis. Vet. Microbiol. 40:125-135. [DOI] [PubMed] [Google Scholar]
- 37.World Organization for Animal Health. 2004. Annual animal disease status, bovine tuberculosis. World Organization for Animal Health, Paris, France.