Abstract
Shigella and pathogenic Escherichia coli are major causes of human infectious diseases and are responsible for millions of cases of diarrhea worldwide every year. A convenient and rapid method to identify highly pathogenic serotypes of Shigella and E. coli is needed for large-scale epidemiologic study, timely clinical diagnosis, and reliable quarantine of the pathogens. In this study, a DNA microarray targeting O-serotype-specific genes was developed to detect 15 serotypes of Shigella and E. coli, including Shigella sonnei; Shigella flexneri type 2a; Shigella boydii types 7, 9, 13, 16, and 18; Shigella dysenteriae types 4, 8, and 10; and E. coli O55, O111, O114, O128, and O157. The microarray was tested against 186 representative strains of all Shigella and E. coli O serotypes, 38 clinical isolates, and 9 strains of other bacterial species that are commonly present in stool samples and was shown to be specific and reproducible. The detection sensitivity was 50 ng genomic DNA or 104 CFU per ml in mock stool specimens. This is the first report of a microarray for serotyping Shigella and pathogenic E. coli. The method has a number of advantages over traditional bacterial culture and antiserum agglutination methods and is promising for applications in basic microbiological research, clinical diagnosis, food safety, and epidemiological surveillance.
Shigella and pathogenic Escherichia coli are the major causative agents of diarrhea. E. coli clones are normally classified by a combination of oligosaccharide (O), flagellar (H), and capsular (K) antigens. Shigella clones are classified by the O antigen only, as they lack H and K antigens (14). There are four named species of Shigella classified on the basis of biochemical and O-antigen serological differences: Shigella dysenteriae (consisting of 13 serotypes), Shigella flexneri (consisting of 14 serotypes [including subtypes]), Shigella boydii (consisting of 18 serotypes), and Shigella sonnei (27). Of the estimated 165 million cases of Shigella diarrhea per year, 69% of the episodes occurred in children under 5 years of age, and 1.1 million deaths were attributed to Shigella infections (24). In the United States, most infections are caused by S. sonnei; S. flexneri is the second most common serotype (4, 24).
The E. coli species consist of various serotypes, ranging from highly pathogenic to nonpathogenic strains, which are from normal intestinal flora and are often used as safe laboratory strains (32). There are five major pathotypes of E. coli strains that cause diarrhea in humans: enteropathogenic, enterotoxigenic, enteroinvasive, enteroaggregative, and enterohemorrhagic (28). These pathotypes consist of genetic clones that often correspond to distinct O:H serotypes (15). Serotypes O55, O111, O114, and O128 belong to the group of enteropathogenic E. coli-associated O serotypes (23). E. coli O157:H7 is one of the enterohemorrhagic E. coli strains, and it causes significant illness and represents a serious public health threat worldwide (5).
The O antigen, which consists of repeats of the O unit, is part of the lipopolysaccharide in the outer membrane of gram-negative bacteria and contributes major antigenic variability to the cell surface. In Shigella and E. coli, genes involved in the biosynthesis of the O antigen are normally clustered in the chromosome between two housekeeping genes, galF and gnd, and are classified into three main classes: the nucleotide sugar biosynthesis pathway genes, glycosyltransferase genes, and O-unit processing genes encoding flippase and polymerase (wzx and wzy) (14). Glycosyltransferase genes encode enzymes for the transfer of sugars to build the O unit (12). The role of Wzx is to translocate, or flip, the O units formed at the cytoplasmic face of the inner membrane to the periplasmic face. The O units are then polymerized by Wzy to form a long-chain O antigen at the periplasmic face of the membrane (19). The diverse forms of O antigen are almost entirely due to genetic variation in the O-antigen gene cluster (29). Due to the relatively low similarity of glycosyltransferase, wzx, and wzy genes among different serotypes, their sequences are normally highly specific to individual O antigens (7, 8, 11, 14, 30).
The highly variable nature of the O antigen provides the basis for serotyping, and more than 160 different serotypes (1, 8, 28, 33, 35) have been recognized in E. coli. Traditional serotyping requires the use of a large panel of antisera; moreover, it is subjective and cross-reactive (16). In recent years, PCR assays based on O-serotype-specific genes have been proposed by us and others for molecular typing of many Shigella and E. coli O serotypes (2, 3, 7, 8, 10, 11, 12, 15, 18, 31, 34, 36). Molecular typing has many advantages over the traditional method. However, it is difficult to quantify PCR products and to differentiate bands of similar size in a multiplex PCR mixture.
The oligonucleotide-based microarray assay is an efficient approach for parallel analyses of a large number of specific sequences (37). In this study, we developed a DNA microarray based on the target genes wzx, wzy, and wfaU (encoding glycosyltransferase) of the 15 Shigella and E. coli serotypes: S. sonnei; S. flexneri type 2a; S. boydii types 7, 9, 13, 16, and 18; S. dysenteriae types 4, 8, and 10; and E. coli O55, O111, O114, O128, and O157. The DNA microarray method described in this communication is specific, sensitive, and reliable and serves as a prototype for an array of all serotypes of Shigella and E. coli in our future work.
MATERIALS AND METHODS
Bacterial strains.
The strains used in this study include 186 representative strains of all Shigella and E. coli O serotypes (15), 38 clinical isolates, and 9 strains of Salmonella enterica, Staphylococcus aureus, Bacillus cereus, and Vibrio cholerae (Table 1). Serotypes of the 38 clinical isolates were identified using commercial antisera from the Chengdu Institute of Biological Products, China (data not shown).
TABLE 1.
Shigella and E. coli clinical isolates and strains of other bacterial species
| Bacterium | Serotype | No. of strains of each source | Total no. | |||
|---|---|---|---|---|---|---|
|
Shigella and E. coli clinical isolates used for blind testing of the microarray (n = 38)
|
||||||
| S. sonnei | 1a, 2b | 3 | ||||
| S. flexneri | Type 2a | 1a, 1b | 2 | |||
| S. boydii | Type 7 | 1a, 1b | 2 | |||
| Type 9 | 1a, 1b | 2 | ||||
| Type 13 | 1a, 1b | 2 | ||||
| Type 16 | 1a, 1b | 2 | ||||
| Type 18 | 1a, 1b | 2 | ||||
| S. dysenteriae | Type 4 | 1a, 1b | 2 | |||
| Type 8 | 1a, 1b | 2 | ||||
| Type 10 | 1a, 1b, 1c | 3 | ||||
| E. coli | O55 | 1a, 1c | 2 | |||
| O111 | 1a | 1 | ||||
| O114 | 1a, 7d | 8 | ||||
| O128 | 1a | 1 | ||||
| O157 | 1a, 3d | 4 | ||||
| Other bacterial species used to test the specificity of the probes (n = 9)
|
||||||
| Salmonella enterica | 3a | 3 | ||||
| Staphylococcus aureus | 1c, 1e | 2 | ||||
| Bacillus cereus | 2e | 2 | ||||
| Vibrio cholerae | 2a | 2 | ||||
Institute of Medical and Veterinary Science, Adelaide, Australia.
Institute of Epidemiology and Microbiology, Chinese Academy of Preventive Medicine, Beijing, China.
National Center for Medical Culture Collection, China.
Robert Koch-Institut, Germany.
Institute of Microbiology, Chinese Academy of Sciences, China.
Genomic-DNA extraction.
Genomic DNA was extracted from 1.5 ml of overnight broth culture (approximately 109 CFU) using a DNA extraction kit (Tiangen, Beijing, China). The mock stool specimens were prepared as follows. A serial dilution of bacterial culture of each of the 15 serotype strains in the range of 101 to 106 CFU per ml was prepared and mixed with 0.3 g of stool specimens from adult volunteers. DNA was extracted with a QIAamp Mini Stool Kit (QIAGEN GmbH, Hilden, Germany). For each strain, at least 10 extractions were repeated to verify the reproducibility of the DNA extraction method.
Identification of O-serotype-specific genes by PCR.
Genomic DNAs were prepared from 186 representative strains of all Shigella and E. coli O serotypes and examined for quality by PCR amplification of the mdh gene coding for malate dehydrogenase, as described previously (30). A total of 13 pools of DNA were made, each containing DNAs from 12 to 19 strains (15). The pools were screened using primers based on the wzx and wzy genes of E. coli O128; S. sonnei; S. flexneri type 2a; S. boydii types 7 and 9; and S. dysenteriae types 4, 8, and 10 (Table 2). The PCR cycles used were as follows: 30 cycles of denaturation at 94°C for 2 min, annealing at 45°C for 30 s, and extension at 72°C for 1 min, with a final extension at 72°C for 5 min. An aliquot of 10 μl of the PCR product was examined on an agarose gel.
TABLE 2.
PCR specificity test for E. coli O128, S. sonnei, S. flexneri type 2a , S. boydii types 7 and 9, and S. dysenteriae types 4, 8, and 10
| Serotype | Specific gene | GenBank accession no. | Forward primer sequences (5′-3′) | Reverse primer sequences (5′-3′) | Product size (bp) |
|---|---|---|---|---|---|
| E. coli O128 | wzx | AY217096 | TCTTGCTTATAGCCAGAATT | AATAAACCGACACCGAAA | 1,353 |
| GTGAATCGCAACACTTAT | GCAAACGATAAAGGAGGC | 1,035 | |||
| wzy | ATGATTTCTTACGGAGTGC | CTCTAACCTAATCCCTCCC | 782 | ||
| ATTCTGGTATGCGGTGTT | ATAATTGCTGGGTACATC | 460 | |||
| S. sonnei | wzx | AF285971 | ATTTCATTAACTCTGCTTGT | ACAACCGCTGCTGACCATT | 967 |
| TTGGTCGGTTTAGATGTG | TCCCTACGAAATAGATGC | 516 | |||
| wzy | TGAGGTTTCACGTTTCTC | AATAATCCCTAACTGAGCC | 817 | ||
| CCACCGCAATTATAGTAGT | TACGTATAAAAACCACCGA | 687 | |||
| S. flexneri type 2a | wzx | AY900451 | CACTTGTTGGGTATGCTGG | CCGGCAAACAGATTAGAAA | 782 |
| CTGAAGGTTTCGGTGTTTA | ATTACTTACTGTCATCCAACC | 584 | |||
| wzy | GTGGTGGAAGATTACTGGA | GCTCCAGAAGTGAGGTTAT | 1,084 | ||
| GTGTCGGTGCGATTATCAG | AAGATAAGCAACATAGGAACT | 418 | |||
| S. boydii type 7 | wzx | Laboratory stock | ATTGCTTCCCTATCTTAC | GAGAACTGAGGCTATTTG | 685 |
| TGTGACGACATCCCTATTG | AGTGCTTTATTCAACGCC | 528 | |||
| wzy | TCCTCGTAGGTCAACTCA | AATACAATCCTGCAACAG | 384 | ||
| GGCTCTGCATTATCTGTAAC | ATAAACTTACGACCTGAC | 365 | |||
| S. boydii type 9 | wzx | AF402315 | TTTGTTGGAGGAATGTTGT | TAAACCTTCAGCGACTACA | 804 |
| CTGTTTCTTCCATTATCTGC | TTGTAGATATTTGAGGGT | 1,069 | |||
| wzy | GCGTTGGTTGGTGAAAGAG | TTCCCACAAATCAAACCA | 877 | ||
| TAAGCCACGCTATGTTGA | CTCTTAATTGATTTCCCACA | 736 | |||
| S. dysenteriae type 4 | wzx | Laboratory stock | AACGATTAGTTGGTTGACA | CAATAAATAGACACGCACC | 1,058 |
| CGACTTTGGGAAATGTGGA | GAAGGGTGGAAAACTGGCC | 505 | |||
| wzy | TGTATGCTGGTGGAGGACC | GAACCGTATAGCGGAAAAA | 263 | ||
| TTATGTGGGATATTGCTTC | CATTTTAACCTTCCTTCAT | 719 | |||
| S. dysenteriae type 8 | wzx | Laboratory stock | CTGGTATTTCAGTTGTCAC | CAGAAGCAGCGCCAACCG | 1,096 |
| TGGGTAGTTGGGCAACG | AACCATTAATACTTGCGCC | 869 | |||
| wzy | ATTGGCAACATTCTTTTTCC | CATTGATATAGTTAACACC | 1,139 | ||
| TACCATGAGTTAAATTAT | GTTATTCCCTAAAGACAC | 870 | |||
| S. dysenteriae type 10 | wzx | Laboratory stock | GGAGCATTGGTGGTGT | AGAACGGAAAGTTGGG | 718 |
| TGGCTTGTTATCTGCAGTAT | CTTTTACCAAAACTGACGTG | 728 | |||
| wzy | GACACTGAAAGACTGGCGTT | AAGAAGGTGTTCCAAGCGTA | 623 | ||
| CGCTGTTTCTATATTAATTG | AATTGAAGTGACCAGATAAC | 707 |
Primer design.
The sources of the O-antigen gene cluster sequences of the 15 serotypes are listed in Table 3. Based on the O-serotype-specific genes of the 15 serotypes, the 15 compatible primer pairs in a multiplex PCR were designed: 10 primer pairs for the 10 serotypes of Shigella and 5 primer pairs for the 5 serotypes of pathogenic E. coli (Table 3). There was also one primer pair for amplifying the 16S rRNA genes of Shigella and E. coli. All 16 primer pairs were contained in one multiplex PCR.
TABLE 3.
Multiplex PCR primers and oligonucleotide probes used in the study
| Serotype | Target gene | GenBank accession no. | Primera sequences (5′-3′) | Product size (bp) | Probeb sequence(s) (5′-3′) |
|---|---|---|---|---|---|
| S. flexneri type 2a | wzx | AY900451 | 1F: CACTTGTTGGGTATGCTGG | 782 | 1-1: GGGCAGTGTTTCCAAGGTTAAGTAACATCAAAGACTTTAA |
| 1R: CCGGCAAACAGATTAGAAA | 1-2: CGGTTGGATGACAGTAAGTAATATCATAAGTCCGGTCATG | ||||
| S. sonnei | wzy | AF285971 | 2F: TGAGGTTTCACGTTTCTC | 817 | 2-1: GGCACTGGATTAGGTGTTGCAAATTATGTAAAGGCTA |
| 2R: AATAATCCCTAACTGAGCC | 2-2: TGATTATCGTCGAGTTGAGTTAGTATTTATTGGGGTTGAT | ||||
| 2-3: TGGTTCTTTTGGTGTCATTTTGCATTGGTAAAGATGAGCT | |||||
| S. boydii type 7 | wzx | Laboratory stock | 3F: GTGTTGACTGCTGGATTTC | 572 | 3-1: ATTCCACTTCCATCAATCATATCGTTGATTATTCCTGCTG |
| 3R: CGATATGATTGATGGAAGTG | 3-2: CCCATTGATTTTGCATCAAATAAGTGGCTTTCTTAAAGGA | ||||
| 3-3: CTTGATACTGATAATGGCGTTGAATAAAGCACTAGTTCCA | |||||
| S. boydii type 13 | wzy | AY369140 | 4F: AAAGATTGGTAGCGTCGG | 847 | 4-1: TTATCTTGAGTTTAGTCGAATGCAACGAGTAGTCGCG |
| 4R: TGAAGCCCTGGTAAAGTGC | 4-2: TTATTCTCATATTCGAATGTTACTTTTACCAGCGCAACTG | ||||
| 4-3: GCTGCGGGGAATAGAAATAAGAATCGTCTGTATTACTTTA | |||||
| S. boydii type 16 | wfaU | DQ371800 | 5F: CCATACGGATAATGTTGAG | 698 | 5-1: TTGTATGTAATCGCAAATACTCGCAGTACATGTGGA |
| 5R: TCTTTGTCTTCTCGGCTA | 5-2: CCTGAGCGATTAGAACAACTGTATCTCGAGTCAAGA | ||||
| 5-3: GACATGGGAGTGGAATGAAAACTAAAATTGCTGAGGCTTT | |||||
| 5-4: GTGAGATTGTTGATTGCTTTAGCCGAGAAGACAAAGATAT | |||||
| S. boydii type 18 | wzx | AY948196 | 6F: CAGGGCACAAACTATCT | 1,096 | 6-1: TTTGTCGTGTCTATTGGTATTGTATTCGGGCAATGG |
| 6R: TAATTCGGAATGTGCT | 6-2: TGGAGTTCCATTATCCGAGTGGTATTTTGGTAACAATAAT | ||||
| 6-3: TACTCATTAGGCTTGCTATTTACTGGGCTATTATCAGTTT | |||||
| S. dysenteriae type 4 | wzy | Laboratory stock | 7F: TTTCTGCTTATTCATTATTG | 946 | 7-1: TCGCATTGCTTGGTATTAGAGCTGGTAGTGTTAATT |
| 7R: ACTACCAGCTCTAATACC | 7-2: GCAGCTTCTCTTTACATACGTGCATTATCAGAAACGGGAA | ||||
| 7-3: ACTAGTCTAGATAAAATGGCTAGCTCAGACAATCTTTCTG | |||||
| 7-4: TGGATATTTAATTCGGCTATTCTCTATAGCTAGTGAGCCT | |||||
| S. dysenteriae type 8 | wzy | Laboratory stock | 8F: TTCCCTCTTGTTGTATTGA | 1106 | 8-1: GAGGGTGTGTATGGTATGATTGATTACATATTGGAGGC |
| 8R: ACCTTTATCAATTGCCTCC | 8-2: ACTTCAGTTTCTGGGAACGATAAATTCACACGTTTGC | ||||
| 8-3: TTCCTAAATAATCATCCATTTACTGAGGGTGTGTATGGTA | |||||
| 8-4: CGCCAATTTATTCTGTGCTATTATCGAAATTCACTTCAGT | |||||
| S. dysenteriae type 10 | wzy | Laboratory stock | 9F: CGCTGTTTCTATATTAATTG | 706 | 9-1: ATGGCACTGATAGTAGCGATAAAACTATTTTGATCGGG |
| 9R: AATTGAAGTGACCAGATAAC | 9-2: CGTGCATTAATTGCTATTGTCGTAATTTCTTTCATTGTGG | ||||
| 9-3: GGAACACCTTCTTGGGATTATTTTACGCAACCACTTATTA | |||||
| E. coli O55 | wzy | AF461121 | 10F: GGGAGGAGTATTATCATTAC | 917 | 10-1: AGAGTGAGACGAATAATTGGGTGTTATATAAGCTCTC |
| 10R: ATCAATCTAAAGGTCGGTA | 10-2: GCTTTTGGGGATAACATTATCGATAATACCGACCTTTAGA | ||||
| 10-3: TGAAGCTTTAAAACGTCTAATTATTAGCGGAACTTTCTTG | |||||
| E. coli O111 | wzy | AF078736 | 11F: TTAATGCGGAGGATCTATT | 934 | 11-1: CGGGGATGATATATTATTTGGTTTCACAGCTTGGTG |
| 11R: GTAAGCCCGCAAATCAATC | 11-2: TTACGGTTCTTTATAAGTATTGGTGTGATAGGAGCATTGG | ||||
| 11-3: GCTCCTTTCATTGTTGTAAGTTGTTTGTTACTGTTACA | |||||
| 11-4: TTAAATAACGGCGGACAATATAAGACGTTATATGGACTTC | |||||
| E. coli O114 | wzy | AF573377 | 12F: ACTTTCCCAAGCCCATTA | 852 | 12-1: TTATTGTATGCTTGTTAGTGCTTGTGCTGATTTGTTTTCT |
| 12R: CAGCACAAGCACTAACAAG | 12-2: TGAGATGCTTAAATTAGGTGGATGGAATGTTAATGGG | ||||
| 12-3: GTTGTTCATATGCTCAGGGGAAAATCAGAAGAGTATCTAT | |||||
| 12-4: TGGATGGAATGTTAATGGGTTATTTATTTCAGAAGCATG | |||||
| E. coli O128 | wzy | AY217096 | 13F: ATGATTTCTTACGGAGTGC | 782 | 13-1: TCTGATCTTGGATTAAGTAAGATGTACCCAGCAA |
| 13R: CTCTAACCTAATCCCTCCC | 13-2: CGGTGTTTTGCAAGAGATATAAAAGAGTTAGCTTTAGCAT | ||||
| 13-3: GCTAGGTATTTAGCAAATTCAACAGATTTGGCTGACTTTG | |||||
| E. coli O157 | wzy | AF061251 | 14F: TTGCTGCTGTAGTTTTATTTCTT | 555 | 14-1: CGATTTCTTTCCGACACCAGAGTTAGAAAAGGAATT |
| 14R: TGATGCTTTATTCCCTGTATTCT | 14-2: TAGAGCAAGTTGAAAGTGTTCCATATGTTGTTTCTGAATC | ||||
| 14-3: GTATGCTCGTTGTTTTATCTAAGTTTAGGACAAGACGGAG | |||||
| S. boydii type 9 | wzy | AF402315 | 15F: GCGTTGGTTGGTGAAAGAG | 877 | 15-1: AACTGAGTTCACTTATGGTTCGAGAACCTTTACTCCATTT |
| 15R: TTCCCACAAATCAAACCA | 15-2: GGATTTTAATACAACTGAGTTCACTTATGGTTCGAGAACC | ||||
| 16S rRNA gene | AE000406 | 16F: GACGGGTGAGTAATGTCTGG | 1,251 | 16-1: CGGGAACTCAAAGGAGACTGCCAGTGATAA | |
| 16R: ATCCACGATTACTAGCGATTCC | 16-2: CGGGAACTCAAAGGAGACTGCCAGTGATAAACTGGAG | ||||
| 17: TTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTT | |||||
| 18: TTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTT_Cy3 |
F, forward primer; R, reverse primer.
16-1 and 16-2, positive control probes; 17, negative control probe; 18, positional reference and printing control probe.
Multiplex PCR and labeling of the target genes.
Each multiplex PCR amplification was performed with 30 μl of reaction mixture consisting of 50 to 100 ng of DNA; 1× PCR buffer (50 mM KCl, 10 mM Tris-HCl [pH 8.3]), 0.5 mM MgCl2, 100 μM concentration of deoxynucleoside triphosphate, 1.5 U Taq DNA polymerase, 0.047 μM each of two primers based on the 16S rRNA gene sequence, 0.14 μM each of primers based on each target gene, and 0.15 nM cyanine dye Cy3-dUTP (Amersham Biosciences UK Ltd., Little Chalfont, England). The reaction parameters were 94°C for 5 min; 35 cycles of 94°C for 30 s, 50°C for 1 min, and 72°C for 1 min; and a final extension at 72°C for 5 min. An aliquot of 2 μl of PCR product was run on an agarose gel to check the amplified DNA, and the rest was stored at −20°C in the dark.
Oligonucleotide probe design.
For each serotype, two to four probes were designed with OligoArray 2.0 based on GenBank and an in-house database of all 33 of the O-antigen gene clusters of Shigella and 175 O-antigen gene clusters of E. coli. Two probes based on the 16S rRNA genes of all known Shigella and E. coli strains were designed as positive controls. A probe containing 40 poly(T) oligonucleotides was used as the negative control. A probe labeled with 3′ Cy3 was used as the positional reference and printing control. Each probe was 5′ amino modified. All of the oligonucleotide probes are listed in Table 3.
DNA array preparation.
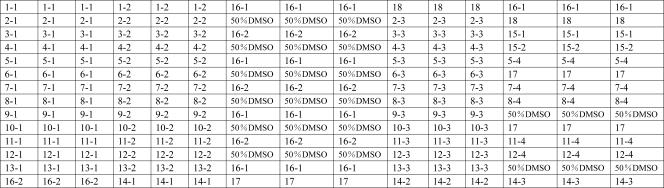
The probes were dissolved in 50% dimethyl sulfoxide at a final concentration of 1 μg/μl and printed onto aldehyde group-modified glass slides (CEL Corporation) using SpotArray72 (Perkin-Elmer Corporation). Each probe was spotted in triplicate. The printed slides were dried for 24 h at room temperature, cross-linked by UV cross-linker (UVP Corporation), and stored at room temperature in the dark. Each slide consisted of four microarrays framed with a 12-μl Geneframe (Beijing Capital Biochip Corporation, Beijing, China), which constituted individual reaction chambers. One of the four microarrays was tested with the positive control standard, 100 ng/μl S. dysenteriae type 8 genomic DNA, to ensure that the reagents were effective. Another was tested with the negative control standard, sterile deionized water, to show that the reagents were uncontaminated. The other two were used to detect samples. A schematic diagram of the probe positions on the microarray is shown in Fig. 1.
FIG. 1.
Probe positions on the slide. Numbers 1-1 to 15-2 are the specific probes for the target strains. Numbers 16-1 and 16-2 are the positive control probes based on the 16S rRNA genes of all Shigella and E. coli strains. Number 17 is the negative control probe. Number 18 is the positional reference and printing control probe.
Hybridization process.
An aliquot of 15 μl of labeled PCR product was baked for about 1.5 h at 65°C until it was dry and diluted in 13 μl of hybridization buffer (25% formamide, 0.1% sodium dodecyl sulfate, 6× SSPE [1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA {pH 7.7}]). After denaturation at 98°C for 5 min, an aliquot of 12 μl of labeled target DNA was hybridized with the probes at 40°C for 16 h. After hybridization, the Geneframe was removed and the slide was washed with solution A (1× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 0.1% sodium dodecyl sulfate) for 3 min, followed by solution B (0.05× SSC) for 3 min, and finally by solution C (95% ethanol) for 1.5 min. The slide was dried under a gentle air stream before it was scanned. For each DNA, at least three hybridization reactions were replicated to verify the reproducibility of the microarray method.
Data acquisition and automated analysis.
The slide was scanned with a laser beam at 532 nm using the GenePix personal 4100A (Axon Instruments), and two files were generated, one for the images, saved as TIF, and the other for the signal intensity, saved as GPR. The signal-to-noise ratio was calculated for each spot using the Bactarray Analyzer 1.0, developed in-house, with the threshold set at 3.0. For each serotype, two to four probes were used, and each probe was printed in triplicate to eliminate any possible physical defects in the glass slide. A serotype was confirmed and reported when the following conditions existed: (i) the positive standard, the negative standard, the two positive control probes, the negative control probe, and the printing control probe all provided the expected signals and (ii) more than half of all the probes of the given serotype generated positive signals above the signal-to-noise ratio threshold.
RESULTS
Identification of specific genes.
The O-serotype-specific genes of each of the S. boydii types 13, 16, and 18 and E. coli O55, O111, O114, and O157 have been reported previously (7, 12, 13, 15, 22, 33, 36). In this study, we identified the specific genes of the other eight serotypes: S. sonnei; S. flexneri type 2a; S. boydii types 7 and 9; S. dysenteriae types 4, 8, and 10; and E. coli O128. For each of them, two primer pairs based on each of the wzx and wzy genes were designed (Table 2) and used to screen DNA pools containing 186 representative strains of all Shigella and E. coli O serotypes. With primer pairs of S. flexneri type 2a, only the pools containing E. coli O13, O129, and O135, which shared the same O antigen with S. flexneri type 2a (9), in addition to S. flexneri 2a, gave PCR products of the expected size. With the other primer pairs, only the bands of correct size were observed in the pool containing the target strains. Therefore, both the wzx and wzy genes were specific to the eight serotypes. For each serotype, one of the O-serotype-specific genes was chosen as a target gene to identify Shigella and E. coli (Table 3).
Multiplex PCR to amplify the target genes.
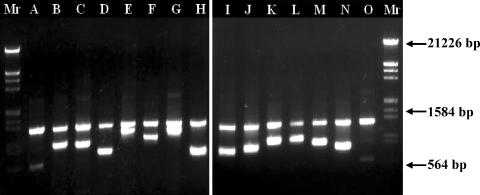
Multiplex PCR was used to streamline the overall test while maintaining specificity for individual amplicons. In the presence of one target strain, only the 16S rRNA gene primer pair and one of the serotype-specific primer pairs performed, while none of the other 14 specific primer pairs annealed with the template DNA. Therefore, for each of the 15 serotype strains, two bands were generated from the PCRs: one was the 16S rRNA gene, and the other was the specific gene (Fig. 2). The amplicon size ranged from 555 bp to about 1.25 kb in length (Table 3). The Shigella and E. coli representative strains belonging to other serotypes (except E. coli O13, O129, and O135 [see below]), S. enterica, and V. cholerae generated only the 16S rRNA gene products. Strains of B. cereus and S. aureus failed to generate any amplicon. The results showed that the 15 primer pairs were specific and compatible in one multiplex PCR.
FIG. 2.
Agrose gel electrophoresis of multiplex PCR products. Lanes: Mr, molecular weight standards (lambda DNA/EcoRI plus HindIII marker); A, S. boydii type 7; B, S. boydii type 9; C, S. boydii type 13; D, S. boydii type 16; E, S. boydii type 18; F, S. dysenteriae type 4; G, S. dysenteriae type 8; H, S. dysenteriae type 10; I, S. flexneri type 2a; J, S. sonnei; K, E. coli O55; L, E. coli O111; M, E. coli O114; N, E. coli O128; O, E. coli O157. Two bands were generated from the PCR products: one was the 16S rRNA gene (1.2 kb), and the other was the gene specific to the individual serotype.
Probe specificity.
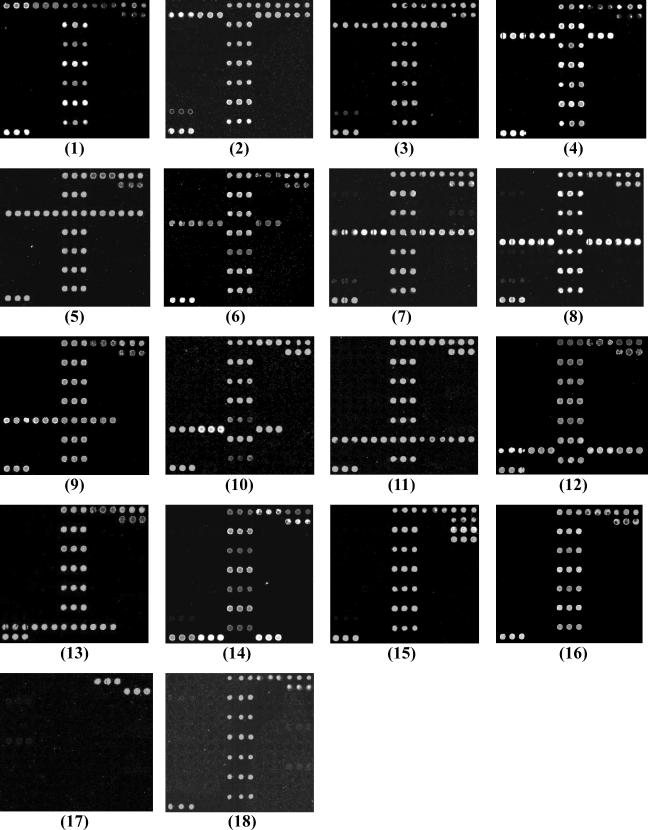
The DNA microarray was tested using 186 representative strains of all Shigella and E. coli O serotypes, 9 strains of other bacterial species (Table 1), and 40 stool specimens. From 141 oligonucleotide probes initially screened, 52 probes were selected for the microarray: 48 probes for specific genes, 2 probes for positive control, 1 for negative control, and 1 for positional reference and printing control (Table 3). All of the representative strains belonging to the 15 serotypes consistently hybridized to their corresponding probes with 100% specificity. The hybridization results are shown in Fig. 3, arrays 1 to 15. Strains of E. coli O13, O129, and O135 gave the same results as S. flexneri type 2a (Fig. 3, array 1), because they had same O antigen (9). None of the 23 representative strains of Shigella and the 148 representative strains of E. coli belonging to other serotypes, or strains of other bacterial species which are likely present in stool samples, hybridized to the serotype-specific probes on the microarray (Fig. 3, arrays 16 and 17). The 40 stool specimens obtained from 20 adult volunteers reacted only to the 16S rRNA gene probes, not the serotype-specific probes (Fig. 3, array 18), a result that was consistent with the fact that large numbers of nonpathogenic E. coli organisms exist in the stools of healthy people.
FIG. 3.
Microarray differentiation of the pathogens. (1) S. flexneri type 2a and E. coli O13, O129, and O135. (2) S. sonnei. (3) S. boydii type 7. (4) S. boydii type 13. (5) S. boydii type 16. (6) S. boydii type 18. (7) S. dysenteriae type 4. (8) S. dysenteriae type 8. (9) S. dysenteriae type 10. (10) E. coli O55. (11) E. coli O111. (12) E. coli O114. (13) E. coli O128. (14) E. coli O157. (15) S. boydii type 9. (16) Other serotype strains of E. coli or Shigella, Salmonella, and V. cholerae. (17) B. cereus and S. aureus. (18) DNAs from healthy-adult stool specimens.
Double-blind test.
A double-blind test was performed in order to verify the stability and specificity of the microarray. A total of 38 clinical isolates of Shigella and E. coli (Table 1) and 70 mock stool samples were selected to hybridize to the microarray without disclosure of their identities during testing. All the detection results were consistent with those of the conventional methods.
Sensitivity of detection with genomic DNA.
Serial dilutions of the genomic DNAs of 15 serotype representative strains in the range of 1 μg, 100 ng, 50 ng, 10 ng, and 1 ng were used as the templates for multiplex PCR to test the sensitivity of the microarray method. The positive signals could be obtained from the dilutions of 1 μg to 10 ng, while the results were negative or the fluorescence signals were very weak with less than 10 ng. We chose 50 ng as the most suitable DNA quantity for this microarray, and all of the 38 clinical isolates belonging to the 15 serotypes could be detected successfully at this level.
Sensitivity of detection with mock stool specimens.
Pure cultures of each of the 15 serotype representative strains were diluted to 101 to 106 CFU per ml, mixed with approximately 0.3 g of fresh stool specimens from healthy people, and tested with the microarray. All of the targets were detected at levels as low as 104 CFU per ml. Some strains, such as E. coli O128, S. sonnei, S. boydii types 16 and 18, and S. dysenteriae types 8 and 10, could be detected successfully at 103 CFU per ml (Table 4).
TABLE 4.
Sensitivity test results for mock stool specimens
| Strain | Test result ata:
|
|||||
|---|---|---|---|---|---|---|
| 106 CFU/ml | 105 CFU/ml | 104 CFU/ml | 103 CFU/ml | 102 CFU/ml | 101 CFU/ml | |
| S. sonnei | + | + | + | + | ± | − |
| S. flexneri type 2a | + | + | + | − | − | − |
| S. boydii type 7 | + | + | + | ± | − | − |
| S. boydii type 9 | + | + | + | − | − | − |
| S. boydii type 13 | + | + | + | ± | ± | − |
| S. boydii type 16 | + | + | + | + | ± | − |
| S. boydii type 18 | + | + | + | + | ± | − |
| S. dysenteriae type 4 | + | + | + | − | − | − |
| S. dysenteriae type 8 | + | + | + | + | ± | − |
| S. dysenteriae type 10 | + | + | + | + | ± | − |
| E. coli O55 | + | + | + | − | − | − |
| E. coli O111 | + | + | + | − | − | − |
| E. coli O114 | + | + | + | ± | − | − |
| E. coli O128 | + | + | + | + | ± | − |
| E. coli O157 | + | + | + | − | − | − |
+, positive signal; −, negative signal; ±, weak or ambiguous signal.
DISCUSSION
Systematic O serotyping of E. coli began in the early 1930s (23), and many studies showed that the O serotypes of E. coli are generally associated with pathogenesis (6, 17, 26, 33, 36). O serotyping became an important tool to classify E. coli in clinical settings. In recent years, some microarray and PCR assays have used toxin genes as targets (20, 23, 25) to identify pathogenic E. coli. However, since mutations, instability, and loss of toxin genes among Shiga-like toxin-producing E. coli are quite common (20, 25), it is not reliable to use the toxin genes, even if a large number of primer pairs are incorporated into the test. wzx, wzy, and glycosyltransferase genes are generally specific to individual O-antigen gene clusters. We previously suggested the application of O-serotype-specific genes for detection and identification of E. coli (14, 30). Here, we demonstrated the feasibility of using a microarray based on wzx, wzy, or transferase genes to identify pathogens from pure culture and more clinically relevant mock stool samples. The DNA microarray described in this communication has paved the way for the establishment of an array of all serotypes of Shigella and E. coli that we are currently working on.
In comparison to the traditional serotyping method, the microarray method is high throughput, specific, and sensitive and also avoids most cross-reactions. Nevertheless, as with traditional serotyping, the microarray has its limits in distinguishing S. sonnei and S. flexneri type 2a from other strains that share the same O-antigen structure and the corresponding O-antigen gene cluster. Among the 46 Shigella serotypes recognized, there are only 33 distinct O antigens. Of the 33 O-antigen forms, 12 are identical to some E. coli O antigens (35). When E. coli O13, O129, or O135 existed in stool samples, the probes for S. flexneri type 2a would have been expected to give positive signals. Similarly, the S. sonnei O-antigen gene cluster is identical in sequence to that of Plesiomonas shigelloides O17 (21). We therefore assumed that when P. shigelloides existed in stool, the probes for S. sonnei would also give false-positive results. Nevertheless, we will try to use other methods or other genes, such as lacZ or cadA (31), which are commonly present in E. coli but absent in Shigella, to differentiate these potential false positives.
The microarray has many advantages over traditional bacterial culture and serotyping methods and is applicable to many fields. First, it facilitates medical detection of Shigella and E. coli from stool samples in a timely fashion. Second, it allows efficient inspection for food contamination. Third, it will be an invaluable tool to sensitively monitor and accurately pinpoint causative bacterial strains to prevent the occurrence or spread of epidemics of bacterial infection.
Acknowledgments
This work was supported by the NSFC Programs (30370023, 30370339, and 30530010), the Tianjin Municipal Special Fund for Science and Technology Innovation (05FZZDSH00800), and funds from the Tianjin Municipal Science and Technology Committee (06YFJZJC02200) to L.W. and L.F.
Footnotes
Published ahead of print on 4 October 2006.
REFERENCES
- 1.Bastin, D. A., and P. R. Reeves. 1995. Sequence and analysis of the O antigen gene (rfb) cluster of Escherichia coli O111. Gene 164:17-23. [DOI] [PubMed] [Google Scholar]
- 2.Beutin, L., Q. Kong, L. Feng, Q. Wang, G. Krause, L. Leomil, Q. Jin, and L. Wang. 2005. Development of PCR assays targeting the genes involved in synthesis and assembly of the new Escherichia coli O174 and O177 O antigens. J. Clin. Microbiol. 43:5143-5149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beutin, L., J. Tao, L. Feng, G. Krause, S. Zimmermann, K. Gleier, Q. Xia, and L. Wang. 2005. Sequence analysis of the Escherichia coli O15 antigen gene cluster and development of a PCR assay for rapid detection of intestinal and extraintestinal pathogenic E. coli O15 strains. J. Clin. Microbiol. 43:703-710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bopp, C. A., F. W. Brenner, P. I. Fields, J. G. Wells, and N. A. Strockbine. 2003. Escherichia, Shigella, and Salmonella, p. 654-671. In P. R. Murray, E. J. Baron, J. H. Jorgensen, M. A. Pfaller, and R. H. Yolken (ed.), Manual of clinical microbiology, 8th ed. American Society for Microbiology, Washington, D.C.
- 5.Call, D. R., F. J. Brockman, and D. P. Chandler. 2001. Detecting and genotyping Escherichia coli O157:H7 using multiplexed PCR and nucleic acid microarrays. Int. J. Food. Microbiol. 67:71-80. [DOI] [PubMed] [Google Scholar]
- 6.Coimbra, R. S., F. Grimont, P. Lenormand, P. Burguiere, L. Beutin, and P. A. Grimont. 2000. Identification of Escherichia coli O-serogroups by restriction of the amplified O-antigen gene cluster (rfb-RFLP). Res. Microbiol. 151:639-654. [DOI] [PubMed] [Google Scholar]
- 7.DebRoy, C., P. M. Fratamico, E. Roberts, M. A. Davis, and Y. Liu. 2005. Development of PCR assays targeting genes in O-antigen gene clusters for detection and identification of Escherichia coli O45 and O55 serogroups. Appl. Environ. Microbiol. 71:4919-4924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DebRoy, C., E. Roberts, J. Kundrat, M. A. Davis, C. E. Briggs, and P. M. Fratamico. 2004. Detection of Escherichia coli serogroups O26 and O113 by PCR amplification of the wzx and wzy genes. Appl. Environ. Microbiol. 70:1830-1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ewing, W. H. 1986. Edwards and Ewing's identification of the Enterobacteriaceae, 4th ed. Elsevier Science Publishers, Amsterdam, The Netherlands.
- 10.Feng, L., W. Han, Q. Wang, D. A. Bastin, and L. Wang. 2005. Characterization of Escherichia coli O86 O-antigen gene cluster and identification of O86-specific genes. Vet. Microbiol. 106:241-248. [DOI] [PubMed] [Google Scholar]
- 11.Feng, L., S. N. Senchenkova, J. Tao, A. S. Shashkov, B. Liu, S. D. Shevelev, P. R. Reeves, J. Xu, Y. A. Knirel, and L. Wang. 2005. Structural and genetic characterization of enterohemorrhagic Escherichia coli O145 O antigen and development of an O145 serogroup-specific PCR assay. J. Bacteriol. 187:758-764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feng, L., S. N. Senchenkova, W. Wang, A. S. Shashkov, B. Liu, S. D. Shevelev, D. Liu, Y. A. Knirel, and L. Wang. 2005. Structural and genetic characterization of the Shigella boydii type 18 O antigen. Gene 355:79-86. [DOI] [PubMed] [Google Scholar]
- 13.Feng, L., S. N. Senchenkova, J. Yang, A. S. Shashkov, J. Tao, H. Guo, G. Zhao, Y. A. Knirel, P. Reeves, and L. Wang. 2004. Structural and genetic characterization of the Shigella boydii type 13 O antigen. J. Bacteriol. 186:383-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feng, L., J. Tao, H. Guo, J. Xu, Y. Li, F. Rezwan, P. Reeves, and L. Wang. 2004. Structure of the Shigella dysenteriae 7 O antigen gene cluster and identification of its antigen specific genes. Microb. Pathog. 36:109-115. [DOI] [PubMed] [Google Scholar]
- 15.Feng, L., W. Wang, J. Tao, H. Guo, G. Krause, L. Beutin, and L. Wang. 2004. Identification of Escherichia coli O114 O-antigen gene cluster and development of an O114 serogroup-specific PCR assay. J. Clin. Microbiol. 42:3799-3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fratamico, P. M., C. E. Briggs, D. Needle, C. Y. Chen, and C. DebRoy. 2003. Sequence of the Escherichia coli O121 O-antigen gene cluster and detection of enterohemorrhagic E. coli O121 by PCR amplification of the wzx and wzy genes. J. Clin. Microbiol. 41:3379-3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gemski, P. J., D. G. Sheahan, O. Washington, and S. B. Formal. 1972. Virulence of Shigella flexneri hybrids expressing Escherichia coli somatic antigens. Infect. Immun. 6:104-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo, H., L. Feng, J. Tao, C. Zhang, and L. Wang. 2004. Identification of Escherichia coli O172 O-antigen gene cluster and development of a serogroup-specific PCR assay. J. Appl. Microbiol. 97:181-190. [DOI] [PubMed] [Google Scholar]
- 19.Guo, H., Q. Kong, J. Cheng, L. Wang, and L. Feng. 2005. Characterization of the Escherichia coli O59 and O155 O-antigen gene clusters: the atypical wzx genes are evolutionary related. FEMS Microbiol. Lett. 248:153-161. [DOI] [PubMed] [Google Scholar]
- 20.Karch, H., T. Meyer, H. Russmann, and J. Heesemann. 1992. Frequent loss of Shiga-like toxin genes in clinical isolates of Escherichia coli upon subcultivation. Infect. Immun. 60:3464-3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lai, V., L. Wang, and P. R. Reeves. 1998. Escherichia coli clone Sonnei (Shigella sonnei) had a chromosomal O-antigen gene cluster prior to gaining its current plasmid-borne O-antigen genes. J. Bacteriol. 180:2983-2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu, B., S. N. Senchenkova, L. Feng, A. V. Perepelov, T. Xu, S. D. Shevelev, Y. Zhu, A. S. Shashkov, M. Zou, Y. A. Knirel, and L. Wang. Structural and molecular characterization of Shigella boydii type 16 O antigen. Gene 380:46-53. [DOI] [PubMed]
- 23.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Niyogi, S. K. 2005. Shigellosis. J. Microbiol. 43:133-143. [PubMed] [Google Scholar]
- 25.Parreira, V. R., and C. L. Gyles. 2002. Shiga toxin genes in avian Escherichia coli. Vet. Microbiol. 87:341-352. [DOI] [PubMed] [Google Scholar]
- 26.Pluschke, G., J. Mayden, M. Achtman, and R. P. Levine. 1983. Role of the capsule and the O-antigen in resistance of O18:K1 Escherichia coli to complement-mediated killing. J. Bacteriol. 42:907-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pupo, G. M., R. Lan, and P. R. Reeves. 2000. Multiple independent origins of Shigella clones of Escherichia coli and convergent evolution of many of their characteristics. Proc. Natl. Acad. Sci. USA 97:10567-10572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scheutz, F., T. Cheasty, D. Woodward, and H. R. Smith. 2004. Designation of O174 and O175 to temporary O groups OX3 and OX7, and six new E. coli O groups that include verocytotoxin-producing E. coli (VTEC): O176, O177, O178, O179, O180 and O181. APMIS 112:569-584. [DOI] [PubMed] [Google Scholar]
- 29.Senchenkova, S. N., L. Feng, J. Yang, A. S. Shashkov, J. Cheng, D. Liu, Y. A. Knirel, P. R. Reeves, Q. Jin, Q. Ye, and L. Wang. 2005. Structural and genetic characterization of the Shigella boydii type 10 and type 6 O antigens. J. Bacteriol. 187:2551-2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tao, J., L. Feng, H. Guo, Y. Li, and L. Wang. 2004. The O-antigen gene cluster of Shigella boydii O11 and functional identification of its wzy gene. FEMS. Microbiol. Lett. 234:125-132. [DOI] [PubMed] [Google Scholar]
- 31.Tao, J., L. Wang, D. Liu, Y. Li, D. A. Bastin, Y. Geng, and L. Feng. 2005. Molecular analysis of Shigella boydii O1 O-antigen gene cluster and its PCR typing. Can. J. Microbiol. 51:387-392. [DOI] [PubMed] [Google Scholar]
- 32.van Ijperen, C., P. Kuhnert, J. Frey, and J. P. Clewley. 2002. Virulence typing of Escherichia coli using microarrays. Mol. Cell Probes 16:371-378. [DOI] [PubMed] [Google Scholar]
- 33.Wang, L., H. Curd, W. Qu, and P. R. Reeves. 1998. Sequencing of Escherichia coli O111 O-antigen gene cluster and identification of O111-specific genes. J. Clin. Microbiol. 36:3182-3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang, L., B. Liu, Q. Kong, H. Steinruck, G. Krause, L. Beutin, and L. Feng. 2005. Molecular markers for detection of pathogenic Escherichia coli strains belonging to serogroups O138 and O139. Vet. Microbiol. 111:181-190. [DOI] [PubMed] [Google Scholar]
- 35.Wang, L., W. Qu, and P. R. Reeves. 2001. Sequence analysis of four Shigella boydii O-antigen loci: implication for Escherichia coli and Shigella relationships. Infect. Immun. 69:6923-6930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang, L., and P. R. Reeves. 1998. Organization of Escherichia coli O157 O antigen gene cluster and identification of its specific genes. Infect. Immun. 66:3545-3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yue, J., W. Shi, J. Xie, Y. Li, E. Zeng, L. Liang, and H. Wang. 2004. Detection of rifampin-resistant Mycobacterium tuberculosis strains by using a specialized oligonucleotide microarray. Diagn. Microbiol. Infect. Dis. 48:47-54. [DOI] [PubMed] [Google Scholar]