Abstract
A high rate of double point mutations in gyrA (56% of 87 ofloxacin-resistant Mycobacterium tuberculosis clinical isolates) indicates the emergence of fluoroquinolone resistance. This is the first report to describe denaturing high-pressure liquid chromatography analysis of mutations in gyrA of M. tuberculosis in a large number of clinical isolates.
Up to the present, fluoroquinolones have been studied as a first-line treatment for tuberculosis (9). However, fluoroquinolone resistance among Mycobacterium tuberculosis strains is emerging, with important implications for treatment (4, 6). Fluoroquinolones have been widely used for tuberculosis treatment in China for more than 10 years and have been given routinely as monotherapy for the empirical treatment of numerous outpatient infections. Thus, China may be one of the countries with the highest rate of fluoroquinolone abuse and resistance in the world. The goal of this work was to identify quinolone resistance-determining regions (QRDRs) of gyrA in ofloxacin-resistant isolates from China by denaturing high-pressure liquid chromatography (DHPLC) and DNA sequencing methods.
(Most of this study was presented at the 41st US-Japan Cooperative Medical Science Program Tuberculosis and Leprosy Research Conference at Kagoshima, Japan, in July 2006.)
The 109 clinical isolates (87 shown to be ofloxacin resistant and 22 shown to be susceptible by a routine proportional method) were collected from patients with pulmonary tuberculosis (65 males and 44 females, aged 17 to 73 years, with 2 to 6 months of fluoroquinolone treatment) over a period of 2 years (2002 to 2003) at the Beijing Tuberculosis and Lung Tumor Research Institute, Tongzhou, China. MICs of ofloxacin were detected by an absolute concentration method in Lowenstein-Jensen medium, and the concentrations were 0.125, 0.25, 1, 2, 4, 8, 10, 16, 20, and 32 μg/ml. For DHPLC analysis, M. tuberculosis H37Rv (ATCC 25618) and M. tuberculosis Erdman (ATCC 35801) were used as reference strains. DHPLC was performed with a WAVE DNA fragment analysis system (Transgenomic Inc.). The melting temperature for gyrA analysis was 67.7°C. The conditions for DNA hybridization and DHPLC analysis have been described in detail elsewhere (10). For DNA sequencing, a 227-bp DNA fragment corresponding to the QRDR was generated by PCR with the following primer set: forward, 5′-GACCGCAGCCACGCCAAG-3′, and reverse, 5′-AGCATCACCATCGCCAACG-3′. After purification, the PCR product (5 ng) was used as a template for TaqCycle sequencing using ABI Prism BigDye Terminator sequencing kits (Applied Biosystems). Cycle sequencing products were subsequently analyzed on an ABI PRISM 310 genetic analyzer (Applied Biosystems).
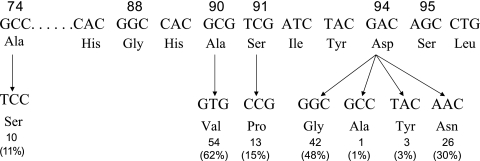
gyrA mutations were found to occur predominantly at codons 90, 91, and 94 and in four types of codon 94 mutation (94Asp→Gly, Ala, Tyr, and Asn) (Fig. 1), largely confirming the findings of other researchers (1, 2, 7, 11, 12). The previously reported mutation involving codon 88 was not found (5). All of the 109 clinical isolates had a codon 95 ACC natural polymorphism, which paralleled the results for 138 other isolates from China (2). However, two new findings were unexpected. One was that 49 of the 87 ofloxacin-resistant isolates (56%) carried double point mutations, and the other was that among these double-mutated isolates, 20% (10/49) harbored an Ala74Ser mutation (Fig. 2), which has not been reported previously for M. tuberculosis. Double point mutation of gyrA is relatively rare (2, 5, 12) and is generally thought to be uncommon in clinical isolates. The Ala74Ser mutation has been reported only for other bacteria (8, 12). This indicates that fluoroquinolone resistance is already emerging in China.
FIG. 1.
Nucleotide sequence and missense mutations within the QRDRs of gyrA. All the isolates contain a naturally occurring polymorphism, codon 95 AGC→ACC. Seventy-three (84%) of the 87 ofloxacin-resistant clinical isolates were found to carry a codon 94 mutation, and 49 (56%) were found to harbor a double point mutation.
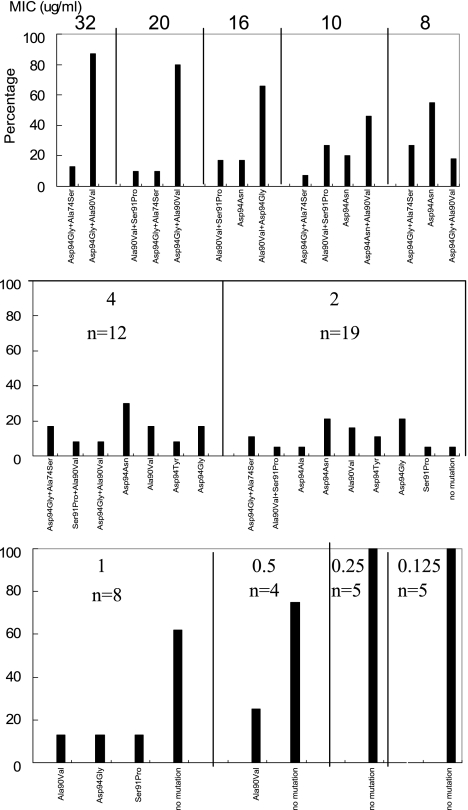
FIG. 2.
Ofloxacin MIC relative to the gyrA QRDR allele spectrum. Ofloxacin MICs are given above each panel. n, number in each MIC group. Bars indicate the percentage represented by each allele.
DHPLC analysis.
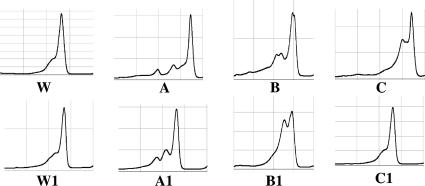
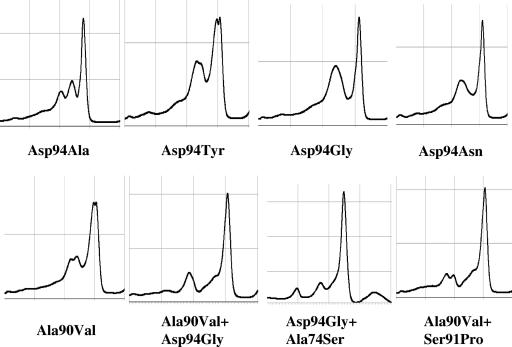
First, M. tuberculosis H37Rv was routinely used as a reference strain, and this revealed that all 109 isolates carried mutations (aberrant peak patterns in Fig. 3). DNA sequencing showed that all the strains possessed a natural codon 95 AGC→ACC (Ser→Thr) polymorphism, which did not have a significant impact on fluoroquinolone susceptibility (4). To improve the DHPLC detection capacity, the other reference strains were selected from H37Ra, M. tuberculosis Kuruno, M. tuberculosis Erdman, and Mycobacterium bovis BCG Pasteur (data not shown). We found that M. tuberculosis Erdman (fluoroquinolone susceptible, with codon 95 ACC in gyrA QRDRs) was the best as the second reference strain in this study. Those isolates with only the codon 95 AGC→ACC polymorphism showed a normal peak (Fig. 3). Thus, the influence of this natural polymorphism was successfully avoided. When M. tuberculosis H37Rv and M. tuberculosis Erdman reference strains were used, a wild-type peak pattern appeared, indicating no point mutation in gyrA QRDRs. Of course, if an isolate carries any point mutation at a codon except codon 95, an aberrant peak pattern will appear. One interesting thing is that most of the isolates with the same mutation showed the same DHPLC patterns. The peak profiles of each mutant are shown in Fig. 4. Asp94Gly and Asp94Asn changes revealed similar patterns that were difficult to distinguish from each other. Other mutations had their own peak patterns. Therefore, it is thought that, to some extent, specific DHPLC patterns may predict the types of resistance.
FIG. 3.
DHPLC patterns of gyrA genes of the 109 clinical isolates when M. tuberculosis H37Rv was used as a reference strain. Patterns A, B, and C are shown as examples (details are shown in Fig. 4). W, H37Rv wild type. When M. tuberculosis Erdman was used as a reference strain, A, B, and C were changed to A1, B1, and C1, respectively. W1 indicates the M. tuberculosis Erdman wild type. Isolates (MIC less than 2 μg/ml) that harbored only the codon 95 ACC natural polymorphism with no other mutation in gyrA QRDRs showed the wild-type pattern C1.
FIG. 4.
Specific DHPLC pattern of each gyrA QRDR mutation type.
DHPLC is a relatively new technique utilizing heteroduplex formation between wild-type and mutated DNA strands to identify mutations and was predicted to be a potentially useful genotypic screening method for gene mutations conferring drug resistance in M. tuberculosis (3, 8, 10). In this study, by introducing M. tuberculosis H37Rv and M. tuberculosis Erdman as two reference strains, the interference from the codon 95 AGC→ACC natural polymorphism was successfully avoided. Since no other polymorphism has been found in gyrA QRDRs except for that in codon 95, and all the point mutations in codons 74, 88, 90, and 91 correlate with fluoroquinolone resistance, the DHPLC method devised in this study can be regarded as a useful and powerful tool for analysis of gyrA mutation in tuberculosis.
Acknowledgments
Ruiru Shi is a recipient of The Japan-China Medical Association Fellowship sponsored by the Sasagawa Memorial Foundation.
Footnotes
Published ahead of print on 11 October 2006.
REFERENCES
- 1.Alangaden, G. J., E. K. Manavathu, S. B. Vakulenko, and S. A. Lerner. 1995. Characterization of fluoroquinolone-resistant mutant strains of Mycobacterium tuberculosis selected in the laboratory and isolated from patients. Antimicrob. Agents Chemother. 39:1700-1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheng, A. F., W. W. Yew, E. W. Chan, and R. C. Chan. 2004. Multiplex PCR amplimer conformation analysis for rapid detection of gyrA mutations in fluoroquinolone-resistant Mycobacterium tuberculosis clinical isolates. Antimicrob. Agents Chemother. 48:596-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cooksey, R. C., G. P. Morlock, B. P. Holloway, J. Limer, and M. Hepburn. 2002. Temperature-mediated heteroduplex analysis performed by using denaturing high-performance liquid chromatography to identify sequence polymorphisms in Mycobacterium tuberculosis complex organisms. J. Clin. Microbiol. 40:1610-1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ginsburg, A. S., J. H. Grosset, and W. R. Bishai. 2003. Fluoroquinolones, tuberculosis, and resistance. Lancet Infect. Dis. 3:432-442. [DOI] [PubMed] [Google Scholar]
- 5.Ginsburg, A. S., R. Sun, H. Calamita, W. R. Bishai, and J. H. Grosset. 2005. Emergence of fluoroquinolone resistance in Mycobacterium tuberculosis during continuously dosed moxifloxacin monotherapy in a mouse model. Antimicrob. Agents Chemother. 49:3977-3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hooper, D., and J. Wolfson. 1993. Mechanisms of bacterial resistance to quinolones, p. 97-118. In D. Hooper and J. Wolfson (ed.), Quinolone antimicrobial agents. American Society for Microbiology, Washington, D.C.
- 7.Kocagöz, T. C., J. Hackbarth, I. Unsal, H. Nikaido, and H. F. Chambers. 1996. Gyrase mutations in laboratory-selected, fluoroquinolone-resistant mutants of Mycobacterium tuberculosis H37Ra. Antimicrob. Agents Chemother. 40:1768-1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mohamed, A. M., D. R. Bastola, G. P. Morlock, R. C. Cooksey, and S. H. Hinrichs. 2004. Temperature-mediated heteroduplex analysis for detection of mutations associated with pyrazinamide resistance and differentiation between Mycobacterium tuberculosis and Mycobacterium bovis by denaturing high-performance liquid chromatography. J. Clin. Microbiol. 42:1016-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O'Brien, R. J. 2003. Development of fluoroquinolones as first-line drugs for tuberculosis—at long last. Am. J. Respir. Crit. Care Med. 168:1266-1268. [DOI] [PubMed] [Google Scholar]
- 10.Shi, R., K. Otomo, H. Yamada, T. Tatsumi, and I. Sugawara. 2006. Temperature-mediated heteroduplex analysis for the detection of drug-resistant gene mutations in clinical isolates of Mycobacterium tuberculosis by denaturing HPLC, SURVEYOR nuclease. Microbes Infect. 8:128-135. [DOI] [PubMed] [Google Scholar]
- 11.Takiff, H., L. Salazar, C. Guerrero, S. T. Cole, and A. Telenti. 1994. Cloning and nucleotide sequence of the Mycobacterium tuberculosis gyrA and gyrB genes and detection of quinolone resistance mutations. Antimicrob. Agents Chemother. 38:773-780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu, C., B. N. Kreiswirth, S. Sreeratsan, J. M. Musser, and K. Drlica. 1996. Fluoroquinolone resistance associated with specific gyrase mutations in clinical isolates of multidrug-resistant Mycobacterium tuberculosis. J. Infect. Dis. 174:1127-1130. [DOI] [PubMed] [Google Scholar]