Abstract
Smallpox, once a devastating disease caused by Variola virus, a member of the Orthopoxvirus genus, was eradicated in 1980. However, the importance of variola virus infections has been stressed widely in the last few years, particularly following recent social events in the world. Today, variola virus is considered to be one of the most significant agents with potential use as a biological weapon. In this study we developed an internally controlled real-time PCR assay for rapid detection and simultaneous differentiation of variola virus from other orthopoxviruses. The assay is based on TaqMan 3′-minor groove binder (MGB) chemistry and uses generic primers, designed in highly conserved genomic regions of the crmB gene, and three TaqMan MGB probes designed to identify orthopoxviruses, variola virus, and an internal control. The results obtained suggest that the assay is rapid, sensitive, specific, and suitable for the generic detection of orthopoxviruses and the identification of variola virus and avoids false-negative results in a single reaction tube.
Variola virus (VARV) is the etiological agent of smallpox, a severe and deadly disease that is considered one of the most serious human infections in the history of mankind. Humans are the only known reservoir of VARV, and no animal or insect reservoirs have been identified. VARV is a member of the family Poxviridae, subfamily Chordopoxvirinae, and genus Orthopoxvirus. Its genome consists of 186 kb of linear double-stranded DNA (25). The Orthopoxvirus genus includes other human pathogen viruses: Vaccinia virus (VACV) (used for immunization against smallpox disease), Monkeypox virus (MPXV), Cowpox virus (CPXV), and Buffalopox virus. Following the eradication of smallpox through a global immunization effort, there are still stocks of VARV held in the United States and the former Soviet Union (7). The importance of smallpox has been stressed widely in past years; moreover, VARV is currently considered to be a major biological weapon (2, 14, 26).
Several molecularly based diagnostic methods to detect the DNA of VARV and differentiate it from other orthopoxviruses were published before September 2001. These methods were based on the analysis of the pattern lengths of the restriction endonuclease digestion products of previously amplified genomes (13, 22).
Soon after this date, as an alternative to these time-consuming methods, several real-time PCR protocols for the detection and identification of orthopoxvirus DNA were described (5, 8, 17, 18, 19, 24).
In this study we describe a real-time PCR based on TaqMan 3′-minor groove binder (MGB) chemistry (Applied Biosystems) to simultaneously detect VARV and differentiate it from other orthopoxviruses in a single reaction tube. Moreover the method uses an internal control to detect false negatives.
A careful review of previously published methods for the real-time amplification of variola virus is also described.
MATERIALS AND METHODS
Cell culture and viral strains.
VACV, Western Reserve strain, was grown for 2 days on CV1 cell monolayers and maintained in Dulbecco's modified Eagle medium (DMEM), supplemented with fetal bovine serum (2%) and a mixture of penicillin and streptomycin (0.5%). When the cytopathic effect was evident, the infected cultures were scraped and the infected cells harvested in DMEM. Titration was undertaken by adding a mixture of 50% agar (Difco) in DMEM. The plaques were visualized by adding the violet crystal solution.
Rabbitpox virus (RPXV), Utrecht strain, ectromelia virus (ECTV), Moscow strain, and CPXV, Brighton strain, were obtained from the American Type Culture Collection.
DNAs from Yersinia enterocolitica, Yersinia pseudotuberculosis, Bacillus anthracis, Salmonella typhi, Burkholderia mallei, Francisella tularensis subsp. holarctica, Brucella melitensis, Brucella suis, and Brucella abortus and from varicella-zoster virus and herpes simplex virus 1 were obtained to ascertain the specificity of the method.
Quality control panels.
A first panel, containing 15 lyophilized human samples, spiked with some orthopoxviruses, was provided by the European Network for Imported Viral Diseases (ENIVD). The samples contained camelpox virus (CMLV), CPXV (81/02 and Brighton strains), VACV (Elsree and modified virus Ankara strains), ECTV, and serial dilutions of MPXV (Lam87 strain) (15). Moreover, 10-fold dilutions of MPXV, CMLV, CPXV, and VACV were obtained from P7, P9, P12, P13, and P15 samples of this panel to determine the sensitivity limits of each orthopoxvirus.
The second orthopoxvirus PCR European quality control panel, similar to the previous one, was sent to our laboratory to form part of a number of quality assurance studies by ENIVD. The panel included serial dilutions of MPXV, CPXV, VACV, tanapox virus (genus Yatapoxvirus), and parapox virus (genus Parapoxvirus). Also, four samples included inhibitor factors to test for possible false-negative results (16).
Primers and probes.
The highly conserved genomic region of the crmB gene was the target used to design a real-time PCR assay. Nucleotide sequences for VARV and other orthopoxviruses were obtained from GenBank databases and then aligned using MEGA 2 software (9) (Fig. 1). Primers (Formia 1 and Formia 3) and probes were selected using the Primer Express software (Applied Biosystems). Each probe contained a different fluorescent report dye at the 5′ end: 6-carboxyfluorescein (FAM; VAR130, specific for VARV), 6-carboxyrhodamine (VIC; OPOX143; generic for orthopoxviruses), and NED (IC; specific for the internal control). MGB and nonfluorescent quencher (NFQ) were added to the 3′ end of each (Table 1).
FIG. 1.
Alignment of orthopoxvirus and probe sequences within the crmB gene. BPXV, buffalopox virus; GBLV, taterapox virus. VIR 130 is specific probe for variola virus; ortopox 143 is generic probe. Accession numbers are shaded.
TABLE 1.
Primer and probe sequences
| Primer or probe | Sequencea |
|---|---|
| Formia 1 | 5′-28593CGTGTAACACGACTCACAATAGAATCT28619 |
| Formia 3 | 5′-28788YGGAAGAGACGGTGTRAGAATATGT28766 |
| VAR130 | 5′-FAM-28722ACACGTCTGTTGGAGACG28739-MGB-NFQ |
| OPOX143 | 5′-VIC-28735AGACGTCATCTGTTCTC28751-MGB-NFQ |
| IC | 5′-NED-CCAGCACACATGTGTCTACT-MGB-NFQ |
| ICSb | 5′-CGTGTAACACGACTCACAATAGAATCTCCAGCACACATGTGTCTACTAATAAAAGTTACAGAATAT |
| ICAb | 5′-CTAATAAAAGTTACAGAATATTTTTCCATAAGTTTTTTAACATATTCTTACACCGTCTCTTCCG |
The sequence positions (subscript numbers) correspond to VARV Somalia-1977 (GenBank accession number U18341).
ICS and ICA, sense and antisense primers, respectively, for constructing an internal control.
Construction of a chimerical DNA crmB gene fragment of VARV.
A 216-bp chimerical fragment of synthetic DNA containing part of the crmB gene of VARV was obtained by three following-overlap-extension amplifications with three pairs of tailed primers designed between nucleotides 28593 and 28808 of VARV Somalia-1977 (GenBank accession number U18341). The fragment of the desired product was subsequently cloned into a pCR4-TOPO TA cloning reagent set (Invitrogen, Carlsbad, California) by following the instructions of the manufacturer. Positive clones were extracted with the QIAprep Spin MINIPREP kit (QIAGEN, Hilden, Germany) and sequenced to show the absence of mutations by using M13 forward, reverse universal primers, and the Big Dye terminator cycle sequencing ready kit (Applied Byosystems, Foster City, CA) on an automatic ABI Prism 3700 DNA analyzer (Applied Biosystems, Foster City, CA). The plasmid was linearized with PstI restriction endonuclease at 37°C for 60 min. The concentration of plasmid was measured at an optical density of 260 nm (OD260) using a spectrophotometer and also by calculating the molar concentration. To determine the sensitivity of the real-time PCR assay, a plasmid stock preparation at 2 × 108 copies/μl was 10-fold diluted to 2 × 10−1 copies/μl and stored at −80°C until use.
Construction of an internal control plasmid.
A DNA fragment was obtained by an overlap extension PCR (18) carried out with ICS and ICA primers (Table 1). The product was cloned and sequenced as previously described. As expected, the cloned DNA contained the binding sites for the Formia 1 and Formia 3 primers, flanking the heterologous sequence selected as the IC probe.
DNA extraction.
DNA from VACV was isolated from the infected-cell cultures from preparations of viruses with a guanidinium thiocyanate lysis buffer by following a previously described procedure (3).
DNA from samples included in the ENIVD quality controls and from RPXV, ECTV, and CPXV was extracted using a QIAamp RNA minikit (QIAGEN, Hilden, Germany) by following the manufacturer's instructions. Each tube used in the assay contained 100 copies of internal control plasmid.
Real-time PCR assay.
Genome amplifications were carried out in triplicate in an ABI PRISM 7000 sequence detection system (Applied Biosystems, Foster City, CA) in a final volume of 50 μl including 25 μl of TaqMan 2× Universal PCR Master Mix, No Amperase (Applied Biosystems), 900 nM of Formia 1 primer, 300 nM of Formia 3 primer, 200 nM of each of three fluorogenic TaqMan MGB probes, and 5 μl of extracted DNA. Thermal cycling was performed as follows: 1 cycle at 95°C for 10 min, followed by 45 cycles of 95°C for 15 s and 60°C for 1 min.
RESULTS
Sensitivity and linearity of the assay.
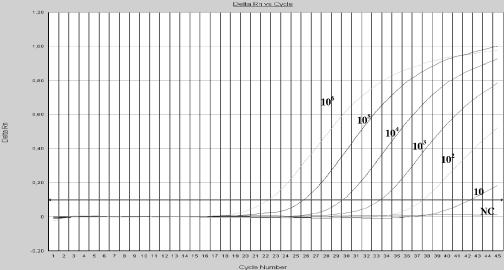
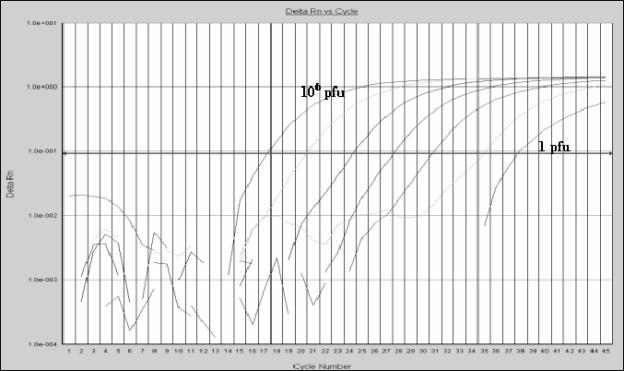
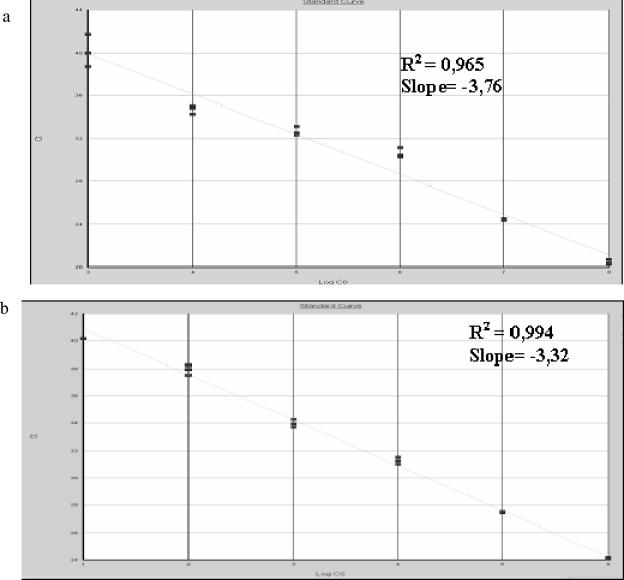
The sensitivity of the method was calculated by amplifying decreasing dilutions of chimerical VARV DNA and genomic VACV DNA. The concentration of TaqMan MGB probes and primers was optimized on the basis of the lowest cycle threshold (Ct). After optimization, the assay detected 10 to 100 copies per tube of VARV DNA using the OPOX143 and VAR130 probes, respectively (Fig. 2). When the genomic VACV DNA was assayed, the lowest dilution detected was 1 PFU per tube (Fig. 3). The data from both chimerical and genomic DNA showed a linear correlation, with a dynamic range of 6 orders of magnitude (Fig. 4).
FIG. 2.
Detection of chimerical VARV DNA by TaqMan real-time PCR. Amplification plots (cycle number versus delta Rn) of serial 10-fold dilutions (106 to 10 copies per tube) of chimerical VARV DNA tested in triplicate in the same experiment. NC is the negative control. Rn, the normalized reporter signal, is the fluorescence signal of the reporter dye divided by the fluorescence signal of the passive, internal reference dye. Delta Rn is determined by the formula Rn+ − Rn−, where Rn+ is the Rn value for a reaction involving all components, including the template, and Rn− is the value for an unreacted sample.
FIG. 3.
Detection of VACV (Western Reserve strain) by TaqMan real-time PCR. The amplification plot was realized on 10-fold dilutions (106 to 1 PFU per tube), assayed in triplicate, of extracted DNA from a VACV stock. See the Fig. 2 legend for Delta Rn definition.
FIG. 4.
Linearity of the TaqMan PCR assay. The standard curve is generated by plotting the observed Ct value, measured in triplicate, against (a) the log10 of the input copy number (106 to 10 copies per tube) of chimerical VARV DNA and (b) the log of serial dilutions (106 to 1 PFU per tube) of VACV.
In addition, we detected 50 copies/ml of MPXV (Lam87 strain), 60 copies/ml of CPXV, 81/01 strain, 240 copies/ml of CPXV, Brighton strain, and 160 copies/ml of VACV (modified virus Ankara strain), calculated by amplifying a 10-fold dilution of extracted DNA from P7, P9, P11, P12, and P15 samples from the first ENIVD quality control panel (15).
Specificity of the assay.
The specificity of the probes was determined by assessing virus-specific fluorescence signals with the FAM, VIC, and NED reporter dyes developed in each reaction and using as templates chimerical VARV DNA and genomic DNA from MPXV (Lam87 strain), CMLV, RPXV (Utrecht strain), CPXV (81/02 and Brighton strains), ECTV (Moscow strain), and VACV (Elstree, Western Reserve, and modified virus Ankara strains). The VAR130 probe can recognize specifically recognize VARV DNA, whereas other different orthopoxvirus DNAs were recognized only by OPOX143. Neither ECTV, an orthopoxvirus not infecting humans that lacks the selected crmB region, nor tanapox virus and parapox virus, other poxviruses not belonging to the Orthopoxvirus genus, were detected. When samples containing amplification inhibitors were assayed, the IC probe showed a negative signal, identifying potential false-negative results (Table 2).
TABLE 2.
Second orthopoxvirus PCR European quality control panel
| Sample no.a or source | Contentsb | Ct valuec for:
|
Results of this study ford:
|
|||
|---|---|---|---|---|---|---|
| OPOX143 | VAR130 | IC | OPOX | VARV | ||
| ENIVD02#07 | MPXV | 24.98 | Undet | Undet | Pos | Neg |
| ENIVD02#10 | MPXV | 29.10 | Undet | Undet | Pos | Neg |
| ENIVD02#08 | MPXV | 32.21 | Undet | 42.94 | Pos | Neg |
| ENIVD02#05 | MPXV | 36.33 | Undet | 41.64 | Pos | Neg |
| ENIVD02#20 | MPXV | 36.70 | Undet | 42.14 | Pos | Neg |
| ENIVD02#01 | MPXV | Undet | Undet | 41.32 | Neg | Neg |
| ENIVD02#14 | Inh f + MPXV | Undet | Undet | Undet | Inh | Inh |
| ENIVD02#15 | Inh f + MPXV | Undet | Undet | Undet | Inh | Inh |
| ENIVD02#17 | CPXV | 41.76 | Undet | Undet | Pos | Neg |
| ENIVD02#13 | VACV | 31.93 | Undet | Undet | Pos | Neg |
| ENIVD02#12 | Tanapox virus | Undet | Undet | 40.53 | Neg | Neg |
| ENIVD02#19 | Parapox virus | Undet | Undet | 40.60 | Neg | Neg |
| ENIVD02#06 | Inh f | Undet | Undet | Undet | Inh | Inh |
| ENIVD02#09 | Inh f | Undet | Undet | Undet | Inh | Inh |
| ENIVD02#02 | Neg | Undet | Undet | 40.69 | Neg | Neg |
| ENIVD02#03 | Neg | Undet | Undet | 40.84 | Neg | Neg |
| ENIVD02#04 | Neg | Undet | Undet | 41.64 | Neg | Neg |
| ENIVD02#11 | Neg | Undet | Undet | 41.35 | Neg | Neg |
| ENIVD02#16 | Neg | Undet | Undet | 40.41 | Neg | Neg |
| ENIVD02#18 | Neg | Undet | Undet | 40.28 | Neg | Neg |
| This study | VARV (1 × 106) | 27.79 | 24.78 | Undet | Pos | Pos |
| This study | VARV(1 × 105) | 31.72 | 28.54 | Undet | Pos | Pos |
| This study | VARV (1 × 104) | 35.59 | 32.21 | Undet | Pos | Pos |
| This study | VARV (1 × 103) | 39.89 | 36.08 | 44.66 | Pos | Pos |
| This study | VARV (1 × 102) | 41.37 | 40.97 | 41.82 | Pos | Pos |
| This study | VARV (1 × 101) | Neg | Neg | 41.01 | Neg | Neg |
| This study | VARV (1) | Neg | Neg | 41.55 | Neg | Neg |
Sample number from the second orthopoxvirus PCR European quality control panel performed by ENIVD.
Inh f, inhibitor factors; Neg, no virus. Concentrations (copies per tube) of chimerical VARV DNA obtained in this study are in parentheses.
Ct is the average cycle at which the fluorescence crossed the threshold; values are geometric means of triplicate experiments. Undet, undetectable.
Results obtained by using this real-time PCR assay. OPOX, orthopoxvirus; pos, positive result; neg, negative result; Inh, inhibition.
Furthermore, we evaluated the specificity of the assay by testing DNA extracted from varicella-zoster virus, herpes simplex virus 1, Salmonella typhi, Yersinia enterocolitica, Y. pseudotuberculosis, Bacillus anthracis, Burkholderia mallei, Francisella tularensis subsp. holarctica, Brucella melitensis, B. suis, and B. abortus; these DNA samples showed only signals for IC, consistent with true-negative results.
Reproducibility of real-time PCR assay.
In order to evaluate the intra-assay reproducibility, a dilution end point standard curve was drawn in triplicate. Ct values were plotted against the log10 of the known input copy number dilutions (107 to 100 copies) of chimerical VARV DNA; the variation coefficient average was 0.4%.
To determine the interassay reproducibility, Ct values were measured in triplicate and in three different days; a variation coefficient average of 0.34% was obtained.
In multiple assays 100 copies of VARV DNA were regularly detected; however, 10 copies were detected in some experiments. Amplification efficiencies were similar, as indicated by the slopes of the curves, the values of which ranged between −3.32 and −3.76.
DISCUSSION
Fear of smallpox has grown in recent years, and smallpox virus is currently considered one of the most powerful biological weapons. Following the World Health Organization's smallpox vaccination program and the eradication of smallpox virus in 1980, recent social events have revived interest in an infectious disease that had been forgotten by recent generations of clinicians and virologists. Furthermore, the cessation of routine smallpox vaccination has made the population susceptible to VARV and zoonotic orthopoxvirus infection. In this regard, the recent outbreak of febrile illness with vesiculopustular eruptions in the United States in summer 2003, caused by an imported MPXV infection and associated with direct contact with ill prairie dogs (20), has emphasized the importance of distinguishing smallpox from similar clinical outbreaks and the need to develop a diagnostic tool capable of rapidly detecting VARV and differentiating it from other orthopoxviruses. The real-time PCR method is suitable for generic detection of orthopoxviruses and simultaneously identifies variola virus in the same reaction tube and could be used in the differential diagnosis of smallpox from other, similar infections (e.g., a monkeypox virus infection outbreak that has a similar clinical onset).
In the past, electron microscopy assays, virus cultures, and serology methods have been used in the laboratory diagnosis of smallpox. Additionally, classical molecular methods such as PCR or nested PCR, DNA endonuclease cleavage, and genome sequencing have proved highly efficient, although they are time-consuming and could generate false positives due to possible contamination events. Recently, an oligonucleotide-based microchip technology (10, 11) capable of detecting and differentiating members of the genus Orthopoxvirus has been described. However, its implementation in other laboratories could be difficult, due to the need for technical experts to prepare this kind of assay. Therefore, the applicability of these methods in antibioterrorism programs is currently being discussed.
A bioterrorism response plan depends on the immediate clinical suspicion of infectious disease and on rapid and accurate laboratory identification of the variola virus. Early diagnosis is essential to control VARV infection, mainly in the initial phases. Based on the rapidity and efficiency of real-time PCR assays, several laboratories involved in biodefense programs are turning to them for the early diagnosis of VARV infection.
Recently, several different real-time PCR assays for orthopoxvirus detection have been described. Most of them use generic detection of orthopoxviruses and posterior melting analysis for VARV identification. Others differentiate VARV from other members of the Orthopoxvirus genus by means of VARV-specific assays. However, it would appear that none of these assays may have actually combined robust and specific VARV identification in the presence of an internal control.
In this study we describe a real-time PCR assay for the rapid detection and simultaneous differentiation of VARV from other orthopoxviruses using a specific VARV probe. We take advantage of the new generation of TaqMan probes with a conjugated MGB group and an NFQ at the 3′ end. Upon hybridization to a complementary target, the MGB molecule folds into a duplex and hyperstabilizes it, allowing the use of a shorter and more specific probe that still meets the high-melting-temperature (Tm) requirement for a PCR to be designed (1, 6). This is very important when the design of a specific probe in a genome as highly conserved as that of the Orthopoxvirus genus is required. After a general analysis of Orthopoxvirus genomes, we identified the crmB gene as an attractive target for our objective; with respect to other genes analyzed, it contains a homologous region for the design of generic primers and probes and a polymorph target region where a specific probe for VARV can be selected.
The primers used in this assay succeeded in amplifying not only VARV but also several orthopoxviruses, including all those capable of causing disease in humans. We detected MPXV (Lam87 strain), CMLV, CPXV (81/02 and Brighton strains), and VACV (Elstree and modified virus Ankara strains) present in the quality control panels. We did not detect ECTV, a virus that infects mice but that is not considered to be an infectious agent for humans (12), since its genome is deleted in the selected target region (21). Parapox and tanapox viruses, included in the second ENIVD panels and not belonging to the Orthopoxvirus genus, rendered negative results (Table 2).
The specificity of the method has also been proved with other DNAs from agents that should be considered in the differential diagnosis of smallpox. None of these produced fluorescent signals with the VAR130 or OPOX143 probe.
Moreover, the assay includes an internal control to detect the potential presence of inhibitors in clinical specimens that may lead to false-negative results. One hundred copies of the internal control plasmid were inserted into the buffer for DNA extraction, amplified in the same reaction tube, and distinguished by the specific probe for the plasmid. The internal control had no influence on the performance of the assay, nor did it decrease sensitivity. Additionally, the robustness of the internal control, which had been shown to be effective in detecting inhibitor effects, was analyzed by testing a panel of 10 negative human samples in triplicate (clinical samples routinely tested for herpesvirus and enterovirus). This was done by using another internal-control PCR (4). The internal control obtained in this study showed very high efficiency, failing in only 1 out of 10 samples (data not shown).
External quality assurance studies were conducted through the ENIVD in order to ascertain the diagnostic proficiency of the laboratories involved in orthopoxvirus diagnosis. We feel that the results obtained in the second ENIVD quality control panel (Table 2) and the good score achieved, based on sensitivity, specificity, rapidity, and ability to detect inhibitor effects of the samples, show that this assay is a sensitive, specific, and rapid tool for the diagnosis of VARV infection. Comparison of results with a conventional generic nested PCR for orthopoxvirus (23), previously designed in our laboratory and confirmed by two ENIVD quality control panels, showed the same sensitivity limit. Also, the possibility of obtaining synthetic DNA with the same properties as genomic DNA to be used as positive controls makes this method easily transferable to other laboratories. This is a very important factor in establishing a rapid response to a possible biological alert.
There is only one previous description of a real-time assay including an internal control (19); nevertheless, this method, based on fluorescence resonance energy transfer probes and Tm analysis, could pose problems of specificity, since other orthopoxviruses (GenBank accession numbers: CPXV, AF375084, AF375085, AF375086, AF377878, AF377879, AF377880, AF377881, AF377882, AF377883, AF377884, AF377885, and AF377886; taterapox virus, AF375093; elephantpox virus, AF375090) have the same sequence as VARV at the probe sites. In addition, it may be the case that many of the previously published methods, based on real-time PCR and the use of specific probes, require reconsideration on a theoretical basis.
Sofi Ibrahim et al. (24) proposed the use of VARV-specific TaqMan probes designed on the hemagglutinin gene, but there are 7 out 32 VARV sequences that present one mismatch with these probes (GenBank accession numbers AF375129, AF375130, AF375137, AF375138, AF375141, AF375142, and Y16780). The use of short probes with an internal mismatch could theoretically dramatically decrease the sensitivity of the method. Recently, Kulesh et al. (8) also proposed two alternative methods based on other genes. One of these involved the use of a specific probe on the B9R gene, but two CPXV strains (GenBank accession numbers AY519982 and AY519984) have the same sequence as the proposed VARV-specific probe; moreover, one VARV strain (GenBank accession number AY552594) has one mismatch with the probe. The first real-time method for identifying VARV published (5) proposes the use of probes with sequences of VARV; however, these sequences are also present in CPXV (GenBank accession numbers AF375084, AF377878, AF377879, AF377880, AF377881, AF377882, AF377883, AF377886, AF375084, AF375086, and AF375087), CMLV (GenBank accession numbers AF375079, AF375080, AF375081, AF375082, AF438165, and AY009089) and taterapox virus (GenBank accession number AF375093).
Another method based on melting analysis (18) also poses theoretical problems because it is based on the presence of one mismatch between the VARV sequences and the probe; unfortunately all the known equivalent sequences of CMLV also have one mismatch (GenBank accession numbers AF438165, AY009089, AY223496, AY299081, AY299082, AY299083, AY299084, AY299085, AY299086, AY299087, and X75156).
Finally, the three methods recently proposed by Nitsche et al. (17) are also based on the melting analysis of probes selected on the rpo18, VETF, and A13L genes; none of them have theoretical problems, but only three VARV sequences are known for these genes.
The method proposed here has been designed on the basis of 129 sequences of orthopoxviruses for the crmB gene, including 15 VARV sequences; the VAR130 probe was selected because it represents a specific sequence for VARV that is present in all the known VARV genomes in the crmB gene but not in the other orthopoxviruses, which show between 2 and more than 10 mismatches with the probe (Fig. 1). Clearly, new knowledge of sequences of VARV or other orthopoxviruses could also affect the proficiency of this design in the future. That is why, as in the case of previous studies, we emphasize the need for conventional molecular methods, such as identification by sequencing the amplified product, in order to confirm a positive results of orthopoxvirus infection.
Acknowledgments
We thank Fiona Westbury for her assistance in the preparation of this manuscript and Matthias Niedrig for providing the ENIVD quality control panels. We are also grateful to R. Escudero, S. Valdezate, M. I. Gimenez, and F. Pozo from our National Center for providing bacterial and viral DNA.
This study was partially funded by the Instituto de Salud Carlos III. A. Negredo and F. Molero are engaged under an agreement between the Spanish Defense Ministry and the Instituto de Salud Carlos III for the development of safe and sensitive molecular tools for the detection of viruses with the potential to be used as biological weapons.
Footnotes
Published ahead of print on 25 October 2005.
REFERENCES
- 1.Afonina, I., M. Zivarts, I. Kutyavin, E. Lukhatanov, H. Gamper, and R. B. Meyer. 1997. Efficient priming of PCR with short oligonucleotides conjugated to a minor grove binder. Nucleic Acids Res. 25:2657-2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atlas, R. M. 1998. The threat of bioterrorism returns the fear of smallpox. Curr. Opin. Microbiol. 1:719-721. [DOI] [PubMed] [Google Scholar]
- 3.Casas, I., L. Powell, P. E. Klapper, and G. M. Cleator. 1995. New method for the extraction of viral RNA and DNA from cerebrospinal fluid for use in the polymerase chain reaction assay. J. Virol. Methods 53:25-36. [DOI] [PubMed] [Google Scholar]
- 4.Casas, I., A. Tenorio, J. M. Echevarria, P. E. Klapper, and G. M. Cleator. 1997. Detection of enteroviral RNA and specific DNA of herpesvirus by multiplex genome amplification. J. Virol. Methods 66:39-50. [DOI] [PubMed] [Google Scholar]
- 5.Espy, M. J., F. R. Cockerill III, R. F. Meyer, M. D. Bowen, G. A. Poland, T. L. Hadfield, and T. F. Smith. 2002. Detection of smallpox virus DNA by LightCycler PCR. J. Clin. Microbiol. 40:1985-1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heid, C. A., J. Stevens, K. J. Livak, and P. M. Willams. 1996. Real time quantitative PCR. Genome Res. 10:986-994. [DOI] [PubMed] [Google Scholar]
- 7.Henderson, D. A., T. V. Inglesby, J. G. Bartlett, M. S. Ascher, E. Eitzen, P. B. Jahrling, J. Hauer, M. Layton, J. McDade, M. T. Osterholm, T. O'Toole, G. Parker, T. Perl, P. K. Russell, and K. Tonat. 1999. Smallpox as a biological weapon: medical and public health management. JAMA 281:2127-2137. [DOI] [PubMed] [Google Scholar]
- 8.Kulesh, D. A., R. O. Baker. B. M. Loveless, D. Norwood, S. H. Zwiers, E. Mucker, E. Hartmann, R. Herrera, D. Miller, D. Christensen, L. P. Wasieloski, Jr., J. Huggins, and P. B. Jahrling. 2004. Smallpox and pan-orthopox virus detection by real-time 3′-minor groove binder TaqMan assays on the Roche LightCycler and the Cepheid Smart Cycler platforms. J. Clin. Microbiol. 42:601-609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei, M. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 10.Laassri, M., V. Chizhikov, M. Mikheev, S. Shchelkunov, and K. Chumakov. 2003. Detection and discrimination of orthopoxviruses using microarrays of immobilized oligonucleotides. J. Virol. Methods 112:6778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lapa, S., M. Mikheev, S. Shchelkunov, V. Mikhsilovich, A. Sobolev, V. Blinov, I. Babkin, A. Guskov, E. Sokunova, A. Zasedatelev, L. Sandakhchiev, and A. Mirzabekov. 2002. Species-level identification of orthopoxviruses with an oligonucleotide microchip. J. Clin. Microbiol. 40:753-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lewis-Jones, S. 2004. Zoonotic poxvirus infections in humans. Curr. Opin. Infect. Dis. 17:81-89. [DOI] [PubMed] [Google Scholar]
- 13.Loparev, V. N., R. F. Massung, J. J. Esposito, and H. Mayer. 2001. Detection and differentiation of old world orthopoxviruses: restriction fragment length polymorphism of the crmB gene region. J. Clin. Microbiol. 39:94-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller, J. M. 2001. Agents of bioterrorism. Preparing for bioterrorism at the community health care level. Infect. Dis. Clin. N. Am. 15:1127-1156. [DOI] [PubMed] [Google Scholar]
- 15.Niedrig, M., H. Schmitz, S. Becker, S. Günther, J. Meulen, H. Meyer, H. Ellerbrok, A. Nitsche, H. R. Gelderblom, and C. Drosten. 2004. First international quality study on the rapid detection of viral agents of bioterrorism. J. Clin. Microbiol. 42:1753-1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Niedrig, M., H. Meyer, M. Panning, and C. Drosten. 2006. Follow up on diagnostic proficiency of laboratories equipped to perform orthopovirus detection and quantification by PCR: the second international external quality assurance study. J. Clin. Microbiol. 44:1283-1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nitsche, A., H. Ellerbrok, and G. Pauli. 2004. Detection of orthopoxvirus DNA by real-time PCR and identification of variola virus DNA by melting analysis. J. Clin. Microbiol. 42:1207-1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Olson, V. A., T. Laue, M. T. Laker, I. V. Babkin, C. Drosten, S. N. Shchelkunov, M. Niedrig, I. K. Damon, and H. Meyer. 2004. Real-time PCR system for detection of orthopoxviruses and simultaneous identification of smallpox virus. J. Clin. Microbiol. 42:1940-1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Panning, M., M. Asper, S. Kramme, H. Schmitz, and C. Drosten. 2004. Rapid detection and differentiation of human pathogenic orthopox viruses by fluorescence resonance energy. Clin. Chem. 50:702-708. [DOI] [PubMed] [Google Scholar]
- 20.Reed, K. D, J. W. Melski, M. B. Graham, R. L. Regnery, M. J. Sotir, M. V. Wegner, J. J. Kazmierczak, E. J. Stratman, Y. Li, J. A. Fairley, G. R. Swain, V. A. Olson, E. K. Sargent, S. C. Kehl, M. A. Frace, R. Kline, S. L. Foldy, J. P. Davis, and I. K. Damon. 2004. The detection of monkeypox in humans in the Western Hemisphere. N. Engl. J. Med. 350:342-350. [DOI] [PubMed] [Google Scholar]
- 21.Ribas, G., J. Rivera, M. Saraiva, R. D. Campbell, and A. Alcami. 2003. Genetic variability of immunomodulatory genes in ectromelia virus isolates detected by denaturing high-performance liquid chromatography. J. Virol. 77:10139-10146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ropp, S. L., Q. Jin, J. C. Knight, R. F. Massung, and J. J. Esposito. 1995. PCR strategy for identification and differentiation of smallpox and other orthopoxviruses. J. Clin. Microbiol. 33:2069-2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanchez-Seco, M. P., L. Hernández, J. M. Eiros, A. Negredo, C. G. Fedele, and A. Tenorio. 2006. Detection and identification of orthopoxviruses using a generic nested-PCR followed by sequencing. Br. J. Biomed. Sci. 63:1-7. [DOI] [PubMed] [Google Scholar]
- 24.Sofi Ibrahim, M., D. A. Kulesh, S. S. Saleh, I. K. Damon, J. J. Esposito, A. L. Schmaljohn, and P. B. Jahrling. 2003. Real-time PCR assay to detect smallpox virus. J. Clin. Microbiol. 41:3835-3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van Regenmortel, M. H. V., C. M. Fauquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. H. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner. 2000. Virus taxonomy: classification and nomenclature of viruses. Seventh report of the International Committee of Taxonomy of Viruses. Academic Press, New York, N.Y.
- 26.Whitley, R. J. 2003. Smallpox: a potential agent of bioterrorism. Antivir. Res. 57:7-12. [DOI] [PubMed] [Google Scholar]