Abstract
We developed a real-time PCR assay combined with melting curve analysis for rapidly genotyping quinolone resistance-determining regions (QRDR) of topoisomerase genes in Streptococcus pneumoniae. This assay was not only accurate for the screening of fluoroquinolone (FQ) resistance but also relevant as an early warning system for detecting preexisting single QRDR mutations.
Streptococcus pneumoniae is the leading cause of community-acquired pneumonia and is also responsible for substantial morbidity and mortality worldwide. Antipneumococcal fluoroquinolones (FQs), such as levofloxacin, gatifloxacin, gemifloxacin, and moxifloxacin, have greater activity against S. pneumoniae than do other drugs, and several are now approved for empirical treatment of community-acquired pneumonia (17, 19). However, recent reports have already noted an increase in the prevalence of FQ-resistant pneumococci (3, 10, 27).
FQ resistance in S. pneumoniae often involves mutation in the quinolone resistance-determining regions (QRDRs) of the parC gene and the gyrA gene, which encode subunits of topoisomerase IV and DNA gyrase, respectively (8, 13, 14, 24). Recent clinical treatment failures of FQ-resistant pneumococcal pneumonia have been reported to be due to strains which acquired FQ resistance as a result of stepwise QRDR mutations (6, 9, 21). Rapid detection of QRDR mutations is required and may constitute a more reliable approach than the current phenotypic method, which can represent susceptibility but cannot detect the potential of S. pneumoniae strains to harbor resistance. PCR-based techniques, such as PCR-restriction fragment length polymorphism analysis (1, 15, 20), single-stranded conformational polymorphism analysis (15), and PCR-oligonucleotide ligation (2), have been developed to detect QRDR mutations; however, these techniques are not appropriate for practical use, as they are complicated and time-consuming. More recently, real-time PCR methods combined with melting curve analysis (PCR-MCA) have been reported to be successful in the detection of key gene mutations associated with drug resistance in various microorganisms (12, 25, 26).
The aim of the present study was to develop and validate a rapid single-step PCR-MCA assay for genotyping S. pneumoniae strains, targeting four QRDR positions (Ser79 and Asp83 of the parC gene and also Ser81 and Gly85 of the gyrA gene) that are most frequently associated with FQ resistance (3, 7, 22).
Seventy-two S. pneumoniae clinical isolates were used. They consisted of 22 levofloxacin (LVX)-resistant strains, with MICs of ≥8 μg/ml, and 50 LVX-susceptible strains, with MICs of ≤2 μg/ml (5). Twenty-two resistant strains were collected from Nagasaki University Hospital (7 strains), Sapporo Medical University School of Medicine (5 strains) (28, 29), and the Levofloxacin Surveillance Group (10 strains) (27) in Japan. Fifty susceptible strains were isolated at Nagasaki University Hospital. Identification of S. pneumoniae was confirmed by optochin susceptibility (11) and by the presence of the autolysine gene (23). The MICs of ciprofloxacin (CIP), LVX, gatifloxacin (GAT), and moxifloxacin (MXF) were determined by a broth dilution method (4, 5). S. pneumoniae ATCC 49619 was used for quality control.
DNA was extracted from each strain by using a QIAamp DNA mini kit (QIAGEN, Hilden, Germany). All of the designed oligonucleotides are shown in Table 1. These refer to the known sequences of the parC and gyrA genes, which were derived from GenBank accession no. AF170996 and AF053121, respectively. PCR was performed in a total volume of 20 μl containing 5 μl of DNA template (average, 5 ng/reaction), 2 μl of LightCycler FastStart reaction mixture (Roche Diagnostics, Basel, Switzerland), 3 mM MgCl2, a 0.2 μM concentration of each probe, and a 0.5 μM concentration of each primer. Thermal cycling was performed with an initial hold for 10 min at 95°C, followed by 35 cycles of 5 s at 95°C, 9 s at 55°C, and 12 s at 72°C. A melting curve was generated by cooling the reaction mixture to 35°C for 10 s, followed by heating to 70°C at a rate of 0.2°C/s. The PCR-MCA assay was performed using LightCycler analysis software 3.5 (Roche Diagnostics, Basel, Switzerland). The total assay time was approximately 1 h. PCR amplification products for all 72 strains were sequenced directly at the nucleotide level, using a BigDye Terminator v.3.1 standard sequencing kit and an ABI PRISM 310 genetic analyzer (both by Applied Biosystems, CA). The QRDR DNA sequencing results were compared with the sequence of strain R6 (GenBank accession no. NC_003098), which was used as the wild-type standard strain.
TABLE 1.
Oligonucleotides used for QRDR mutation detection with PCR-MCA assay
| Target gene or codon | Primer or probeb | Sequence(5′-3′)a | Position(s) (nt) | Amplicon size (bp) |
|---|---|---|---|---|
| Genes | ||||
| parC | Forward | GTTCAACGCCGTATTCTT | 138-155 | 246 |
| Reverse | TGCCTCAGTATAACGCATAG | 364-383 | ||
| gyrA | Forward | GAATGAATTGGGTGTGAC | 282-299 | 225 |
| Reverse | ATACGTGCCTCGGTATAA | 489-506 | ||
| Codons | ||||
| Codon 79 of parC | Anchor | GTCGGCCAAGTCAGTCGGGAACATCATGGGGAATTTCCACCC-FITC | 203-244 | |
| Sensor | LCRed640-CACGGGGATTCTTCTATC-P | 246-264 | ||
| Codon 83 of parC | Anchor | TATCTATGATGCCATGGT-FITC | 260-277 | |
| Sensor | LCRed640-CGTATGTCACAGAACTGGAAAAATCGTGAGATTCTAGTTGAAATGCACGG-P | 279-328 | ||
| Codon 81 of gyrA | Anchor | TAAATAGAGGAATCCCC-FITC | 367-383 | |
| Sensor | LCRed640-TGTGGGTGGTATTTACCCATGACATCCCCTGTAATACGAGCAGATT-P | 320-365 | ||
| Codon 85 of gyrA | Anchor | TCTATTTATGAAGCCATG-FITC | 376-394 | |
| Sensor | LCRed640-CCGTATGGCTCAATGGTGGAGCTACCGTTACATGCTTGTAGATGGTCATG-P | 396-445 |
Abbreviations: FITC, fluorescein isothiocyanate; LCRed 640, LightCyclerRed 640. LCRed 640 is a fluorophore. P, the 3′ end of the probe was phosphorylated to prevent probe elongation by Taq polymerase during PCR.
Primers were used to target genes, and probes were used to target amino acids.
Establishment of MCA for genotyping mutant and wild-type strains. (i) MCA for LVX-resistant S. pneumoniae strains and sequence analysis of QRDRs.
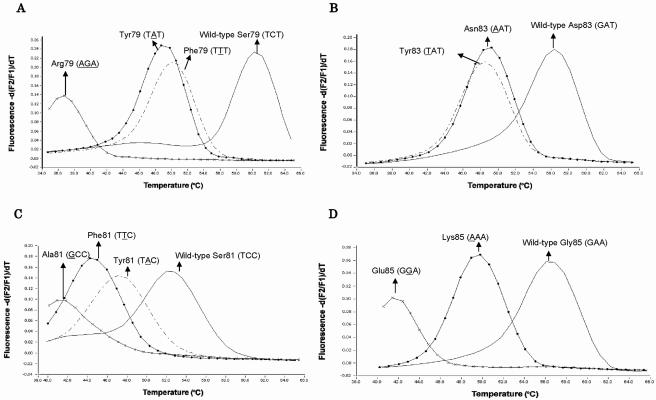
From sequencing results, all 22 LVX-resistant strains had at least one single amino acid substitution at four QRDR positions (Table 2). No silent mutations were detected in any of these 22 strains. Initially, to establish the PCR-MCA method for genotyping QRDRs, ATCC 49619 was selected as the wild-type strain. Strains DR22 (TTT237-239 [Ser79Phe] in parC and GGA255-257 [Gly85Glu] in gyrA), L007 (TAT237-239 [Ser79Tyr] in parC), SR68 (AGA237-239 [Ser79Arg] in parC and TTC243-245 [Ser81Phe] in gyrA), SR27 (TAT249-251 [Asp83Tyr] in parC), SR248 (AAT249-251 [Asp83Asn] in parC and GCC243-245 [Ser81Ala] in gyrA), L001 (TAC243-245 [Ser81Tyr] in gyrA), and L003 (AAA255-257 [Gly85Lys] in gyrA) were chosen as mutant control strains. Using probes specific for the wild-type strain, all of the control strains showed characteristic melting peaks, with a distinct Tm value corresponding to each mutant, as shown in Fig. 1. The MCA for parC codon 79 produced four different peaks, with the following Tm values: TCT237-239 (wild type), 60.4°C; TTT237-239 mutant, 50.0°C; TAT237-239 mutant, 49.3°C; and AGA237-239 mutant, 36.8°C (Fig. 1A). Similarly, MCA produced three different peaks for parC codon 83 (Fig. 1B), four different peaks for gyrA codon 81 (Fig. 1C), and three different peaks for gyrA codon 85 (Fig. 1D). The minimum Tm shifts for mutant strains relative to the wild-type strain were 10.0°C for parC codon 79, 7.7°C for parC codon 83, 4.5°C for gyrA codon 81, and 6.2°C for gyrA codon 85, with an acceptable Tm reproducibility with a <0.8% coefficient of variation (CV) (MCA was performed five times for each strain). The differences in Tm values between the mutants with TTT237-239 (Ser79Phe) and TAT237-239 (Ser79Tyr) at parC codon 79 (Fig. 1A) and between those with TAT249-251 (Asp83Tyr) and AAT249-251 (Asp83Asn) at parC codon 83 (Fig. 1B) were both very similar, and thus these mutations are impossible to differentiate but can be detected as being present. The PCR-MCA assay correctly genotyped 22 LVX-resistant strains, as determined by comparison with sequencing results (Table 2).
TABLE 2.
Melting peak (Tm) and direct DNA sequencing results for four QRDR positions in 22 LVX-resistant S. pneumoniae strains
| Straina | LVX MIC (μg/ml) | ParC codon 79
|
ParC codon 83
|
GyrA codon 81
|
GyrA codon 85
|
||||
|---|---|---|---|---|---|---|---|---|---|
| Tm (°C) | Sequenceb | Tm (°C) | Sequenceb | Tm (°C) | Sequenceb | Tm (°C) | Sequenceb | ||
| ATCC 49619 | 0.5 | 60.4 | TCT/Ser | 56.7 | GAT/Asp | 52.3 | TCC/Ser | 56.8 | GAA/Glu |
| N001 | 8 | 50.2 | TTT/Phe | 56.6 | WTf | 52.3 | WT | 56.8 | WT |
| N002 | 8 | 61.0 | WT | 56.8 | WT | 45.4 | TTC/Phe | 56.0 | WT |
| N003 | 8 | 50.1 | TTT/Phe | 56.7 | WT | 52.3 | WT | 50.2 | AAA/Lys |
| N004 | 16 | 50.8 | TTT/Phe | 56.4 | WT | 51.4 | WT | 49.7 | AAA/Lys |
| N005 | 16 | 50.1 | TTT/Phe | 56.8 | WT | 52.1 | WT | 50.2 | AAA/Lys |
| N006 | 8 | 50.1 | TTT/Phe | 56.8 | WT | 52.1 | WT | 56.4 | WT |
| N007 | 16 | 50.4 | TTT/Phe | 56.8 | WT | 52.1 | WT | 50.3 | AAA/LysMC |
| L001 | 32 | 50.1 | TTT/Phe | 56.7 | WT | 47.7 | TAC/TyrMC | 56.6 | WT |
| L002 | 8 | 50.0 | TTT/Phe | 56.4 | WT | 52.2 | WT | 56.8 | WT |
| L003 | ≥64 | 50.1 | TTT/Phe | 49.0 | AAT/Asn | 52.2 | WT | 50.3 | AAA/Lys |
| L004 | 16 | 50.0 | TTT/Phe | 56.6 | WT | 44.9 | TTC/Phe | 56.8 | WT |
| L005 | 32 | 49.5 | TAT/Tyr | 56.6 | WT | 52.2 | WT | 50.4 | AAA/Lys |
| L006 | 16 | 50.1 | TTT/Phe | 56.8 | WT | 45.1 | TTC/Phe | 56.8 | WT |
| L007 | 16 | 49.3 | TAT/TyrMC | 56.7 | WT | 52.3 | WT | 56.8 | WT |
| L008 | 8 | 60.4 | WT | 56.7 | WT | 45.1 | TTC/Phe | 56.8 | WT |
| L009 | 16 | 49.9 | TTT/Phe | 56.8 | WT | 45.0 | TTC/Phe | 56.8 | WT |
| L010 | 32 | 49.9 | TTT/Phe | 56.7 | WT | 45.1 | TTC/Phe | 56.8 | WT |
| DR22d | 32 | 50.0 | TTT/PheMC | 56.5 | WT | 41.3 | GCC/Phe | 41.7 | GGA/GluMC |
| SR27c | 32 | 60.7 | WT | 48.5 | TAT/TyrMC | 44.5 | TTC/Phe | 56.6 | WT |
| SR68c | 32 | 36.8 | AGA/ArgMC | 56.5 | WT | 44.4 | TTC/PheMC | 56.8 | WT |
| SR179c | 8 | 36.6 | AGA/Arg | 56.6 | WT | 44.5 | TTC/Phe | 56.8 | WT |
| SR248d | 8 | 60.5 | WT | 48.7 | AAT/AsnMC | 41.2 | GCC/AlaMC | 56.6 | WT |
Strains N001 to N007 were isolated at Nagasaki University Hospital, and strains L001 to L010 were supplied by the Levofloxacin Surveillance Group.
Nucleotide sequence/amino acid sequence. Changed nucletides are shown in bold. WT, identical to the nucleotide distribution of the wild type, ATCC 49619. MC, mutant control.
Strains sourced from reference 22, supplied by Sapporo Medical University.
Strains sourced from reference 23, supplied by Sapporo Medical University.
FIG. 1.
Melting peak patterns for QRDR mutants with substitutions in parC and gyrA. Melting curve analysis was performed with the 246-bp amplicon of the parC gene and the 225-bp amplicon of the gyrA gene obtained by real-time PCR from wild-type and mutant control strains. Panels A, B, C, and D show melting peak patterns for codon 79 of the parC gene, codon 83 of the parC gene, codon 81 of the gyrA gene, and codon 85 of the gyrA gene, respectively. Each value on the y axis represents the ratio of the first negative derivative of the change in fluorescence (dF) to the variation in temperature. Point mismatches in the QRDRs resulted in lower Tm values for mutant strains than for the wild-type strain.
(ii) MCA of LVX-susceptible S. pneumoniae strains and sequence analysis of QRDRs.
Forty-seven of the 50 LVX-susceptible S. pneumoniae strains had similar Tm values to that of the wild-type strain. The mean Tm values for the 47 strains were 60.4°C (CV, 0.41%) for parC codon 79, 56.7°C (CV, 0.52%) for parC codon 83, 56.5°C (CV, 0.87%) for gyrA codon 81, and 56.7°C (CV, 0.34%) for gyrA codon 85, and sequencing results confirmed that these were in fact wild-type strains. Two of the remaining three strains had Tm values of 49.8°C and 50.0°C, which were lower than that (60.4°C) of the wild-type strain for parC codon 79, while the other strain had a Tm of 48.8°C, which differed from that of the wild-type strain (56.7°C) for parC codon 83. Indeed, direct sequencing revealed that the former two strains had a TTT237-239 (Ser79Phe) mutation, while the latter strain had an AAT249-251 (Asp83Asn) mutation in the parC gene.
Comparison of outcomes between conventional phenotyping and MCA genotyping.
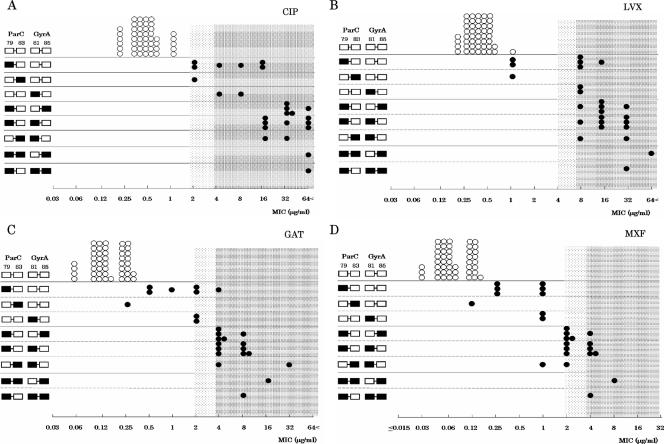
We compared the ability of the present PCR-MCA assay to detect FQ susceptibility in 72 S. pneumoniae strains with that of the conventional phenotypic method, as shown in Fig. 2 and Table 3. All 47 LVX-susceptible strains with no mutations had susceptibility MICs of ≤2 μg/ml, while 22 of the remaining 25 mutation-containing strains (88%) had resistance MICs of ≥8 μg/ml (Fig. 2B; Table 3). Interestingly, the mutation profiles for the QRDRs in the parC and gyrA genes revealed a close relationship between MIC level and the number of QRDR mutations. We observed that six of the nine strains with a single QRDR mutation were highly resistant to LVX (Fig. 2B; Table 3). This can be explained by the fact that three of the six strains harbored additional parE mutations and performed active efflux, two of the six strains harbored additional parE mutations, and the remaining strain harbored an additional gyrB mutation (data not shown). Compared with the conventional phenotypic method, the diagnostic sensitivity and specificity of the PCR-MCA assay for detecting non-FQ-susceptible strains were 100% (25/25) and 100% (47/47), respectively, for CIP, 100% (22/22) and 94% (47/50), respectively, for LVX, 100% (21/21) and 92% (47/51), respectively, for GAT, and 100% (15/15) and 82% (47/57), respectively, for MXF. Assuming that QRDR mutation provides a positive diagnosis of the presence of non-FQ-susceptible S. pneumoniae, this assay is particularly useful because it has a negative predictive value of 100% for all four FQs and positive predictive values of 100% for CIP, LVX, and GAT and 93.7% for MXF (Fig. 2; Table 3).
FIG. 2.
Comparison of outcomes between conventional FQ susceptibility testing and the PCR-MCA assay for genotyping QRDR mutants. Panels A, B, C, and D show results with CIP, LVX, GAT, and MXF, respectively. White squares represent the wild type, and black squares represent the mutant types for four positions of the QRDRs in the parC gene and gyrA gene obtained by the PCR-MCA assay. White circles represent wild-type strains, and black circles represent mutation-containing strains. The horizontal axis represents the MIC of each strain. The dark mesh area represents resistance, and the light mesh area represents intermediate susceptibility to each FQ. The CLSI MIC breakpoints (26) were used for the following drugs: LVX (susceptible, ≤2 μg/ml; intermediate, 4 μg/ml; and resistant, ≥8 μg/ml), GAT (susceptible, ≤1 μg/ml; intermediate, 2 μg/ml; and resistant, ≥4 μg/ml), and MXF (susceptible, ≤1 μg/ml; intermediate, 2 μg/ml; and resistant, ≥4 μg/ml). The breakpoint standard for CIP was obtained from the interpretive guideline supplied by the Japanese Society of Chemotherapy (susceptible, ≤1 μg/ml; intermediate, 2 μg/ml; and resistant, ≥4 μg/ml).
TABLE 3.
Relationship between QRDR mutations detected by MCA and MICs of FQs for 72 S. pneumoniae strains
| FQ and QRDR mutation | No. of isolates with MICa
|
||
|---|---|---|---|
| S | I | R | |
| CIP | |||
| None | 47 | 0 | 0 |
| Single parC or gyrA mutation | 0 | 3 | 6 |
| parC and gyrA mutations | 0 | 0 | 16 |
| LVX | |||
| None | 47 | 0 | 0 |
| Single parC or gyrA mutation | 3 | 0 | 6 |
| parC and gyrA mutations | 0 | 0 | 16 |
| GAT | |||
| None | 47 | 0 | 0 |
| Single parC or gyrA mutation | 4 | 4 | 1 |
| parC and gyrA mutations | 0 | 0 | 16 |
| MXF | |||
| None | 47 | 0 | 0 |
| Single parC or gyrA mutation | 9 | 0 | 0 |
| parC and gyrA mutations | 1 | 8 | 7 |
Abbreviations: S, susceptible; I, intermediate; and R, resistant. The CLSI MIC breakpoints were used for LVX (susceptible, ≤2 μg/ml; intermediate, 4 μg/ml; and resistant, ≥8 μg/ml), GAT (susceptible, ≤1 μg/ml; intermediate, 2 μg/ml; and resistant, ≥4 μg/ml), and MXF (susceptible, ≤1 μg/ml; intermediate, 2 μg/ml; and resistant, ≥4 μg/ml). The breakpoint standard for CIP was obtained from the interpretive guidelines supplied by the Japanese Society of Chemotherapy (susceptible, ≤1 μg/ml; intermediate, 2 μg/ml; and resistant, ≥4 μg/ml).
The discrepancies observed in the specificity of the assay were instructive because the conventional phenotypic method failed to pick up strains which have high proportions of a single QRDR mutation. Several reports have noted that a significant number of isolates already have a single mutation but are still considered fully susceptible (7, 22). Although our study lacks a collection of moderately FQ-resistant (LVX MIC of 2 μg/ml or 4 μg/ml) S. pneumoniae strains, Lim et al. (16) reported that about 60% of S. pneumoniae isolates with an LVX MIC of 2 μg/ml carry preexisting parC mutations. Typically, stepwise mutation starts with the parC gene, which frequently leads to secondary mutations in the gyrA gene, eventually resulting in high-level resistance to all FQs (18). This has also been supported by clinical reports of an FQ-susceptible S. pneumoniae strain carrying a first-step mutation that evolved into a second-step QRDR mutation during FQ treatment, resulting in treatment failure (6). We emphasize the clinical importance of the detection of first-step QRDR mutations in either gyrA or parC for attempting to predict evolution into FQ resistance. There are some limitations of this assay that remain to be ironed out; for instance, any mutations outside the sensor probe would be undetectable. Resistance as a result of other mechanisms (such as parE or gyrB mutation and efflux) also cannot be detected. However, these mechanisms provide resistance potential but are not conclusive indicators of high-level resistance to FQs compared with parC and gyrA mutations (7, 22).
In conclusion, the PCR-MCA assay was easily and quickly performed and had an accuracy which was at least as satisfactory as that of the conventional phenotypic method. Moreover, single QRDR mutations which harbored potential for FQ resistance could be detected. This assay is also useful for surveillance studies in the screening of FQ resistance as an alternative to DNA nucleotide sequencing. The application of this novel method would be a valuable tool for achieving rapid screening of QRDR mutations and as an early warning system for the emergence of FQ-resistant S. pneumoniae.
Acknowledgments
We thank Shin-ichi Yokota for providing strains DR22, SR27, SR68, SR179, and SR248, the Levofloxacin Surveillance Group for the gift of strains L001 to L010, and Maiko Motoshima for help with antibiotic susceptibility testing.
Footnotes
Published ahead of print on 4 October 2006.
REFERENCES
- 1.Alonso, R., M. Galimand, and P. Courvalin. 2004. An extended PCR-RFLP assay for detection of parC, parE, and gyrA mutations in fluoroquinolone-resistant Streptococcus pneumoniae. J. Antimicrob. Chemother. 53:682-683. [DOI] [PubMed] [Google Scholar]
- 2.Bui, M. H., G. G. Stone, A. M. Nilius, L. Almer, and R. K. Flamm. 2003. PCR-oligonucleotide ligation assay for detection of point mutations associated with quinolone resistance in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 47:1456-1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Canton, R., M. Morosini, M. C. Enright, and I. Morrissey. 2003. Worldwide incidence, molecular epidemiology and mutations implicated in fluoroquinolone-resistant Streptococcus pneumoniae: data from the global PROTEKT surveillance programme. J. Antimicrob. Chemother. 52:944-952. [DOI] [PubMed] [Google Scholar]
- 4.Clinical and Laboratory Standards Institute. 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 7th edition. CLSI document M7-A7. Clinical and Laboratory Standards Institute, Wayne, Pa.
- 5.Clinical and Laboratory Standards Institute. 2006. Performance standards for antimicrobial susceptibility testing; 16th informational supplement. CLSI document M100-S16. Clinical and Laboratory Standards Institute, Wayne, Pa.
- 6.Davidson, R., R. Cavalcanti, J. L. Brunton, D. J. Bast, J. C. Azavedo, P. Kibsey, C. Fleming, and D. E. Low. 2002. Resistance to levofloxacin and failure of treatment of pneumococcal pneumonia. N. Engl. J. Med. 346:747-750. [DOI] [PubMed] [Google Scholar]
- 7.Doern, G. V., S. S. Richter, A. Miller, N. Miller, C. Rise, K. Heilmann, and S. Beekmann. 2005. Antimicrobial resistance among Streptococcus pneumoniae in the United States: have we begun to turn the corner on resistance to certain antimicrobial classes? Clin. Infect. Dis. 41:139-148. [DOI] [PubMed] [Google Scholar]
- 8.Eliopoulos, G. M. 2004. Quinolone resistance mechanisms in pnemococci. Clin. Infect. Dis. 38(Suppl. 4):S350-S356. [DOI] [PubMed] [Google Scholar]
- 9.Endimiani, A., G. Brigante, A. A. Bettaccini, F. Luzzaro, P. Grossi, and A. Q. Toniolo. 2005. Failure of levofloxacin treatment in community-acquired pneumococcal pneumonia. BMC Infect. Dis. 5:106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Felmingham, D., R. R. Reinert, Y. Hirakata, and A. Rodloff. 2002. Increasing prevalence of antimicrobial resistance among isolates of Streptococcus pneumoniae from PROTEKT surveillance study, and comparative in vitro activity of the ketolide, telithromycin. J. Antimicrob. Chemother. 50(Suppl. S1):25-37. [DOI] [PubMed] [Google Scholar]
- 11.Gardam, M. A., and M. A. Miller. 1998. Optochin revisited: defining the optimal type of blood agar for presumptive identification of Streptococcus pneumoniae. J. Clin. Microbiol. 36:833-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glocker, E., M. Berning, M. M. Gerrits, J. G. Kusters, and M. Kist. 2005. Real-time PCR screening for 16S rRNA mutations associated with resistance to tetracycline in Helicobacter pylori. Antimicrob. Agents Chemother. 49:3166-3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hooper, D. C. 2002. Fluoroquinolone resistance among gram positive cocci. Lancet Infect. Dis. 2:530-538. [DOI] [PubMed] [Google Scholar]
- 14.Hooper, D. C. 1999. Mechanisms of fluoroquinolone resistance. Drug Resist. Updat. 2:38-55. [DOI] [PubMed] [Google Scholar]
- 15.Ip, M., S. S. Chau, F. Chi, A. Qi, and R. W. Lai. 2006. Rapid screening of fluoroquinolone resistance determinants in Streptococcus pneumoniae by PCR-restriction fragment length polymorphism and single-strand conformational polymorphism. J. Clin. Microbiol. 44:970-975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lim, S., D. Bast, A. McGeer, J. de Azavedo, and D. E. Low. 2003. Antimicrobial susceptibility breakpoints and first-step parC mutations in Streptococcus pneumoniae: redefining fluoroquinolone resistance. Emerg. Infect. Dis. 9:833-837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mandell, L. A., J. G. Bartlett, S. F. Dowell, T. M. File, Jr., D. M. Musher, and C. Whitney. 2003. Update of practice guidelines for the management of community-acquired pneumonia in immunocompetent adults. Clin. Infect. Dis. 37:1405-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muńoz, R., and A. G. de la Campa. 1996. ParC subunit of DNA topoisomerase IV of Streptococcus pneumoniae is a primary target of fluoroquinolones and cooperates with DNA gyrase A subunit in forming resistance phenotype. Antimicrob. Agents Chemother. 40:2252-2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Niederman, M. S., L. A. Mandell, A. Anzueto, J. B. Bass, W. A. Broughton, G. D. Campbell, N. Dean, T. File, M. J. Fine, P. A. Gross, F. Martinez, T. J. Marrie, J. F. Plouffe, J. Ramirez, G. A. Sarosi, A. Torres, R. Wilson, and V. L. Yu. 2001. Guidelines for the management of adults with community-acquired pneumonia. Am. J. Respir. Crit. Care Med. 163:1730-1754. [DOI] [PubMed] [Google Scholar]
- 20.Pan, X. S., J. Ambler, S. Mehtar, and M. Fisher. 1996. Involvement of topoisomerase IV and DNA gyrase as ciprofloxacin targets in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 40:2321-2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perez-Trallero, E., J. M. Marimon, L. Iglesias, and J. Larruskain. 2003. Fluoroquinolone and macrolide treatment failure in pneumococcal pneumonia and selection of multidrug-resistant isolates. Emerg. Infect. Dis. 9:1159-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Richter, S. S., K. P. Heilmann, S. E. Beekman, N. J. Miller, C. L. Rice, and G. V. Doern. 2005. The molecular epidemiology of Streptococcus pneumoniae with quinolone resistance mutations. Clin. Infect. Dis. 40:225-235. [DOI] [PubMed] [Google Scholar]
- 23.Sheppard, C. L., T. G. Harrison, R. Morris, A. Horgan, and R. C. George. 2004. Autolysine-targeted LightCycler assay including internal process control for detection of Streptococcus pneumoniae DNA in clinical samples. J. Med. Microbiol. 53:189-195. [DOI] [PubMed] [Google Scholar]
- 24.Tankovic, J., B. Perichon, J. Duval, and P. Courvalin. 1996. Contribution of mutations in gyrA and parC genes to fluoroquinolone resistance of mutants of Streptococcus pneumoniae obtained in vivo and in vitro. Antimicrob. Agents Chemother. 40:2505-2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vacher, S., A. Menard, E. Bernard, A. Santos, and F. Megraud. 2005. Detection of mutations associated with macrolide resistance in thermophilic Campylobacter spp. by real-time PCR. Microb. Drug Resist. 11:40-47. [DOI] [PubMed] [Google Scholar]
- 26.Vernel-Pauillac, F., V. Falcot, D. Whiley, and F. Merien. 2006. Rapid detec-tion of a chromosomally mediated penicillin resistance-associated ponA mutation in Neisseria gonorrhoeae using a real-time PCR assay. FEMS Microbiol. Lett. 255:66-74. [DOI] [PubMed] [Google Scholar]
- 27.Yamaguchi, K., and A. Ohno. 2005. Investigation of the susceptibility trends in Japan to fluoroquinolones and other antimicrobial agents in a nationwide collection of clinical isolates: a longitudinal analysis from 1994 to 2002. Diagn. Microbiol. Infect. Dis. 52:135-143. [DOI] [PubMed] [Google Scholar]
- 28.Yokota, S., K. Sato, O. Kuwahara, S. Habadera, N. Tsukamoto, H. Ohuchi, H. Akizawa, T. Himi, and N. Fujii. 2002. Fluoroquinolone-resistant Streptococcus pneumoniae strains occur frequency in elderly patients in Japan. Antimicrob. Agents Chemother. 46:3311-3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yokota, S., K. Sato, S. Yoshida, and N. Fujii. 2004. Molecular epidemiology of fluoroquinolone-resistant Streptococcus pneumoniae in Japan. Kansenshogaku Zasshi 78:428-434. (In Japanese.) [DOI] [PubMed] [Google Scholar]