Abstract
Escherichia coli is a diverse bacterial species which is widely distributed in the environment but also exists as a commensal and pathogen of different host species. Human intestinal pathogenic E. coli causes over 160 million cases of diarrhea and an estimated 1 million deaths per year. The majority of deaths are attributable to one pathovar of E. coli, namely, enterotoxigenic E. coli. The pathogenesis of enterotoxigenic E. coli is dependent on the production of a colonization factor to promote adhesion to the intestinal epithelium and the elaboration of heat-labile or heat-stable toxins which induce a secretory diarrhea. Despite the high morbidity and mortality associated with enterotoxigenic E. coli infection, little is known of the genetic background of this global pathogen. Here we demonstrate by multilocus sequence typing that enterotoxigenic E. coli isolates are present in all phylogenetic lineages of E. coli, indicating that acquisition of the toxin genes may be sufficient to generate an enterotoxigenic E. coli strain. In addition, screening of diarrheal isolates for the presence of additional genes previously associated with the virulence of enterotoxigenic E. coli revealed that they were not abundant. These observations have significant implications for disease epidemiology and for the design of effective vaccines.
The species of Escherichia coli, together with the now indisputably derivative and polyphyletic Shigella spp., represent an extraordinarily diverse group of pathogenic and commensal organisms (29). Numerous studies of both pathogenic and nonpathogenic E. coli strains derived from different mammalian hosts suggest that this versatile organism is a clonal organism which can be separated into five major evolutionary groups: A, B1, B2, D, and E (sometimes termed “unclassified”) (26). These groups have been supported across several studies using a variety of methods of phylogenetic reconstruction, although there have been disagreements regarding the relationships between groups (2, 26, 31, 52). However, comparisons of three published E. coli genomes revealed that only 39.2% of the genes were present in all strains, highlighting the extraordinary degree of genomic diversity within this species (66). These data suggest that the phenotype and niche adaptation of a particular E. coli strain are determined by periodic selection resulting in clonal expansion and by the horizontal acquisition of mobile DNA, such as plasmids and phages, of different evolutionary origins. Indeed, investigation of E. coli K-12 suggests that one-quarter of the genome has been acquired horizontally in over 200 separate events (30). It is these latter genetic acquisition events that are thought to provide the repertoire of virulence factors which distinguish commensal and pathogenic E. coli.
In contrast to the phylogenetic groupings, intestinal pathogenic E. coli strains have been classified into six pathovars based upon the repertoire of virulence factors and the clinical manifestations of infection, namely, enterohemorrhagic (EHEC), enteropathogenic (EPEC), enteroaggregative (EAEC), diffuse-adhering, enteroinvasive, and enterotoxigenic (ETEC) E. coli (reviewed in reference 41). ETEC is the most common cause of E. coli-mediated human diarrhea worldwide (22). There are estimated to be about 650 million cases of ETEC infection each year in developing countries, with around 800,000 deaths occurring, mostly in young children (50, 71). It also poses a significant problem for travelers and military personnel visiting countries where ETEC is endemic (41). In addition to the high levels of morbidity and mortality associated with human infection, ETEC also has important financial implications for the farming industry, where it is a major pathogen of cattle and neonatal and postweaning piglets (22, 39). The importance of ETEC as a pathogen is highlighted by the fact that of the six pathovars of diarrheagenic E. coli it is alone in being the subject of a World Health Organization initiative to find an efficacious vaccine (71).
Vaccine studies for ETEC have focused on the plasmid-encoded virulence factors, namely, the colonization factors (CFs), which are required for adherence to the small intestine, and the heat-stable (ST) or heat-labile (LT) toxins, which induce the secretory diarrhea typical of an ETEC infection (6). Despite ETEC being a major cause of diarrhea worldwide, very little is known about the chromosomal determinants of enteropathogenicity. However, investigations of a variety of ETEC strains, including the type strain E. coli H10407, have identified a small number of putative virulence factors, other than the CFs and toxins, which could be involved in ETEC virulence. These include (i) LeoA, implicated in LT secretion; (ii) TibA, a glycosylated autotransporter involved in adhesion to epithelial cells; (iii) Tia, an outer membrane adhesin and invasin; (iv) ClyA, a hemolysin which requires positive transcriptional activation; and (v) EatA, a serine protease autotransporter associated with fluid accumulation in the ileal loop model (18-20, 35, 48, 58, 67). While it remains a subject of debate whether these additional virulence factors contribute significantly to ETEC-mediated disease, it is clear that a CF and at least one of the toxins are essential virulence factors for the disease process (22, 40, 41, 56).
Previously, it was suggested that for pathogenic E. coli, including ETEC, a specific genetic background was necessary for the expression and maintenance of certain virulence factors (15). Indeed, phylogenetic comparisons of strains within the EAEC, diffuse-adhering E. coli, EPEC, and EHEC pathovars suggest that pathogenic isolates within these pathovars have arisen through clonal expansion and thus possess a common genetic background (3, 4, 10, 44, 54, 68). Such inferences are further supported by observations that some virulence factors associated with a particular pathovar are found only within particular phylogenetic groups, despite their distribution being sporadic and consistent with horizontal transfer rather than vertical inheritance (47, 52, 53). Similar detailed phylogenetic analyses have been lacking for ETEC; thus, we wished to determine the clonal relatedness of ETEC strains derived from multiple geographic locations and mammalian hosts and to determine whether the virulence factors that were previously associated with ETEC infection were common to strains that were associated with disease.
MATERIALS AND METHODS
Bacterial isolates.
A total of 209 E. coli strains were studied (see Table S1 in the supplemental material). These included 116 ETEC strains isolated from humans with diarrhea from around the globe and in different years, 34 ETEC strains isolated from cattle, and 59 of the Escherichia coli Reference Collection (ECOR) collection including strains representing all the major phylogenetic branches as previously determined by multilocus enzyme electrophoresis (MLEE) (26). The human diarrheal isolates were collected over a period of several years in separate studies in Mexico, Guatemala, and India.
MLST analysis.
The housekeeping genes adk, fumC, gyrB, icd, mdh, purA, and recA, selected from the multilocus sequence typing (MLST) website for E. coli (http://web.mpiib-berlin.mpg.de/mlst/dbs/Ecoli), were amplified for typing by using the primers listed in Table S2 in the supplemental material. The template for PCR was 5 μl cell lysate extracted from a culture of the strain by being boiled for 10 min. Bio-X-act Short mix (Bioline) with proofreading was used in reaction conditions consisting of 10 min of denaturation at 94°C followed by 30 cycles of 30 s of denaturation at 94°C, 30 s at the specific annealing temperature for the primer, and 3 min of extension at 68°C. Purified PCR products were sequenced using nested sequencing primers listed in Table S2 in the supplemental material. Sequences were compared with the MLST database alleles to obtain allele numbers and sequence types. To confirm the phylogenetic groups determined by MLST, all strains were also analyzed by triplex PCR as previously described by Clermont et al. (9).
Phylogenetic analysis.
The sequences obtained from MLST analyses were supplemented by equivalent regions from all available complete and draft E. coli or Shigella genome sequences. The sequences were aligned using ClustalW version 1.8 (62), with manual inspection using Seaview (23), and concatenated to give an alignment of 3,405 bp. To circumvent the problem of inappropriate or computationally intensive substitution models for coding sequences (57), phylogenetic analysis was performed only on third codon positions, using the HKY85 model of DNA substitution with a gamma distribution of rates across sites. However, these results were similar to those obtained for analysis of sequences including all three positions within the codon. The Bayesian Metropolis-coupled Markov chain Monte Carlo method of tree reconstruction, as implemented in MrBayes version 3.1.2 (55), was employed. Four chains were run with default heating parameters, and trees sampled from the posterior probability distribution were used to generate a 50% majority rule consensus tree.
Virulence factor screening.
The presence of the toxins was confirmed by the use of oligonucleotides labeled with polynucleotide T4 kinase and [32P]ATP, as previously described (38). Primers were designed to amplify small fragments (500 bp to 1 kb) of putative ETEC virulence genes and are listed in Table S2 in the supplemental material. The leoA, tia, and tibC primers were designed based upon the E. coli H10407 sequence. clyA primers were designed to correspond to highly conserved regions of the gene which were determined from comparison of the clyA sequences available from different E. coli isolates (34). Primers specific to eatA were designed after inspection of a ClustalW multiple alignment of all the known serine protease autotransporter proteins. The presence or absence of the virulence gene was determined by PCR using two pairs of primers for each locus.
Genomic comparisons.
In an attempt to identify chromosomal genes potentially involved in enterotoxigenicity, genomic comparisons of the draft ETEC genome sequences from strains E24377A (GenBank accession number AAJZ01000001), B7A (GenBank accession numbers AAJT01000001 to AAJT01000198), and H10407 (available from http://www.sanger.ac.uk/Projects/E_coli_H10407) were performed. Orthologues were identified using the mutual best hits procedure, as previously described (7). Genes that had copies in all three ETEC genomes were further compared in the same manner with a representative set of complete and draft E. coli genome sequences (laboratory strain K-12 MG1655, GenBank accession number U00096; EHEC strain O157:H7 Sakai, GenBank accession number BA000007; uropathogenic strain CFT073, GenBank accession number AE014075; commensal strain HS, GenBank accession AAJY01000001; enteroinvasive E. coli strain 53638, GenBank accession numbers AAKB01000001 to AAKB01000119; EPEC1 strain E22, GenBank accession numbers AAJV01000001 to AAJV01000109; EPEC2 strain B171, GenBank accession numbers AAKB01000001 to AAKB01000119; and EAEC strain 042, available from http://www.sanger.ac.uk/Projects/Escherichia_Shigella).
RESULTS
Is MLST appropriate for phylogenetic investigations of diarrheagenic E. coli?
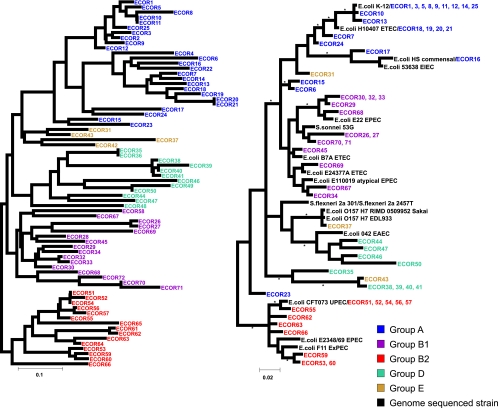
Previous phylogenetic analysis of the ECOR collection based on the application of the neighbor-joining algorithm to MLEE data (26) defined five major phylogenetic groupings (Fig. 1, left). Application of Bayesian phylogenetic analysis to the MLST data obtained in this study (Fig. 1, right) broadly confirmed these groupings, but in line with previous studies (2, 11, 15, 31, 49), there was some disagreement in branching order relative to the MLEE tree, both within and between groups. Groups A and B1 clustered together based on the MLST, contrary to the grouping of B1 and B2 seen in the MLEE tree but consistent with ancillary characteristics such as the distribution of the ETT2 pathogenicity island (53). The group E (or unclassified) sequences from the MLEE tree did not cluster together in the current study, and group D was not monophyletic. The MLST method is less discriminatory than MLEE; identical sequences were obtained for a number of ECOR strains that could be distinguished based on MLEE. The groupings obtained by Bayesian analysis of the MLST data were in agreement with those found using neighbor joining and BURST (data not shown).
FIG. 1.
Comparison of the phylogenetic relationships of the ECOR collection using MLEE and MLST. (Left) Phylogeny of the ECOR strains, as determined by neighbor-joining analysis of the MLEE data available at http://foodsafe.msu.edu/Whittam/ecor. Analysis was carried out using Neighbor from the PHYLIP package, and the tree was rooted on group B2, which has been shown by several studies to be the most basal. (Right) Analysis was performed based on MLST sequence data, using MrBayes (see Materials and Methods). ECOR strains are colored according to their group designation based upon the MLEE analysis. Asterisks indicate branches with a posterior probability of >95%.
A small number of strains were placed in different phylogenetic groups relative to MLEE, possibly due to the effects of horizontal gene transfer. ECOR 23 was originally classified as a group A strain, but work from our lab showed that it possesses virulence genes associated with B2 strains, particularly those associated with pathogenicity island II of the prototype uropathogenic E. coli strain CFT073 (26, 47). Analysis of the genetic backbone of this strain by MLST places it between group D/E and B2, the two groups more commonly associated with pathogenic isolates. ECOR 31, which was unclassified by the original analysis and more recently associated with group E, was the only other strain significantly “misplaced” by our analysis and was associated with group A. All remaining previously unclassified strains were found in the group assigned as D/E in this study, which is not monophyletic and confirms the close relationship of strains in these two groups. Ecor70, a strain found classified as B1 by MLEE, which had been placed in A by triplex PCR in the study by Clermont et al. (9) and in an additional group C in the study by Escobar-Páramo et al. (16), was found to be identical to ECOR 71 and located in group B1. It is noteworthy that the only two misassigned strains in the initial triplex PCR study (9) were designated as group A strains where the previous designation was B1; this was the most common discrepancy observed in our data, which otherwise showed good agreement. This is likely to be a result of the triplex PCR protocol using three genetic markers that are closely linked on the chromosome compared to the widely distributed housekeeping loci. However, the differences are minor, and the results confirm the suitability of this MLST protocol for phylogenetic analysis of E. coli.
ETEC demonstrates a polyphyletic origin.
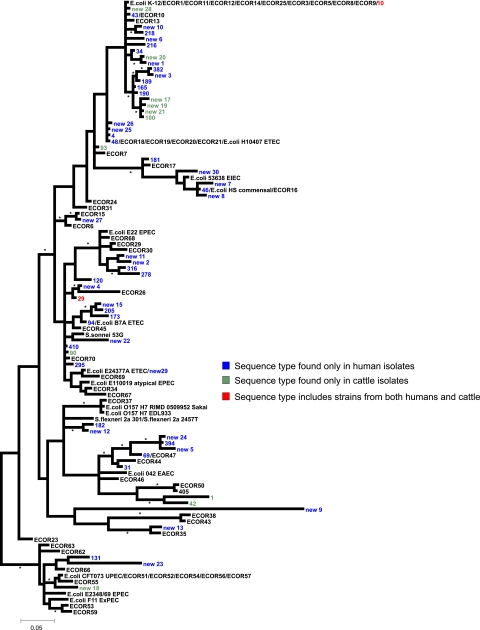
A further phylogenetic analysis was performed, expanding the data set to include a collection of 150 ETEC strains. The MLST database currently contains information for 735 strains, with 457 distinct sequence types. Twenty-eight of the sequence types obtained in the current study had not been identified previously and have been submitted to the MLST database. A total of 61 sequence types were identified among the ETEC strains tested, and of these only 21 arose more than once in the collection (see Table S1 in the supplemental material). This observation is consistent with the absence of a specific ETEC clonal lineage.
The phylogeny demonstrates that the ETEC strains with both characterized and novel sequence types are widely dispersed throughout the tree and not restricted to any particular phylogenetic group (Fig. 2). As seen in previous studies the ETEC type strain E. coli H10407 is located in group A (15). Figure 2 is rooted using group B2 as an outgroup, based on ancillary data and previous phylogenetic studies (15, 31). However, if the tree is midpoint rooted, or rooted using an outgroup such as Citrobacter species (data not shown), then the ETEC sequence type new 9 becomes the most basal, falling outside the E. coli diversity as represented by the ECOR strain collection. Blast analysis of the individual sequences for this strain reveal that it is most likely a recombinant, since three of the seven loci have typical E. coli sequences, while the remaining four are more similar to the equivalent regions from Escherichia fergusonii. The sequence may be partially derived from an ancestral-type E. coli outside the recognized E. coli diversity (69).
FIG. 2.
Phylogeny of ETEC clinical isolates. Based on MLST data the phylogenetic relationships of a collection of ETEC strains from humans and animals and a selection of ECOR strains are depicted. The phylogenetic tree is supplemented by data from the available genome-sequenced E. coli strains. Colored labels depict different sequence types depending on the source of strains harbored within each sequence type. The data demonstrate a polyphyletic distribution of the ETEC strains. The phylogenetic tree was constructed as for the right side of Fig. 1.
To add additional support to the groupings identified by MLST analyses, the ETEC isolates were tested using the rapid triplex PCR protocol described by Clermont et al. (see Table S1 in the supplemental material) (9). Overall there was 84% agreement with the group assignment based on our analysis between the triplex PCR protocol and the MLST analyses. Over half of the anomalies were sequence type 90, 14 animal isolates, which were assigned as group B1 by MLST and A by triplex PCR. If these isolates are excluded, congruency between the two data sets is over 92%, a figure similar to that noted by Clermont et al. (9). Previous analysis of ETEC strains had not detected any strains in group B2 or D (15). The triplex PCR showed agreement for strains assigned to these two groups, further confirming the observation that ETEC strains have arisen multiple times and can arise in any E. coli lineage.
Phylogeny of ETEC isolates from different host organisms.
In addition to infecting humans ETEC is also an important animal pathogen (17, 24, 39). The host specificity of ETEC strains is thought to be defined by specific CFs which are encoded on mobile genetic elements (5). Human and animal isolates were detected in all major branches of the tree including the B2 and D/E groups (Fig. 2). Furthermore, sequence types 10 and 29 contain isolates from both humans and animals (see Table S1 in the supplemental material). These data demonstrate that specific lineages of ETEC are not absolutely associated with one particular species and suggest that a specific genetic background is not important for species specificity.
Phylogeny of ETEC strains isolated from different geographic locations.
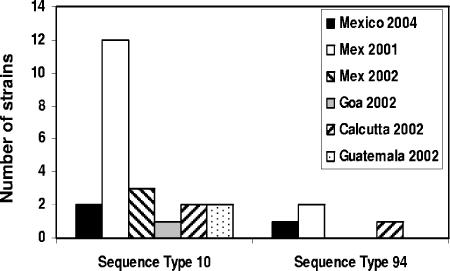
Previous phylogenetic analyses of both commensal and pathogenic isolates has demonstrated that E. coli is diverse and that this diversity is widely distributed, with closely related strains occurring in distinct geographic locations (10, 13, 16, 68). Similarly, the diversity of ETEC strains through the ETEC lineage is not restricted by geographic location, with ETEC strains from different locations occurring in all major phylogenetic groups (Fig. 2; see also Table S1 in the supplemental material). Furthermore, the frequently arising sequence type 10 was detected from four separate regions of the world at different time points (Fig. 3; see Table S1 in the supplemental material). The remaining sequence types also show variation in location.
FIG. 3.
Geographic distribution of ETEC sequence types. The graph depicts the geographic distribution and year of isolation for ETEC strains from representative MLST sequence types. The number of isolates in this study which was derived from each location is indicated. Strains belonging to the same sequence type have been identified in different regions of the world and in different years.
Distribution of the toxin genes among ETEC isolates.
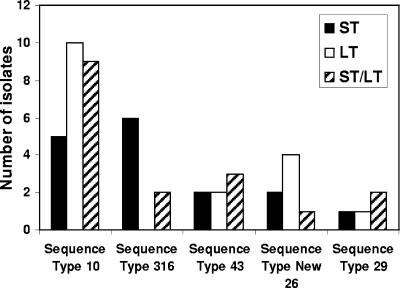
Strains belonging to the ETEC pathovar are traditionally defined by the presence of one or more of the toxin (ST and LT) genes and a specific CF (41). Previous investigations suggested that a specific genetic background was required for the acquisition of the genes encoding these toxins (15). The toxin genotypes were determined for each of the ETEC isolates included in this study (see Table S1 in the supplemental material). In agreement with previous studies (5, 27, 28, 50, 51, 59, 61, 70) multiple toxin genotypes, i.e., genes encoding either or both of the toxins, were detected among the clinical isolates. There was no apparent association of any one toxin genotype with any of the major phylogenetic groups (Fig. 2; see also Table S1 in the supplemental material). Furthermore, multiple toxin genotypes were detected within isolates of a single sequence type (Fig. 4; see also Table S1 in the supplemental material). These data suggest that the presence of the toxins is not dependent on clonal expansion and indicate that acquisition of the toxin genes is not dependent on a specific genetic background.
FIG. 4.
Distribution of toxin genotypes among ETEC isolates. The chart depicts the distribution of the three toxin genotypes (LT, ST, and LT/ST) among ETEC strains from the representative MLST sequence types. The number of isolates within a given sequence type which were positive for ST (black), LT (white), or both toxins (diagonally striped bars) is shown. The distribution of the toxins is not confined to a particular phylogenetic lineage or sequence type.
Distribution of accessory virulence factors among ETEC isolates.
In addition to the well-characterized toxin and CFs, a number of other proteins have been implicated in the pathogenicity of ETEC. These include ClyA, EatA, TibA, Tia, and LeoA (14, 19, 20, 46, 48). With the exception of TibA, the presence within the collection of ETEC isolates of the genes encoding these virulence factors was determined by amplification of a small specific PCR fragment within the coding region of the gene. TibA is encoded on a four-gene operon (tibABCD) (14, 32, 37), and to detect the presence of this locus a fragment of tibC, the gene encoding the TibA glycosylase, was amplified. This avoided the problems associated with nucleotide sequence similarity between homologous autotransporter proteins, and also, as the gene is not exposed to the selective pressure of the immune system, its sequence is less likely to vary. Very little is known about the abundance of these additional virulence genes within ETEC isolates. Despite being present in E. coli H10407, with the exception of clyA, the additional virulence factors are not abundant, although they are widely distributed and not confined to a phylogenetic group (Table 1; see also Table S1 in the supplemental material). This suggests that they are all mobile and that a specific genetic background is not required for their transmission; however, their rarity suggests that they have little consequence for the disease. Although clyA is widely distributed, it requires positive regulation from a lysR family regulator to induce a hemolytic phenotype; therefore, its role in pathogenicity is still unclear (46). Unlike E. coli H10407, none of the ETEC isolates, including those closely related to E. coli H10407, possessed all five of the additional virulence factors.
TABLE 1.
Function, prevalence and phylogenetic grouping of the additional ETEC virulence factors
| Gene | Protein/operon function | % Prevalence in ETEC isolates | Phylogenetic groups |
|---|---|---|---|
| clyA | Hemolysin | 93 | A, B1, B2, D/E |
| eatA | Serine protease autotransporter | 26 | A, B1, D/E |
| leoA | LT secretion | 3 | A, B1, D/E |
| tia | Outer membrane adhesin/invasin | 5 | A, B1, D/E |
| tib | Adhesin autotransporter | 16 | A, B1, B2, D/E |
As a further step, in an attempt to identify other potential virulence factors that may be essential for ETEC pathogenicity, we performed a comparison of the available complete and almost-complete E. coli genome sequences. Orthologous genes were identified using pairwise all-against-all BLAST searches. We looked for chromosomal genes that were shared among the three phylogenetically distinct ETEC genome sequences that are available but absent from all other E. coli genome sequences. Such genes would be strong candidates for a potential role in enterotoxigenicity. However, no such genes were identified. This is consistent with the hypothesis that there are no specific chromosomal factors required for enterotoxigenicity.
DISCUSSION
Understanding the phylogeny of pathogenic bacteria is essential to comprehending how pathogenic bacteria have evolved. MLST is increasingly being applied to large collections of bacterial isolates for phylogenetic studies (1, 36, 63). MLST uses protocols which were developed for experimental simplicity and the potential for automation (63). The increasing use of the Internet for the transfer of data has enabled large collections of MLST data to be collated and made available for public scrutiny. However, application of the MLST analysis methods to the well-studied ECOR collection revealed that in the case of E. coli, MLST was less discriminatory than MLEE; a number of isolates appear identical by MLST but are separated by MLEE. This observation may be explained by the fact that MLEE combines data derived from 38 enzyme loci, rather than the seven loci used for MLST, and presumably increasing the number of loci used in the MLST protocols would increase the discriminatory power of the technique.
Despite not possessing the discriminatory power of MLEE, MLST analysis of the ECOR collection showed accurate major branching of strains into phylogenetic groups, indicating that it was a suitable method for analyzing the phylogenetics of diarrheagenic E. coli. Extension of the study to incorporate a comprehensive collection of ETEC strains indicated that these strains are distributed throughout the E. coli lineage, in all major phylogenetic groups. While it is apparent from the MLST profiles of the ETEC strains that some clonal expansion may have occurred, the polyphyletic distribution of ETEC strains within the E. coli lineage indicates that ETEC strains have arisen multiple times and that there is no common clonal lineage. Previous phylogenetic investigations of ETEC using a variety of techniques have demonstrated similar results even though they were not placed within the context of the larger E. coli lineage. Thus, ETEC strains possessing the same CF-toxin genotypes often clustered together, suggesting that these strains had arisen by clonal expansion (33, 42, 43, 51, 59, 61, 64, 65). However, in these same studies the CF-toxin genotypes were not strictly associated with phylogenetic clusters and closely related strains were also found to possess different CF-toxin profiles. In combination with the MLST data, these observations indicate multiple and independent acquisitions of the CF and toxin genes by both phylogenetically similar and diverse strains and support the notion that ETEC strains do not have specific clonal lineages. The occurrence of such events may not be surprising if one considers that the CF and toxin genes are encoded on plasmids which may easily be transferred between E. coli strains from different phylogenetic groups.
Proportionally, more ETEC strains were detected in the A and B1 lineages than in the B2 and D phylogenetic groups. Although not restrictive, this could reflect a predilection of the plasmids encoding the toxin and CFs for a particular genetic background. Indeed, phylogenetic analyses of a diverse array of pathogenic and commensal E. coli isolates by Escobar-Paramo et al. appeared to suggest that a specific background was required for the acquisition of the ETEC virulence factors (15). However, this study was based on analysis of only a small number of ETEC isolates, and given the widespread phylogenetic differences in the ETEC strains analyzed in this study the argument that ETEC strains require a particular genetic background for enterotoxigenicity seems improbable.
An alternative, and perhaps more likely, explanation for the association of ETEC strains with the A and B1 phylogenetic clusters is that such strains are simply more abundant in nature. Support for this latter theory is derived from several studies which indicate that group A and B1 strains are more prevalent as commensal organisms in both animals and humans (12, 13, 16). Furthermore, one study specifically highlighted the fact that the majority of commensal E. coli strains isolated from individuals living in tropical regions belonged to the A and B1 phylogenetic groups (16). This is an important observation given that ETEC tends to be endemic in tropical areas and that all of our isolates were derived from tropical regions. Interestingly, a recent MLST analysis of a large collection of E. coli strains suggested that bacteria belonging to the A group, and in particular the sequence type 10 complex, were not pathogenic (69). However, in this study we found that sequence type 10 possessed the highest number of ETEC isolates. These data pose the tantalizing suggestion that nonpathogenic bacteria may become pathogenic simply by the acquisition of plasmids encoding a CF and one of the toxins. Further support for this hypothesis can be derived from a recent study which demonstrated that the laboratory-adapted commensal E. coli MG1655 strain and the prototypical ETEC strain (E. coli H10407) were highly similar and were predicted to be 96% identical; the main differences between the two strains could in large part be accounted for by genetic regions encoding the putative E. coli H10407 virulence factors identified previously in several different studies, viz., the clyA, tia, leoA, and tib loci (8).
A question which arose during the course of this study was whether any of the previously identified putative virulence factors contributed significantly to the pathogenesis of ETEC. The clyA gene was detected in the majority of ETEC strains, which would appear to suggest a role for ClyA in pathogenesis. However, clyA has also been detected in commensal E. coli as well as pathogenic strains of the other E. coli pathovars (35, 45, 46). Thus, even if ClyA plays a role in pathogenesis it is not specifically associated with ETEC-mediated disease. Fleckenstein et al. identified leoA as part of a four-gene operon present on a pathogenicity island of E. coli H10407, which played an undefined role in LT secretion (20). However, it is clear from the current study that leoA is not abundantly present within ETEC and numerous isolates possessing only the LT toxin were devoid of leoA, suggesting that it is not required for ETEC pathogenicity. Similarly, the relative rarity of the tia and tib loci suggest that their impact on ETEC-mediated disease is minimal. Comparative genomic analysis of the three ETEC strains for which genomic data are available supports these observations and furthermore demonstrates that no single gene is present within the chromosomal background which is not also present within commensal strains. These observations support the hypothesis that acquisition of a toxin and CF is sufficient for the emergence of a pathogenic ETEC strain. Nevertheless, it remains possible that these virulence factors perform roles in E. coli H10407 pathogenesis that are performed by functional analogues in other ETEC strains or that they contribute to the various degrees of disease severity which have been observed in infected individuals (50). However, it should be pointed out that the absence of chromosomally encoded virulence factors does not rule out the possibility that additional virulence factors, other than the CF and toxins, are encoded on the virulence-associated plasmids. In this respect, Froehlich et al. recently demonstrated the presence of loci encoding VirK and the serine protease autotransporter EatA on the large pCoo virulence plasmid, homologues of which have previously been implicated in the virulence of pathogenic E. coli (21, 25, 48).
The absence of conserved chromosomal antigens and the genetic variation in the chromosomal background of ETEC may explain the lack of complete protection against infection with ETEC possessing the same CF-toxin profiles. It was noted previously that that ETEC infection with a particular CF-toxin profile induced partial protection (47%) against strains with exactly the same CF-toxin profile and that the CFs and toxins did not contribute significantly to this protection (60, 64). The results presented here suggest that the lack of complete protection against strains with the same CF-toxin profile may be due to the fact that the strains responsible for subsequent infection possess genetically distinct backgrounds and therefore a different repertoire of antigens which are not recognized by the immune system. Such lack of cross protection and the genetic variation of ETEC strains pose significant problems for development of a broadly efficacious ETEC vaccine. Purified CFs, LT B subunits, inactivated whole cells, and live-attenuated organisms have all been investigated as potential vaccines (for reviews see references 6 and 50) but have met with only marginal success. If immunity to ETEC infection is based solely on common chromosomally encoded factors, ETEC vaccines will have to include a repertoire of different protective antigens from across the breadth of the E. coli lineage in order to be efficacious. The collection of strains examined in this study, which we refer to as the ETEC collection, represents an invaluable resource for such studies.
In conclusion we have demonstrated that ETEC strains have a polyphyletic origin, implying that the genetic background of ETEC strains is not conserved. ETEC is one of the last pathovars of E. coli to be sequenced, which has limited our understanding of factors other than the toxins and CFs which might be involved in virulence. With further completed ETEC genomes it is possible that new ETEC virulence genes will be identified. However, the diversity observed here suggests that efforts in the search for novel virulence factors and potential vaccine candidates should be concentrated on sequencing the virulence-associated plasmids of a wide range of isolates.
Supplementary Material
Acknowledgments
S.M.T. and R.R.C. are supported by BBSRC grants RRAF10791 and EGA16107, respectively.
Sequencing was performed by the Functional Genomics Laboratory, The University of Birmingham. Data from several ongoing E. coli genome projects were made available by the Wellcome Trust Sanger Institute (http://www.sanger.ac.uk).
Footnotes
Published ahead of print on 18 October 2006.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Aanensen, D. M., and B. G. Spratt. 2005. The multilocus sequence typing network: mlst.net. Nucleic Acids Res. 33:W728-W733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnold, C., L. Metherell, G. Willshaw, A. Maggs, and J. Stanley. 1999. Predictive fluorescent amplified-fragment length polymorphism analysis of Escherichia coli: high-resolution typing method with phylogenetic significance. J. Clin. Microbiol. 37:1274-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beutin, L., S. Kaulfuss, S. Herold, E. Oswald, and H. Schmidt. 2005. Genetic analysis of enteropathogenic and enterohemorrhagic Escherichia coli serogroup O103 strains by molecular typing of virulence and housekeeping genes and pulsed-field gel electrophoresis. J. Clin. Microbiol. 43:1552-1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beutin, L., I. Orskov, F. Orskov, S. Zimmermann, J. Prada, H. Gelderblom, R. Stephan, and T. S. Whittam. 1990. Clonal diversity and virulence factors in strains of Escherichia coli of the classic enteropathogenic serogroup O114. J. Infect. Dis. 162:1329-1334. [DOI] [PubMed] [Google Scholar]
- 5.Blanco, J., M. Blanco, J. I. Garabal, and E. A. Gonzalez. 1991. Enterotoxins, colonization factors and serotypes of enterotoxigenic Escherichia coli from humans and animals. Microbiologia 7:57-73. [PubMed] [Google Scholar]
- 6.Boedeker, E. C. 2005. Vaccines for enterotoxigenic Escherichia coli: current status. Curr. Opin. Gastroenterol. 21:15-19. [PubMed] [Google Scholar]
- 7.Chaudhuri, R. R., A. M. Khan, and M. J. Pallen. 2004. coliBASE: an online database for Escherichia coli, Shigella and Salmonella comparative genomics. Nucleic Acids Res. 32:D296-D299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, Q., S. J. Savarino, and M. M. Venkatesan. 2006. Subtractive hybridization and optical mapping of the enterotoxigenic Escherichia coli H10407 chromosome: isolation of unique sequences and demonstration of significant similarity to the chromosome of E. coli K-12. Microbiology 152:1041-1054. [DOI] [PubMed] [Google Scholar]
- 9.Clermont, O., S. Bonacorsi, and E. Bingen. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66:4555-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Czeczulin, J. R., T. S. Whittam, I. R. Henderson, F. Navarro-Garcia, and J. P. Nataro. 1999. Phylogenetic analysis of enteroaggregative and diffusely adherent Escherichia coli. Infect. Immun. 67:2692-2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diamant, E., Y. Palti, R. Gur-Arie, H. Cohen, E. M. Hallerman, and Y. Kashi. 2004. Phylogeny and strain typing of Escherichia coli, inferred from variation at mononucleotide repeat loci. Appl. Environ. Microbiol. 70:2464-2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dixit, S. M., D. M. Gordon, X. Wu, T. Chapman, K. Kailasapathy, and J. J. Chin. 2004. Diversity analysis of commensal porcine Escherichia coli—associations between genotypes and habitat in the porcine gastrointestinal tract. Microbiology 150:1735-1740. [DOI] [PubMed] [Google Scholar]
- 13.Duriez, P., O. Clermont, S. Bonacorsi, E. Bingen, A. Chaventre, J. Elion, B. Picard, and E. Denamur. 2001. Commensal Escherichia coli isolates are phylogenetically distributed among geographically distinct human populations. Microbiology 147:1671-1676. [DOI] [PubMed] [Google Scholar]
- 14.Elsinghorst, E. A., and J. A. Weitz. 1994. Epithelial cell invasion and adherence directed by the enterotoxigenic Escherichia coli tib locus is associated with a 104-kilodalton outer membrane protein. Infect. Immun. 62:3463-3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Escobar-Paramo, P., O. Clermont, A. B. Blanc-Potard, H. Bui, C. Le Bouguenec, and E. Denamur. 2004. A specific genetic background is required for acquisition and expression of virulence factors in Escherichia coli. Mol. Biol. Evol. 21:1085-1094. [DOI] [PubMed] [Google Scholar]
- 16.Escobar-Páramo, P., K. Grenet, A. Le Menac'h, L. Rode, E. Salgado, C. Amorin, S. Gouriou, B. Picard, M. C. Rahimy, A. Andremont, E. Denamur, and R. Ruimy. 2004. Large-scale population structure of human commensal Escherichia coli isolates. Appl. Environ. Microbiol. 70:5698-5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fairbrother, J. M., E. Nadeau, and C. L. Gyles. 2005. Escherichia coli in postweaning diarrhea in pigs: an update on bacterial types, pathogenesis, and prevention strategies. Anim. Health Res. Rev. 6:17-39. [DOI] [PubMed] [Google Scholar]
- 18.Fleckenstein, J. M., J. T. Holland, and D. L. Hasty. 2002. Interaction of an outer membrane protein of enterotoxigenic Escherichia coli with cell surface heparan sulfate proteoglycans. Infect. Immun. 70:1530-1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fleckenstein, J. M., D. J. Kopecko, R. L. Warren, and E. A. Elsinghorst. 1996. Molecular characterization of the tia invasion locus from enterotoxigenic Escherichia coli. Infect. Immun. 64:2256-2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fleckenstein, J. M., L. E. Lindler, E. A. Elsinghorst, and J. B. Dale. 2000. Identification of a gene within a pathogenicity island of enterotoxigenic Escherichia coli H10407 required for maximal secretion of the heat-labile enterotoxin. Infect. Immun. 68:2766-2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Froehlich, B., J. Parkhill, M. Sanders, M. A. Quail, and J. R. Scott. 2005. The pCoo plasmid of enterotoxigenic Escherichia coli is a mosaic cointegrate. J. Bacteriol. 187:6509-6516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gaastra, W., and A. M. Svennerholm. 1996. Colonization factors of human enterotoxigenic Escherichia coli (ETEC). Trends Microbiol. 4:444-452. [DOI] [PubMed] [Google Scholar]
- 23.Galtier, N., M. Gouy, and C. Gautier. 1996. SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Comput. Appl. Biosci. 12:543-548. [DOI] [PubMed] [Google Scholar]
- 24.Harnett, N. M., and C. L. Gyles. 1985. Enterotoxin plasmids in bovine and porcine enterotoxigenic Escherichia coli of O groups 9, 20, 64 and 101. Can. J. Comp. Med. 49:79-87. [PMC free article] [PubMed] [Google Scholar]
- 25.Henderson, I. R., F. Navarro-Garcia, M. Desvaux, R. C. Fernandez, and D. Ala'Aldeen. 2004. Type V protein secretion pathway: the autotransporter story. Microbiol. Mol. Biol. Rev. 68:692-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Herzer, P. J., S. Inouye, M. Inouye, and T. S. Whittam. 1990. Phylogenetic distribution of branched RNA-linked multicopy single-stranded DNA among natural isolates of Escherichia coli. J. Bacteriol. 172:6175-6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang, Z. D., B. Lowe, M. P. Verenkar, D. Ashley, R. Steffen, N. Tornieporth, F. von Sonnenburg, P. Waiyaki, and H. L. DuPont. 2002. Prevalence of enteric pathogens among international travelers with diarrhea acquired in Kenya (Mombasa), India (Goa), or Jamaica (Montego Bay). J. Infect. Dis. 185:497-502. [DOI] [PubMed] [Google Scholar]
- 28.Jiang, Z. D., J. J. Mathewson, C. D. Ericsson, A. M. Svennerholm, C. Pulido, and H. L. DuPont. 2000. Characterisation of enterotoxigenic Escherichia coli strains in patients with travelers' diarrhea acquired in Guadalajara, Mexico, 1992-1997. J. Infect. Dis. 181:779-782. [DOI] [PubMed] [Google Scholar]
- 29.Lan, R., and P. R. Reeves. 2002. Escherichia coli in disguise: molecular origins of Shigella. Microbes Infect. 4:1125-1132. [DOI] [PubMed] [Google Scholar]
- 30.Lawrence, J. G., and H. Ochman. 2002. Reconciling the many faces of lateral gene transfer. Trends Microbiol. 10:1-4. [DOI] [PubMed] [Google Scholar]
- 31.Lecointre, G., L. Rachdi, P. Darlu, and E. Denamur. 1998. Escherichia coli molecular phylogeny using the incongruence length difference test. Mol. Biol. Evol. 15:1685-1695. [DOI] [PubMed] [Google Scholar]
- 32.Lindenthal, C., and E. A. Elsinghorst. 1999. Identification of a glycoprotein produced by enterotoxigenic Escherichia coli. Infect. Immun. 67:4084-4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lortie, L. A., J. D. Dubreuil, and J. Harel. 1991. Characterization of Escherichia coli strains producing heat-stable enterotoxin b (STb) isolated from humans with diarrhea. J. Clin. Microbiol. 29:656-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ludwig, A., S. Bauer, R. Benz, B. Bergmann, and W. Goebel. 1999. Analysis of the SlyA-controlled expression, subcellular localization and pore-forming activity of a 34 kDa haemolysin (ClyA) from Escherichia coli K-12. Mol. Microbiol. 31:557-567. [DOI] [PubMed] [Google Scholar]
- 35.Ludwig, A., C. von Rhein, S. Bauer, C. Huttinger, and W. Goebel. 2004. Molecular analysis of cytolysin A (ClyA) in pathogenic Escherichia coli strains. J. Bacteriol. 186:5311-5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maiden, M. C., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moormann, C., I. Benz, and M. A. Schmidt. 2002. Functional substitution of the TibC protein of enterotoxigenic Escherichia coli strains for the autotransporter adhesin heptosyltransferase of the AIDA system. Infect. Immun. 70:2264-2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murray, B. E., J. J. Mathewson, H. L. DuPont, and W. E. Hill. 1987. Utility of oligodeoxyribonucleotide probes for detecting enterotoxigenic Escherichia coli. J. Infect. Dis. 155:809-811. [DOI] [PubMed] [Google Scholar]
- 39.Nagy, B., and P. Z. Fekete. 1999. Enterotoxigenic Escherichia coli (ETEC) in farm animals. Vet. Res. 30:259-284. [PubMed] [Google Scholar]
- 40.Nair, G. B., and Y. Takeda. 1998. The heat-stable enterotoxins. Microb. Pathog. 24:123-131. [DOI] [PubMed] [Google Scholar]
- 41.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nishikawa, Y., A. Helander, J. Ogasawara, N. P. Moyer, M. Hanaoka, A. Hase, and A. Yasukawa. 1998. Epidemiology and properties of heat-stable enterotoxin-producing Escherichia coli serotype O169:H41. Epidemiol. Infect. 121:31-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Okamoto, K., Y. Fujii, N. Akashi, S. Hitotsubashi, H. Kurazono, T. Karasawa, and Y. Takeda. 1993. Identification and characterization of heat-stable enterotoxin II-producing Escherichia coli from patients with diarrhea. Microbiol. Immunol. 37:411-414. [DOI] [PubMed] [Google Scholar]
- 44.Orskov, F., T. S. Whittam, A. Cravioto, and I. Orskov. 1990. Clonal relationships among classic enteropathogenic Escherichia coli (EPEC) belong to different O groups. J. Infect. Dis. 162:76-81. [DOI] [PubMed] [Google Scholar]
- 45.Oscarsson, J., Y. Mizunoe, L. Li, X. H. Lai, A. Wieslander, and B. E. Uhlin. 1999. Molecular analysis of the cytolytic protein ClyA (SheA) from Escherichia coli. Mol. Microbiol. 32:1226-1238. [DOI] [PubMed] [Google Scholar]
- 46.Oscarsson, J., Y. Mizunoe, B. E. Uhlin, and D. J. Haydon. 1996. Induction of haemolytic activity in Escherichia coli by the slyA gene product. Mol. Microbiol. 20:191-199. [DOI] [PubMed] [Google Scholar]
- 47.Parham, N. J., S. J. Pollard, R. R. Chaudhuri, S. A. Beatson, M. Desvaux, M. A. Russell, J. Ruiz, A. Fivian, J. Vila, and I. R. Henderson. 2005. Prevalence of pathogenicity island IICFT073 genes among extraintestinal clinical isolates of Escherichia coli. J. Clin. Microbiol. 43:2425-2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Patel, S. K., J. Dotson, K. P. Allen, and J. M. Fleckenstein. 2004. Identification and molecular characterization of EatA, an autotransporter protein of enterotoxigenic Escherichia coli. Infect. Immun. 72:1786-1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Picard, B., J. S. Garcia, S. Gouriou, P. Duriez, N. Brahimi, E. Bingen, J. Elion, and E. Denamur. 1999. The link between phylogeny and virulence in Escherichia coli extraintestinal infection. Infect. Immun. 67:546-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qadri, F., A. M. Svennerholm, A. S. Faruque, and R. B. Sack. 2005. Enterotoxigenic Escherichia coli in developing countries: epidemiology, microbiology, clinical features, treatment, and prevention. Clin. Microbiol. Rev. 18:465-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Regua-Mangia, A. H., B. E. Guth, J. R. da Costa Andrade, K. Irino, A. B. Pacheco, L. C. Ferreira, V. Zaharia, and L. M. Teixeira. 2004. Genotypic and phenotypic characterisation of enterotoxigenic Escherichia coli (ETEC) strains isolated in Rio de Janeiro city, Brazil. FEMS Immunol. Med. Microbiol. 40:155-162. [DOI] [PubMed] [Google Scholar]
- 52.Ren, C. P., S. A. Beatson, J. Parkhill, and M. J. Pallen. 2005. The Flag-2 locus, an ancestral gene cluster, is potentially associated with a novel flagellar system from Escherichia coli. J. Bacteriol. 187:1430-1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ren, C. P., R. R. Chaudhuri, A. Fivian, C. M. Bailey, M. Antonio, W. M. Barnes, and M. J. Pallen. 2004. The ETT2 gene cluster, encoding a second type III secretion system from Escherichia coli, is present in the majority of strains but has undergone widespread mutational attrition. J. Bacteriol. 186:3547-3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rodrigues, J., I. C. Scaletsky, L. C. Campos, T. A. Gomes, T. S. Whittam, and L. R. Trabulsi. 1996. Clonal structure and virulence factors in strains of Escherichia coli of the classic serogroup O55. Infect. Immun. 64:2680-2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ronquist, F., and J. P. Huelsenbeck. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572-1574. [DOI] [PubMed] [Google Scholar]
- 56.Sears, C. L., and J. B. Kaper. 1996. Enteric bacterial toxins: mechanisms of action and linkage to intestinal secretion. Microbiol. Rev. 60:167-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shapiro, B., A. Rambaut, and A. J. Drummond. 2006. Choosing appropriate substitution models for the phylogenetic analysis of protein-coding sequences. Mol. Biol. Evol. 23:7-9. [DOI] [PubMed] [Google Scholar]
- 58.Sherlock, O., R. M. Vejborg, and P. Klemm. 2005. The TibA adhesin/invasin from enterotoxigenic Escherichia coli is self recognizing and induces bacterial aggregation and biofilm formation. Infect. Immun. 73:1954-1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Steinsland, H., P. Valentiner-Branth, P. Aaby, K. Molbak, and H. Sommerfelt. 2004. Clonal relatedness of enterotoxigenic Escherichia coli strains isolated from a cohort of young children in Guinea-Bissau. J. Clin. Microbiol. 42:3100-3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Steinsland, H., P. Valentiner-Branth, H. K. Gjessing, P. Aaby, K. Molbak, and H. Sommerfelt. 2003. Protection from natural infections with enterotoxigenic Escherichia coli: longitudinal study. Lancet 362:286-291. [DOI] [PubMed] [Google Scholar]
- 61.Steinsland, H., P. Valentiner-Branth, M. Perch, F. Dias, T. K. Fischer, P. Aaby, K. Molbak, and H. Sommerfelt. 2002. Enterotoxigenic Escherichia coli infections and diarrhea in a cohort of young children in Guinea-Bissau. J Infect. Dis. 186:1740-1747. [DOI] [PubMed] [Google Scholar]
- 62.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Urwin, R., and M. C. Maiden. 2003. Multi-locus sequence typing: a tool for global epidemiology. Trends Microbiol. 11:479-487. [DOI] [PubMed] [Google Scholar]
- 64.Valentiner-Branth, P., H. Steinsland, T. K. Fischer, M. Perch, F. Scheutz, F. Dias, P. Aaby, K. Molbak, and H. Sommerfelt. 2003. Cohort study of Guinean children: incidence, pathogenicity, conferred protection, and attributable risk for enteropathogens during the first 2 years of life. J. Clin. Microbiol. 41:4238-4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Valvatne, H., H. Steinsland, and H. Sommerfelt. 2002. Clonal clustering and colonization factors among thermolabile and porcine thermostable enterotoxin-producing Escherichia coli. APMIS 110:665-672. [DOI] [PubMed] [Google Scholar]
- 66.Welch, R. A., V. Burland, G. Plunkett III, P. Redford, P. Roesch, D. Rasko, E. L. Buckles, S. R. Liou, A. Boutin, J. Hackett, D. Stroud, G. F. Mayhew, D. J. Rose, S. Zhou, D. C. Schwartz, N. T. Perna, H. L. Mobley, M. S. Donnenberg, and F. R. Blattner. 2002. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 99:17020-17024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Westermark, M., J. Oscarsson, Y. Mizunoe, J. Urbonaviciene, and B. E. Uhlin. 2000. Silencing and activation of ClyA cytotoxin expression in Escherichia coli. J. Bacteriol. 182:6347-6357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Whittam, T. S., M. L. Wolfe, I. K. Wachsmuth, F. Orskov, I. Orskov, and R. A. Wilson. 1993. Clonal relationships among Escherichia coli strains that cause hemorrhagic colitis and infantile diarrhea. Infect. Immun. 61:1619-1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wirth, T., D. Falush, R. Lan, F. Colles, P. Mensa, L. H. Wieler, H. Karch, P. R. Reeves, M. C. Maiden, H. Ochman, and M. Achtman. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol. Microbiol. 60:1136-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wolf, M. K. 1997. Occurrence, distribution, and association of O and H serogroups, colonization factor antigens, and toxins of enterotoxigenic Escherichia coli. Clin. Microbiol. Rev. 10:569-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.World Health Organization. 1999. The world health report 1999: making a difference. World Health Organization, Geneva, Switzerland.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.