Abstract
Nontyphoidal salmonellae are among the most common causes of bacterial gastroenteritis worldwide. They are also notable causes of extraintestinal infections, including bacteremia and vascular infections. Salmonella enterica serotype Choleraesuis is typically associated with invasive infections. We report a patient who had an infected intra-abdominal aortic aneurysm due to an unusually mucoid strain of Salmonella enterica serotype Choleraesuis. The isolate was erroneously identified as Hafnia alvei by the Vitek GNI+ card system. A blood culture isolate taken from the same patient 9 months earlier was also identified as H. alvei by the Vitek GNI+ card system. Despite an apparent cure with intravenous amoxicillin-clavulanic acid at that time, the Salmonella infection had not been cleared and manifested as a ruptured infected abdominal aortic aneurysm. Repeated passage of the strain yielded nonmucoid colonies, which were correctly identified by the API and PHOENIX systems. The isolates from the aneurysm and the former bacteremic episode were found to be identical using pulsed field gel electrophoresis. The fallibility of automated bacterial identification systems is highlighted. Such errors are especially important for isolates in which in vitro antibiotic susceptibility testing does not correlate with the clinical success of treatment, as illustrated by Salmonella infections.
CASE REPORT
A 75-year-old man was admitted to our hospital in March 2006 because of persistent back pain and abdominal distention lasting for 2 days. He had a history of hypertension, transient ischemic attack, and pulmonary tuberculosis complicated with bronchiectasis. At the time of admission, his oral temperature was 37.5°C, with a blood pressure of 163/87 mmHg and heart rate of 85 beats per minute. A physical examination revealed distension and tenderness in the lower abdomen. His total leukocyte count was 13.6 × 109/liter (neutrophils, 11.8 × 109/liter; lymphocytes, 0.8 × 109/liter), and his platelet count was 306 × 109/liter. The hemoglobin level dropped from 13.7 to 10.7 g/dl within 10 h after admission, but the clotting profile and liver and renal function tests were all within normal limits. An abdominal radiograph revealed widening of the left psoas shadow. An urgent computed tomography scan revealed an aneurysmal dilatation of the infrarenal aorta extending to the aortic bifurcation, with intramural thrombus and retroperitoneal hematoma, compatible with ruptured infrarenal abdominal aortic aneurysm. Suturing of the aneurismal sac with interposition tube graft bypass was performed. The intramural thrombus was removed and sent for culture. A gram-negative bacillus was recovered from the thrombus culture, which was initially identified as Hafnia alvei. Review of the patient's hospital record showed that he had had an episode of fever and H. alvei bacteremia 9 months before, which resolved with 2 weeks of intravenous amoxicillin-clavulanic acid. He remained afebrile postoperatively. In view of the unusual thrombus culture result, blood was sent for testing by bacterial culture and Widal test (a serological test for typhoidal salmonellosis) while the definitive identification of the bacterial isolates was being performed. He was given intravenous ceftriaxone and discharged after 22 days of hospitalization.
All clinical specimens were collected and handled according to standard protocols (11). A Gram smear of the intramural thrombus revealed the presence of gram-negative rods, which grew on blood agar, chocolate agar, and MacConkey agar as mucoid colonies 4 mm in diameter after 24 h of incubation at 37°C in ambient air (Fig. 1A). The strain fermented glucose, reduced nitrate, and did not produce cytochrome oxidase, which is typical for a member of the family Enterobacteriaceae. Both standard conventional biochemical tests and the Vitek system (GNI+) with software version R09.01 (bioMerieux Vitek, Durham, NC) and the PHOENIX system with software version 3.34A/3.52F (BD, Sparks, MD) failed to identify the mucoid strain, which did not fit the typical profiles of known bacterial species (11). The former identified the strain as an inactive Escherichia coli isolate (normalized percent probability, 31%), and the latter identified it as CDC group EF-4a (confidence value, 90%). The isolate did not agglutinate with poly(O) and poly(H) Salmonella antisera (Murex Biotech Ltd., Temple Hill, Dartford, United Kingdom). API 20E (bioMerieux Vitek, Hazelwood, MO) testing was not performed on the mucoid strain. The bacterium was then repeatedly passaged on blood agar until it reverted to a nonmucoid form (Fig. 1B). Conventional biochemical testing showed that the nonmucoid strain was indole negative and lactose nonfermenting, as well as that it decarboxylated lysine and ornithine and did not utilize citrate or produce hydrogen sulfide. The Vitek GNI+ card system identified the nonmucoid strain as Hafnia alvei (normalized percent probability, 97%), identical to the strain isolated from the patient's blood 9 months earlier, which was not mucoid. No additional tests were recommended. Nonetheless, in view of the atypical clinical presentation, testing of the nonmucoid strain with the API 20E and PHOENIX systems identified the bacterium as Salmonella enterica serotype Choleraesuis. Serotyping with Salmonella antisera was performed, giving an antigenic formula of 6,7:c:1,5 and a negative result for the Vi antigen. Widal testing performed on a serum sample obtained 20 days after admission showed antibody titers of 200 for serotype Paratyphi C-H phase 1 flagellar antigen c and <50 for serotype Typhi O group D somatic antigens, serotype Typhi H flagellar antigen d, serotype Paratyphi A-H flagellar antigen b, and serotype Paratyphi B-H phase 1 flagellar antigen b. As the antigenic formula of Salmonella enterica serotype Choleraesuis (6,7:c:1,5) is similar to that of Salmonella enterica serotype Paratyphi C (6,7,[Vi]:c:1,5), the elevated serotype Paratyphi C-H phase 1 flagellar antigen c titer was likely to be due to cross-reactivity between the two serotypes. The strain isolated from the patient is not Salmonella enterica serotype Paratyphi C because it is negative for the Vi antigen and does not ferment arabinose and trehalose. Ceftriaxone was continued for a total of 21 days. The patient has remained asymptomatic up to the time of this writing, 3 months after discharge.
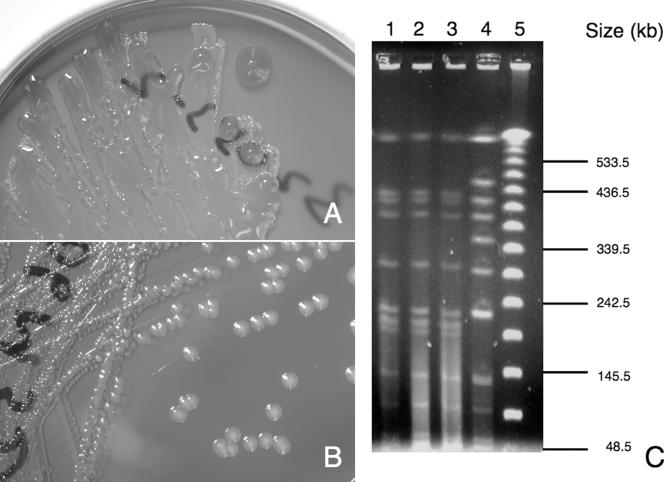
FIG. 1.
Characteristics of the isolate from thrombus. (A) Mucoid colonies on blood agar after 24 h of incubation at 37°C in ambient air. (B) Change of colony morphology to nonmucoid colonies on blood agar after repeated passaging. (C) PFGE patterns of Salmonella enterica serotype Choleraesuis from patients, using XbaI for chromosomal DNA digestion. Lane 1 was the mucoid strain isolated from this patient's intramural thrombus; lane 2 was the nonmucoid strain transformed from the mucoid one; lane 3 was the blood isolate from the same patient 9 months ago; lane 4 was another strain of Salmonella enterica serotype Choleraesuis isolated from blood of another unrelated patient, which served as a control for comparison, and lane 5 was 48.5 kb-lambda ladder.
In view of the discrepancy between the Vitek system and other commercial systems and the agglutination test result, 16S rRNA sequencing using the MicroSeq 500 16S rRNA bacterial identification kit (Perkin-Elmer Applied Biosystems Division, Foster City, CA) was performed according to the manufacturer's instructions. The DNA sequence was compared with known 16S rRNA gene sequences in GenBank by multiple sequence alignment using the CLUSTAL W program (14). The strain was identified as S. enterica, with 1% discrepancy compared to the isolate sequences of S. enterica serotype Choleraesuis.
The so-called H. alvei strain isolated in July 2005 was examined by conventional biochemical tests, the Vitek GNI+ system, and the PHOENIX system. A discrepancy between the Vitek and PHOENIX systems in identification of the strain was noted again. Agglutination with Salmonella was performed, which gave an antigenic formula identical to that of the strain isolated from the thrombus. The susceptibility testing by disk diffusion test on this bacteremic strain of S. enterica serotype Choleraesuis and the nonmucoid thrombus strain from the current admission was performed according to the Clinical and Laboratory Standards Institute standard (3), both of which were susceptible to ampicillin, cefotaxime, ceftriaxone, chloramphenicol, ciprofloxacin, cotrimoxazole, and nalidixic acid. Pulsed-field gel electrophoresis (PFGE) of this patient's S. enterica serotype Choleraesuis isolates and a strain from another patient was performed. The protocol for PFGE analysis has been described previously (9). XbaI was used for digestion of DNA in the PFGE analysis. The result, as demonstrated in Fig. 1C, confirmed that the strains isolated from the blood and the intramural thrombus (both mucoid and nonmucoid phenotypes) in this patient were identical but different from an unrelated strain.
Nontyphoidal salmonellae are associated with self-limiting gastrointestinal tract infection and other extraintestinal infections, such as intravascular infections, arthritis, osteomyelitis, and bacteremia. Severe or recurrent infections are often seen in the elderly or immunocompromised hosts. The Choleraesuis serotype is notable in that it is typically associated with invasive extraintestinal infections rather than gastroenteritis (2). The infection is frequently characterized by bacteremia and severe sepsis with localized pyogenic infections, such as osteomyelitis, arthritis, and vascular infections, such as aortitis and mycotic aneurysm. These focal infections require prolonged and high doses of antibiotics for treatment. Correct identification of this serotype is therefore crucial to alert the clinician to search for an infective focus and for proper management of the patients. On the other hand, the role of Hafnia alvei as a human pathogen is less well established. Sites most likely to yield Hafnia spp. as the sole pathogen include urine and blood samples. H. alvei has been reported to be associated with bacteremia, gastroenteritis, nosocomial pneumonia, and urinary tract infection (7) but has never been reported to cause intravascular infections.
This report is the first documented case of S. enterica serotype Choleraesuis being misidentified as H. alvei by the Vitek GNI+ system. Previously, the accuracy of the system in identifying species of the Enterobacteriaceae family ranged from 75% to 94.4% (1, 5, 12, 13). Table 1 summarizes the current case and four other reports of the Vitek GNI+ card system being unable to identify or incorrectly identifying nontyphoidal salmonellae. Even though Knight et al. (8) reported the accuracy of the Vitek system in identifying Salmonella spp. from food to be as high as 96.7%, the accuracy of identification of isolates from humans in the studies was only 72%. The most common form of misidentification is incorrect identification of Salmonella spp. as E. coli. There was also one isolate of S. enterica subsp. arizonae being mistaken as H. alvei. We noticed that the reason for misidentification was associated with aberrant results of four tests: acid formation from sorbitol and arabinose, fermentation of O-nitrophenyl-beta-d-galacto-pyranoside, and hydrogen sulfide production (Table 2). We suggest that these tests be repeated by conventional methods if identification of strains is doubtful or atypical.
TABLE 1.
Outcome of Vitek GNI+ card on identification of Salmonella spp. from previous and current studies
| Reference | Software version of Vitek system | Organism | No. of isolates
|
|||
|---|---|---|---|---|---|---|
| Total tested | Correctly identified | Not identified | Misidentified (species identified) | |||
| Bourbeau and Heiter (1) | R05.01 | Salmonella spp. | 5 | 5 | 0 | 0 |
| Hansen et al. (5) | R06.01 | Salmonella spp. | 11 | 6 | 4 | 1 (Citrobacter freundii) |
| O'Hara and Miller (12) | Not mentioned (VT2-R02.02 | Salmonella enterica subsp. arizonae | 7 | 5 | 0 | 2 (Escherichia coli) |
| for ID-GNB cards) | Salmonella enterica serotype Choleraesuis | 2 | 1 | 1 | 0 | |
| Salmonella enterica serotype Typhimurium | 1 | 1 | 0 | 0 | ||
| O'Hara et al. (13) | R05.01 | Salmonella enterica subsp. arizonae | 10 | 8 | 0 | 2 (1 Hafnia alvei, 1 Escherichia coli) |
| Salmonella spp. | 16 | 12 | 0 | 4 (3 Escherichia coli, 1 Serratia plymuthica) | ||
| This study | R09.01 | Salmonella enterica serotype Choleraesuis | 1 | 0 | 0 | 1 (Hafnia alvei) |
| Total | 53 | 38 (71.7%) | 5 (9.4%) | 10 (18.9%) | ||
TABLE 2.
Results of four biochemical tests of the patient isolate (nonmucoid strain), Salmonella enterica serotype Choleraesuis, and Hafnia alvei
| Strain or parameter | Biochemical reaction tested
|
|||
|---|---|---|---|---|
| Acid formation from sorbitol | Acid formation from arabinose | ONPGa fermentation | H2S production | |
| Patient isolate (nonmucoid strain) as tested by Vitek GNI+ card system | − | − | − | − |
| Expected result for Salmonella enterica serotype Choleraesuis | + | − | − | + |
| Expected result for Hafnia alvei | − | + | + | − |
ONPG, O-nitrophenyl-beta-d-galacto-pyranoside.
Misidentification of Salmonella spp. is of great concern, especially when the bacterium is isolated from extraintestinal sites. When incorrectly identified as E. coli and H. alvei, the choice of antibiotic treatment may be inappropriate. Treatment with ampicillin and third-generation cephalosporins (such as ceftriaxone) should clear up the bacteremia. However, it is known that using antibiotics such as the first- or second-generation cephalosporins—despite the organisms' apparent in vitro susceptibility—is prone to failure and relapse owing to low intracellular antibiotic concentrations within the phagocytes (6). The third-generation cephalosporins and fluoroquinolones are therefore generally considered to be the drugs of choice because of better intracellular penetration. Whether the aorta had already been infected or not during the earlier episode of bacteremia is unknown. The atherosclerotic aorta might have been seeded by Salmonella during that earlier episode of bacteremia, or it might have been infected prior to that. To address the problem of misidentification, we suggest that all Enterobacteriaceae isolates with unusual biochemical profiles being identified as H. alvei or E. coli should be correlated with patients' clinical pictures. The biochemical profile should be confirmed with conventional testing methods, and alternative identification tests should be performed. Serotyping of these isolates should be performed in situations where Salmonella infections are suspected.
Isolation of mucoid variants of Salmonella from clinical specimens is unusual, although they have been found among survivors of selective agents or detrimental conditions (4). The presence of large amounts of capsular material in mucoid isolates may interfere with the slide agglutination test. A correct bacterial inoculum is essential for antibiotic susceptibility testing and biochemical tests. It is notoriously difficult to prepare an accurate inoculum for highly mucoid strains, and this may result in aberrant biochemical profiles, especially when tested by automated systems (10). These strains must be handled with care. The increased utilization of 16S rRNA sequencing in clinical laboratories could allow more accurate identification of these unusual isolates.
Acknowledgments
We thank J. Y. C. Lo, Consultant Microbiologist, Public Health Laboratory Centre, Hong Kong, for assistance in confirming the identity of the Salmonella isolates.
Footnotes
Published ahead of print on 18 October 2006.
REFERENCES
- 1.Bourbeau, P. P., and B. J. Heiter. 1998. Comparison of Vitek GNI and GNI+ cards for identification of gram-negative bacteria. J. Clin. Microbiol. 36:2775-2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chiu, C. H., L. H. Su, and C. Chu. 2004. Salmonella enterica serotype Choleraesuis: epidemiology, pathogenesis, clinical disease, and treatment. Clin. Microbiol. Rev. 17:311-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clinical and Laboratory Standards Institute. 2006. Performance standards for antimicrobial disk susceptibility tests. Approved standard, 9th ed. Performance standards for antimicrobial susceptibility testing; 16th informational supplement, 7th ed. Clinical and Laboratory Standards Institute, Wayne, Pa.
- 4.Costa, C. S., and D. N. Anton. 2001. Role of the ftsA1p promoter in the resistance of mucoid mutants of Salmonella enterica to mecillinam: characterization of a new type of mucoid mutant. FEMS Microbiol. Lett. 200:201-205. [DOI] [PubMed] [Google Scholar]
- 5.Hansen, D. S., A. G. Jensen, N. Norskov-Lauritsen, R. Skov, and B. Bruun. 2002. Direct identification and susceptibility testing of enteric bacilli from positive blood cultures using VITEK (GNI+/GNS-GA). Clin. Microbiol. Infect. 8:38-44. [DOI] [PubMed] [Google Scholar]
- 6.Hohmann, E. 2001. Non-typhoidal salmonellosis. Clin. Infect. Dis. 32:263-269. [DOI] [PubMed] [Google Scholar]
- 7.Janda, J. M., and S. L. Abbott. 2006. The genus Hafnia: from soup to nuts. Clin. Microbiol. Rev. 19:12-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Knight, M. T., D. W. Wood, J. F. Black, G. Gosney, R. O. Rigney, J. R. Agin, C. K. Gravens, and S. M. Farnham. 1990. Gram-negative identification card for identification of Salmonella, Escherichia coli, and other Enterobacteriaceae isolated from foods: collaborative study. J. Assoc. Off. Anal. Chem. 73:729-733. [PubMed] [Google Scholar]
- 9.Kotetishvili, M., O. C. Stine, A. Kreger, J. G. Morris, Jr., and A. Sulakvelidze. 2002. Multilocus sequence typing for characterization of clinical and environmental Salmonella strains. J. Clin. Microbiol. 40:1626-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morgan, H. R., and T. D. Beckwith. 1939. Immunological relationships of polysaccharides of mucoid organisms of the typhoid-Salmonella group. J Bacteriol. 37:389-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murray, P. R., E. J. Baro, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.). 2003. Manual of clinical microbiology, 8th ed. American Society for Microbiology, Washington, D.C.
- 12.O'Hara, C. M., and J. M. Miller. 2003. Evaluation of the Vitek 2 ID-GNB assay for identification of members of the family Enterobacteriaceae and other nonenteric gram-negative bacilli and comparison with the Vitek GNI+ card. J. Clin. Microbiol. 41:2096-2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O'Hara, C. M., G. L. Westbrook, and J. M. Miller. 1997. Evaluation of Vitek GNI+ and Becton Dickinson Microbiology Systems Crystal E/NF identification systems for identification of members of the family Enterobacteriaceae and other gram-negative, glucose-fermenting and non-glucose-fermenting bacilli. J. Clin. Microbiol. 35:3269-3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]