For infection control and prevention initiatives to be effective in preventing the spread of methicillin-resistant Staphylococcus aureus (MRSA) within health care facilities, prompt identification of this pathogen from clinical samples is fundamental. MRSASelect (Bio-Rad, Marnes-la-Coquette, France) is a new chromogenic agar that permits presumptive identification of MRSA directly from clinical specimens such as nasal swabs. Robust colonies with strong pink coloration appearing on MRSASelect agar within 24 h of incubation are considered MRSA and do not require supplemental testing before reporting the result. The selectivity of MRSASelect is based upon the combined suppressive effects of a high NaCl concentration and a proprietary mixture of antibacterial and antifungal agents that inhibit the majority of normal flora and potentially pathogenic microorganisms other than MRSA.
Here we report a clinical isolate of Staphylococcus cohnii which produced strongly pink colonies on MRSASelect within 24 h of growth. The isolate was obtained from a swab of the anterior nares submitted to the clinical microbiology laboratory for MRSA screening of a patient who was known to be previously colonized with MRSA. The swab was inoculated directly onto MRSASelect and also into an enrichment broth (brain heart infusion broth supplemented with ceftizoxime, 5 μg/ml, and aztreonam, 75 μg/ml) (6) and incubated overnight at 35°C, and then 100 μl was plated to MRSASelect. The directly plated MRSASelect revealed no pink colonies at 24 h. The MRSASelect inoculated with the aliquot of enrichment broth grew many strongly pink colonies (0.8 mm in diameter) at 24 h (Fig. 1). This result was reported as positive for MRSA. Prior to collection of this specimen, the patient had undergone decolonization using topical and systemic antibiotics (mupirocin, 2% cream, applied three times daily; trimethoprim-sulfamethoxazole, 160 mg/800 mg orally, twice daily; and doxycycline, 100 mg orally, twice daily) for 14 days, with five subsequent weekly negative MRSA screening cultures. Because of the “recolonization” of the patient with MRSA, the infection control service requested pulsed-field gel electrophoresis (PFGE) of the isolate to determine its relationship to the patient's previous isolate. As well, screening specimens from all patients on the same ward as the MRSA-positive patient were submitted to the clinical microbiology laboratory. In preparation for PFGE, the initial isolate was subcultured onto a blood agar plate. After 24 h of incubation, off-white, nonhemolytic colonies appeared that were slide coagulase and tube coagulase negative. The isolate was initially identified as Staphylococcus xylosus by Vitek2 (bioMerieux, Marcy l'Etoile, France). Since this organism is only occasionally isolated from human specimens (3), PCR-based 16S rRNA gene sequencing was undertaken to confirm its identity; the latter test identified the isolate as Staphylococcus cohnii (100% identity, BLAST search). Methicillin resistance was confirmed by Vitek2 antimicrobial susceptibility testing, cefoxitin disk diffusion testing, latex agglutination for PBP2a (Oxoid, Hampshire, United Kingdom), and mecA PCR. None of the swabs submitted from concomitant ward patients produced suspicious pink colonies within 24 h. Isolates of other methicillin-resistant coagulase-negative staphylococci from unrelated patient specimens (two isolates of Staphylococcus epidermidis, one isolate of Staphylococcus hominis, and one isolate of Staphylococcus warneri) were directly and indirectly (after overnight incubation in enrichment broth) inoculated onto MRSASelect plates of the same lot; none harbored suspicious colonies within 24 h of incubation.
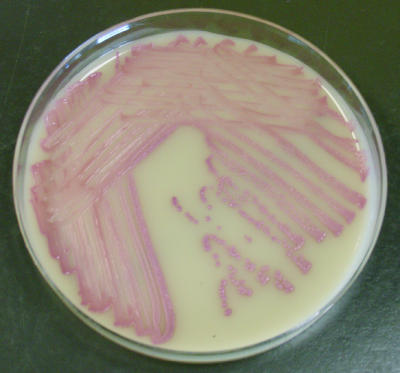
FIG. 1.
S. cohnii on MRSASelect following 24 h of incubation, demonstrating pink colonies suggestive of MRSA.
S. cohnii is a novobiocin-resistant, coagulase-negative Staphylococcus that is known to colonize human skin; it has also been frequently identified in the hospital environment (4). The organism is typically methicillin resistant and frequently harbors plasmids mediating resistance to multiple other antibiotics (5). As with other coagulase-negative staphylococci, S. cohnii is not usually pathogenic.
Users of chromogenic media, such as MRSASelect, should be aware of rare false-positive results with certain methicillin-resistant organisms. Gaillot et al., using CHROMagar Staph aureus (CHROMagar Microbiology, Paris, France) on 2,000 clinical isolates of Staphylococcus spp., reported three suspicious colonies later identified as S. cohnii (2). Louie et al. used MRSASelect to identify 178 isolates producing pink colonies after overnight incubation; one was not an MRSA, but rather a gram-negative bacillus that was not further identified (L. Louie, D. Soares, H. Gardner, H. Meaney, M. Vearncombe, and A. E. Simor, Abstr. Assoc. Med. Microbiol. Infect. Dis. [AMMI] Can.-Can. Assoc. Clin. Microbiol. Infect. Dis [CACMID] 2006 Annu. Conf., abstr. B5, 2006). Ben Nsira et al. (1) reported a coagulase-negative isolate not further identified that produced pink colonies after 18 to 24 h of incubation. The present study is the first to identify S. cohnii as having the ability to mimic the colony appearance of MRSA on MRSASelect.
Although the new chromogenic media have excellent sensitivity (1) and provide the opportunity to rapidly identify potential MRSA carriers by nasal swabs, this report emphasizes that dependence solely on colony color to provide a positive result may occasionally be misleading. Microbiologists should remain vigilant for other methicillin-resistant organisms capable of producing colonies of the “appropriate” color. The few similar reports by others confirm that this appearance is an inherent feature of certain organisms on these proprietary media. In this case, we excluded a faulty lot of plates or altered metabolism induced by the enrichment broth as possible causes for the false-positive result. Furthermore, this case suggests that, in a patient who has previously been decolonized and microbiologically confirmed culture negative for MRSA, the “reappearance” of a colony suspected of being MRSA should probably be confirmed by alternative tests.
Footnotes
Published ahead of print on 11 October 2006.
REFERENCES
- 1.Ben Nsira, S., M. Dupuis, and R. Leclercq. 2006. Evaluation of MRSASelect, a new chromogenic medium for the detection of nasal carriage of methicillin-resistant Staphylococcus aureus. Int. J. Antimicrob. Agents 27:561-564. [DOI] [PubMed] [Google Scholar]
- 2.Gaillot, O., M. Wetsch, N. Fortineau, and P. Berche. 2000. Evaluation of CHROMagar Staph. aureus, a new chromogenic medium, for isolation and presumptive identification of Staphylococcus aureus from human clinical specimens. J. Clin. Microbiol. 38:1587-1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nagase, N., A. Sasaki, K. Yamashita, A. Shimizu, Y. Wakita, S. Kitai, and J. Kawano. 2002. Isolation and species distribution of staphylococci from animal and human skin. J. Vet. Med. Sci. 64:245-250. [DOI] [PubMed] [Google Scholar]
- 4.Szewczyk, E. M., A. Piotrowski, and M. Rozalska. 2000. Predominant staphylococci in the intensive care unit of a paediatric hospital. J. Hosp. Infect. 45:145-154. [DOI] [PubMed] [Google Scholar]
- 5.Szewczyk, E. M., M. Rozalska, T. Cieslikowski, and T. Nowak. 2004. Plasmids of Staphylococcus cohnii isolated from the intensive-care unit. Folia Microbiol. 49:123-131. [DOI] [PubMed] [Google Scholar]
- 6.Wertheim, H., H. A. Verbrugh, C. Van Pelt, P. De Man, A. Van Belkum, and M. C. Vos. 2001. Improved detection of methicillin-resistant Staphylococcus aureus using phenyl mannitol broth containing aztreonam and ceftizoxime. J. Clin. Microbiol. 39:2660-2662. [DOI] [PMC free article] [PubMed] [Google Scholar]