Abstract
The fixation of a new allele can be driven by Darwinian positive selection or can be due to random genetic drift. Identifying instances of positive selection is a difficult task, because its impact is routinely obscured by the action of negative selection. The nature of the genetic code dictates that positive selection in favor of an amino acid replacement should often cause a burst of two or three nucleotide substitutions at a single codon site, because a large fraction of amino acid replacements cannot be achieved after just one nucleotide substitution. Here, we study pairs of successive nonsynonymous substitutions at one codon in the course of evolution of HIV-1 genes within HIV-1 populations inhabiting infected individuals. Such pairs are more numerous and more clumped than expected if different substitutions were independent and than what is observed for pairs of successive synonymous substitutions. Bursts of nonsynonymous substitutions in HIV-1 evolution cannot be explained by mutational biases and must, therefore, be due to positive selection. Both reversals, exact or imprecise, of fixed deleterious mutations and acquisitions of amino acids with new properties are responsible for the bursts. Temporal clumping is strongest at codon sites with a low overall rate of nonsynonymous evolution, implying that a substantial fraction of replacements of conservative amino acids are driven by positive selection. We identified many conservative sites of HIV-1 proteins that occasionally experience positive selection.
Keywords: clumping, phylogeny, reversals, fitness landscape, genetic code
Darwinian positive selection favoring new alleles drives adaptive evolution and thus is of paramount importance (1). However, positive selection always acts over the background of pervasive negative selection, which maintains status quo (2). Even at the simplest level of DNA and protein sequences, disentangling the two remains a major challenge.
Because positive selection promotes change, a variety of methods seek to detect it from a higher rate of evolution, relative to that of selectively neutral sites (3). In particular, positive selection for nonsynonymous substitutions may lead to dN > dS, where dN and dS are per-site rates of nonsynonymous and synonymous substitutions, respectively, so long as synonymous nucleotide substitutions are approximately neutral. However, there are a number of problems with this approach.
Indeed, positive selection is usually less common than negative selection and acts only on some sites during only some intervals of evolutionary time. Thus, if we consider the whole protein-coding gene, dN < dS in a majority of cases (4). The problem can be alleviated if a large number of orthologous genes are compared, which makes it possible to attribute a specific value of dN to each codon site individually (5). Then, positive selection at one or several adjacent codons can be detected through a locally elevated dN, even if negative selection is more common in the whole protein. Still, even at an individual codon site, positive selection does not necessarily act throughout the whole phylogeny of a set of species. Thus, it is preferable to look for lineage-specific episodes of elevated dN, which can lead to detection of the corresponding episodes of positive selection at the site, even if this site was under negative selection for most of its evolutionary history. The locations, on the phylogenetic tree, of these episodes of positive selection can either be assumed a priori (6, 7) or derived from the pattern of substitutions on the tree (8).
Data on within-population variability can also be used. Positive selection may be inferred not only when dN/dS > 1, but also when dN/dS > pN/pS, where pN and pS are the levels of nonsynonymous and synonymous polymorphism (McDonald–Kreitman test; refs. 9 and 10). Indeed, pN/pS may reveal the fraction of sites under negative selection, and using this ratio facilitates detection of positive selection, so long as pN/pS < 1. However, the sensitivity of the McDonald–Kreitman test declines if, at a substantial fraction of sites, negative selection is strong enough to prevent fixations of nonsynonymous mutations but is still insufficient to suppress nonsynonymous polymorphism (10). Also, because levels of polymorphism at individual sites are subject to strong random drift, the McDonald–Kreitman test has so far been applied only to large classes of sites.
We pursue a different approach to detecting positive selection, which does not rely on a high dN/dS ratio. According to the structure of the genetic code, replacing amino acid X with amino acid Z often requires two or even three nonsynonymous substitutions, because, in only 75 out of 190 unordered amino acid pairs, the members can be converted into each other by only a single nucleotide substitution. Thus, when positive selection favors a particular amino acid replacement, in a large fraction of cases, a burst of two or three successive nucleotide substitutions should occur, even if most of the time the site evolves slowly because of negative selection. The idea that bursts of nonsynonymous substitutions can be a signature of positive selection has been proposed by Gillespie (11).
Comparison of rat, mouse, and human orthologous proteins demonstrated that, at codons where rat and mouse differ by two nonsynonymous substitutions, both substitutions tend to occur after the rat–mouse divergence in the same lineage, either rat or mouse (12). This result indicates that bursts of successive nonsynonymous substitutions are common. However, because rat and mouse are tightly related to each other, codons at which they differ by two nucleotide substitutions are rare and must mostly come from the subset of generally rapidly evolving codons.
Here, we analyze the evolution of four genes of HIV-1 by using sets of hundreds of genomes. Different HIV-1 lineages, represented by these genomes, evolved essentially independently of each other. These data make it possible to look for bursts of nonsynonymous substitutions at both rapidly evolving and slowly evolving codon sites.
Results
Phylogenetic trees of 343, 218, 193, and 674 full-length sequences of env, gag, pol, and nef genes from HIV-1 genomes are presented in supporting information (SI) Figs. 5–8. These trees were constructed by using the data on all substitutions. However, to analyze the distribution of nonsynonymous substitutions relative to that of synonymous substitutions, we expressed the lengths of edges of the already constructed trees in the units of synonymous substitutions per codon. Table 1 presents some characteristics of these trees.
Table 1.
Characteristics of the four gene-specific phylogenetic trees
| Total length of all edges* | Distance from the root to a leaf, average and range* | Numbers of nodes between the root and a leaf, average and range | Average dN/dS value | |
|---|---|---|---|---|
| env | 16.12 | 0.214 (0.141–0.285) | 15.10 (3–29) | 0.686 |
| gag | 10.07 | 0.216 (0.124–0.323) | 13.78 (4–22) | 0.325 |
| pol | 9.16 | 0.186 (0.121–0.254) | 11.54 (3–17) | 0.249 |
| nef | 33.30 | 0.237 (0.092–0.423) | 23.10 (3–54) | 0.508 |
*Distances on a tree and edge lengths are measured in average numbers of synonymous substitutions per codon.
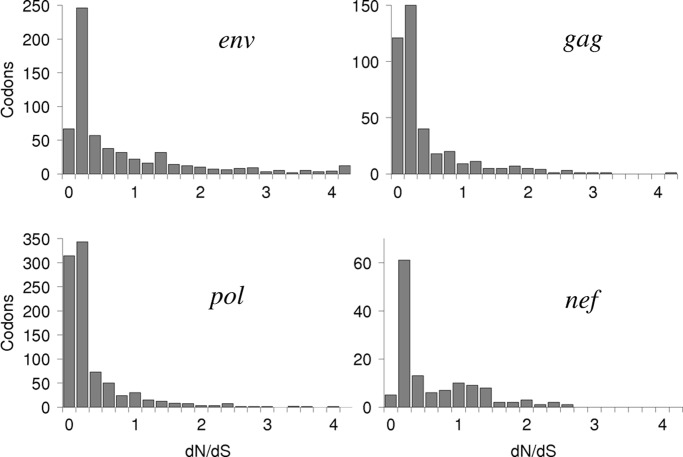
Fig. 1 displays, for each gene, the distribution of codons by their dN/dS values, estimated by using a direct-counting procedure. In agreement with previous studies (13–17), we see that, although on average dN/dS < 1 for every gene, there is a substantial fraction of codons with dN/dS > 1, indicating that positive selection is relatively common in the HIV-1 proteome.
Fig. 1.
Distribution of codon sites by their dN/dS values (the values of dS are assumed to be gene-specific; see SI Supporting Text for details).
Nonsynonymous and synonymous substitutions are not distributed uniformly over any of the four trees (SI Figs. 5–9). Instead, when we approach the leaves of a tree, nonsynonymous substitutions tend to become relatively more common, which is consistent with weak negative selection against a large fraction of nonsynonymous substitutions (18).
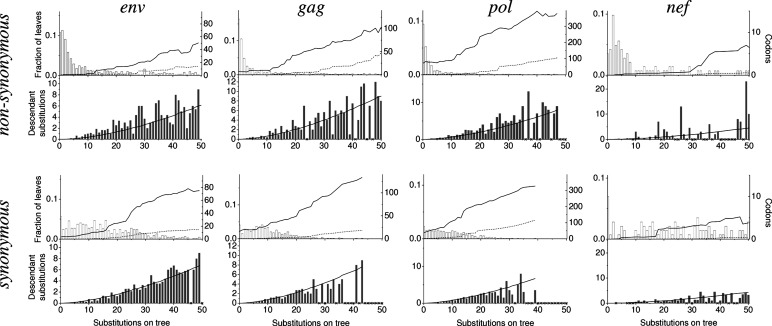
Let us now consider how nonsynonymous substitutions are distributed relative to each other. A nonsynonymous substitution is more likely to have a descendant nonsynonymous substitution at the same codon, located between it and one or more of the leaves of the tree, than what is expected if the same number of nonsynonymous substitutions at the codon were distributed over the tree independently. In contrast, the probability of having a synonymous descendant for a synonymous substitution is much closer to the random expectation (Table 2; Fig. 2).
Table 2.
Mean of the ratios of the actual number of substitutions with at least one descendant substitution over the number of such substitutions obtained in simulations of independent substitutions, at each site
| Nonsynonymous | Synonymous | |
|---|---|---|
| env | 2.95 | 1.55 |
| gag | 3.27 | 1.77 |
| pol | 4.37 | 1.68 |
| nef | 2.64 | 1.21 |
Fig. 2.
Distributions of nonsynonymous and synonymous substitutions on the phylogenetic trees. For each possible number of substitutions at a codon site (within the range 0–50), we present the number of sites with this number of substitutions (Top, empty bars); the fractions of leaves of the phylogenetic tree, for which the number of substitutions at these sites, on the path from the leaf to the root, is two (Top, solid line) or three or more (Top, dotted line) within each sliding window of length 30; the actual average per-site number of substitutions with at least one descendant substitution (Bottom, filled bars); and the average per-site number of substitutions with at least one descendant substitution obtained in simulations of independent substitutions (Bottom, solid line). Data on only nonsynonymous (top row of graphs) and on only synonymous (bottom row of graphs) substitutions are shown. Numbers of substitutions that were not encountered at any site are marked by crosses on the x axis.
Because we cannot detect multiple substitutions that occurred at the same nucleotide site within an edge of a tree, all 1,394 pairs of nonsynonymous substitutions detected in env gene within the same edge occurred at different sites. Among the 7,424 observed pairs of successive nonsynonymous substitutions that occurred in different edges in env, 2,932 (39%) were amino acid reversals, 686 (9%) were nonreversing substitutions at the same nucleotide site, and 1,903 (26%) were nonreversing substitutions affecting different nucleotide sites of the same codon. For the remaining substitutions, categorization was ambiguous, because of intervening synonymous substitutions or multiple nonsynonymous substitutions within an edge. Similar patterns were observed for the other three genes.
In addition to the excess of pairs of successive substitutions (Table 2), nonsynonymous substitutions also display 1.5- to 3-fold reduced distances between the members of a pair relative to the expectation for independent substitutions and to the distances between successive synonymous substitutions. In contrast, the average distance between successive synonymous substitutions was within 20% of that predicted in a simulation that assumed independence (Table 3). Substitutions in pairs were similarly clumped, even if we excluded from the analysis the pairs of substitutions that occurred in the same edge (Table 3).
Table 3.
Average evolutionary distances between pairs of successive substitutions
|
env |
gag |
pol |
nef |
|||||
|---|---|---|---|---|---|---|---|---|
| Nonsyn. | Syn. | Nonsyn. | Syn. | Nonsyn. | Syn. | Nonsyn. | Syn. | |
| Total | 4.91 (8.01) | 8.12 (8.98) | 6.14 (8.58) | 8.20 (9.29) | 5.22 (7.19) | 7.70 (7.74) | 3.28 (9.91) | 8.98 (10.58) |
| Multiple substitutions within edge excluded | 6.54 (11.22) | 8.21 (10.18) | 6.94 (11.63) | 8.19 (10.56) | 6.45 (10.03) | 8.09 (9.01) | 4.39 (13.52) | 9.57 (11.99) |
| Pairs classified by total number of substitutions on tree: | ||||||||
| 2–10 | 2.57 (8.27) | 6.44 (9.47) | 5.43 (8.98) | 6.77 (9.84) | 2.73 (7.47) | 7.60 (8.07) | 0.67 (10.08) | — |
| 11–20 | 3.68 (8.12) | 8.30 (9.29) | 6.80 (8.71) | 9.52 (9.43) | 5.96 (7.28) | 8.14 (7.86) | 0.86 (10.19) | 9.02 (10.85) |
| 21–40 | 5.77 (8.00) | 9.09 (8.96) | 5.57 (8.30) | 7.98 (9.00) | 5.64 (6.97) | 7.45 (7.50) | 3.05 (10.06) | 10.14 (10.67) |
| 41–80 | 6.40 (7.73) | 7.44 (8.48) | 6.49 (7.68) | 9.19 (8.31) | 5.07 (6.65) | — | 3.54 (9.65) | 10.31 (10.32) |
| > 80 | 4.88 (6.99) | 5.52 (7.97) | 5.21 (7.13) | — | 6.22 (6.27) | — | 6.66 (9.31) | 7.70 (9.64) |
| Pairs classified by direction: | ||||||||
| Substitutions in the same nucleotide | ||||||||
| Reversing | 5.98 | — | 6.78 | — | 5.48 | — | 5.32 | — |
| Nonreversing | 5.54 | — | 5.99 | — | 5.50 | — | 5.12 | — |
| Substitutions in different nucleotides | ||||||||
| Progressive | 5.89 | — | 5.93 | — | 6.23 | — | 5.68 | — |
| Nonprogressive | 6.04 | — | 5.73 | — | 7.42 | — | 6.31 | — |
Evolutionary distances and edge lengths are presented as average numbers of synonymous substitutions per 100 codons. The presented numbers were obtained by first calculating the average distance for sites with a particular number of substitutions and then by averaging these averages for all numbers of substitutions at a site. The values obtained in simulations of independent substitutions are given in parentheses. Pairs of substitutions within one edge, which often could not be classified unambiguously, were excluded from the classification of pairs by their direction. Nonsyn., nonsynonomous; syn., synonomous.
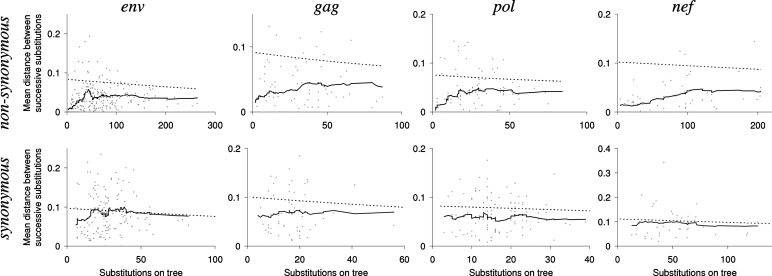
Temporal clumping of successive nonsynonymous substitutions is strongest at highly conservative codons with the smallest total number of nonsynonymous substitutions (Table 3; Fig. 3). For example, in the most conserved sites of env (with 10 or fewer nonsynonymous substitutions on the whole tree), the distance within a pair of nonsynonymous substitutions is more than three times shorter than in simulations (Table 3). The distance between successive nonsynonymous substitutions is similarly reduced when they occurred at the same nucleotide site and at different nucleotide sites within the same codon (Table 3). For same-nucleotide site pairs of substitutions, distance was similarly reduced when the second substitution was a reversal of the first one, and when it was not.
Fig. 3.
Distances between successive nonsynonymous (Upper) and synonymous (Lower) substitutions on the phylogenetic trees. The mean distance between successive nonreversing substitutions is shown for codon sites with each total number of substitutions on the tree (dots). Solid lines present mean distances between successive substitutions within each sliding 30-site window. Dashed lines show mean distances between independent substitutions obtained in simulations.
For pairs of substitutions at different nucleotide sites, distance was similarly reduced when the second substitution was progressive, i.e., when it created a new amino acid unreachable by a single-nucleotide substitution from the original amino acid, and when it was not progressive (Table 3). We analyzed separately all pairs of progressive substitutions with high (>8) directedness index, i.e., those consisting of a very conservative substitution followed by a radical substitution, namely: Leu→Ile→Thr, Ile→Leu→Gln, Ile→Leu→Pro, Ile→Leu→Ser, Leu→Ile→Lys, Leu→Ile→Ser, Met→Ile→Asn, and Lys→Arg→Gly. Within such pairs, the average evolutionary distance was 5.74, whereas 5.93 was observed in all pairs of progressive substitutions, and 11.60 was obtained in simulation of independent substitutions. Therefore, clumping is strongest in pairs with a high-directedness index.
To study the possible impact of the mutational biases (SI Supporting Text) on the observed clumping, we examine the correlation of the distance between the two successive substitutions of given type (synonymous or nonsynonymous) with the opportunity for a substitution of this type at the codon created by the first substitution. These correlations are very weak (Spearman R2 ≤ 0.02; SI Fig. 10), which argues against a mutational explanation of the clumping.
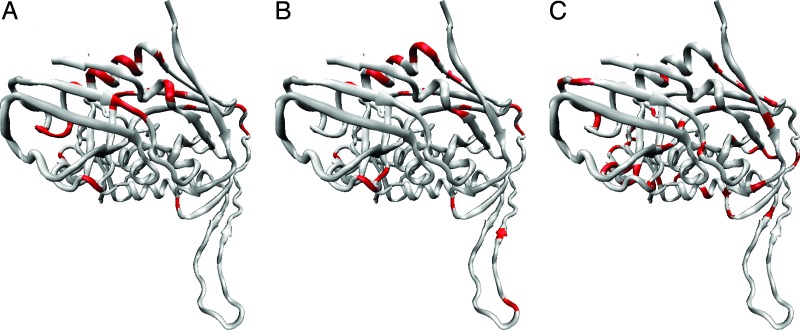
Fig. 4 displays the locations of amino acid sites with strongly clumped nonsynonymous substitutions on the solved (19) structure of HIV-1 gp120 envelope glycoprotein. Most conservative sites with strongly clumped substitutions lie outside the regions where strong positive selection has been detected previously (SI Tables 4–7).
Fig. 4.
Amino acid sites inferred to be under positive selection in HIV-1 gp120. (A) Amino acid sites with >80 replacements. (B) Rapidly evolving sites previously inferred to be under positive selection (27). (C) Conservative sites (<80 replacements) with strongly clumped substitutions. Protein structure was visualized with the VMD package (28).
At conservative sites with dN/dS < 1, some clumping of nonsynonymous substitutions can be caused by alternating episodes of strong negative selection and of neutral evolution (12). This effect can be substantial when the characteristic length of the episodes of neutral evolution is ≈10 times smaller than the average distance of a leaf of the tree from its root; still, for no parameters can this effect explain the observed extent of clumping (SI Fig. 11). In contrast, if evolution during postulated episodes of absent negative selection proceeds much faster than neutrally, the observed clumping can be obtained for some sets of parameter values (SI Fig. 12).
Within nonreversing pairs of nonsynonymous substitutions, the first and the second substitutions can have different effects on the chemical properties of the amino acid. In most cases, the amino acid encoded after the second substitution deviates chemically from the original amino acid more than the amino acid encoded after the first substitution. Among nonreversing pairs of nonsynonymous substitutions, this pattern is observed for 56% of pairs of substitutions at the same nucleotide site, for 71% of pairs of nonprogressive substitutions at different sites, and for 69% of pairs of progressive substitutions at different sites (SI Figs. 13–16). The patterns in the strongly clumped and all other pairs of substitutions are similar.
Discussion
We analyzed the evolution of HIV-1 lineages that correspond to viral populations living in different infected individuals. The effective size of an intraindividual population of HIV-1 is only 103 or 104, and HIV-1 transmission between individuals involves severe bottlenecks (20, 21). Thus, allele substitutions within HIV-1 lineages must occur very rapidly and essentially independently of slow processes that affect the whole metapopulation of HIV-1.
Our analysis reveals a contrast between the dynamics of synonymous vs. nonsynonymous substitutions in the evolution of HIV-1. Although synonymous substitutions occur more or less independently of each other, successive nonsynonymous substitutions are clumped on the phylogenetic trees (Figs. 2 and 3). This clumping is not an artifact of phylogenetic reconstruction and is robust to the choice of a particular tree among the highly parsimonious trees and to the method of reconstruction of the states in the internal nodes (data not shown). Clumping is also not a result of mutational events (or an artifact of sequencing errors) affecting several adjacent nucleotides, because even pairs of successive nonsynonymous substitutions that occurred in different edges of the tree are strongly clumped.
Further, clumping cannot be an artifact of our approach to inferring the timing of individual substitutions. If two or more substitutions occurred at the same nucleotide site between two adjacent nodes, we observe at most only a single-nucleotide difference. Therefore, some substitutions closely following each other must have been missed, inflating the average distance between pairs of successive substitutions. This limitation of our analysis certainly cannot lead to artifactual clumping. Also, when only pairs of substitutions at different nucleotide sites were considered, clumping remained equally strong (Table 3).
Therefore, nonsynonymous substitutions in the evolution of HIV-1 do tend to occur in bursts. Some of these bursts may consist of three or perhaps of even a larger number of successive substitutions. However, the phylogenies of HIV-1 are rather shallow, and the number of nonsynonymous substitutions on the path from the root of a tree to its leaf exceeds two for a substantial fraction of leaves only at very rapidly evolving sites (Fig. 2). Thus, our analysis was limited to only pairs of successive substitutions. A two-substitution burst occurs when the first substitution somehow facilitates the second one.
A feasible mechanism of such facilitation is the increase, by the first substitution, of the general propensity for nonsynonymous mutations at the codon site. Indeed, different codons have different opportunities for nonsynonymous mutations, because of the structure of the genetic code and the peculiarities of the mutational substitution matrix. However, these differences do not explain the observed clumping, because the waiting time for the second substitution is essentially independent of the opportunity for nonsynonymous mutation created by the first substitution (SI Fig. 10). The observed clumping of nonsynonymous substitutions is also not because of alternating episodes of neutral evolution and negative selection (SI Fig. 11). Therefore, we cannot explain the data without invoking positive selection favoring at least the second nonsynonymous substitution in a pair. Three causes of such positive selection are feasible.
One possibility is positive selection favoring the second substitution, which is the reversal of a slightly deleterious first substitution. HIV-1 must be prone to fixation of slightly deleterious mutations because of low effective population size and frequent bottlenecks (20, 21). Indeed, amino acid reversals constitute almost half of all the pairs of successive nonsynonymous substitutions and thus make a large contribution to the observed clumping. Still, reversals are not exclusively responsible for it; the average distance within pairs of successive substitutions such that the second substitution reverses the first one is very close to that within other types of pairs of substitutions (Table 3).
Even in some pairs where the second substitution leads to a new amino acid, this substitution could be favored because it reverses the deleterious effect of the first substitution. If so, we should generally expect that the final amino acid is more similar in its properties to the original than to the intermediate amino acid. However, such pairs of “imperfectly reversing” substitutions constitute only a minority of all pairs of nonsynonymous substitutions and do not display an elevated level of clumping (SI Figs. 13–16). In fact, clumping is the strongest in pairs with high directedness, when the second (radical) substitution (e.g., Ile→Lys) can hardly be even an imperfect reversal of the first (conservative) substitution (e.g., Leu→Ile). Therefore, positive selection for the second nonsynonymous substitution within a pair cannot always be due to exact or imperfect reversals of the first substitution. Instead, selection must often favor a deviation of the properties of the final from the properties of the original amino acid and thus must lead to adaptive evolution. Still, there may be two substantially different mechanisms of such selection.
First, the fitness landscape of a codon site can undergo repeated changes within some time intervals in the evolution of HIV-1 while remaining constant outside these intervals. Indeed, occasional episodes of continuing positive selection can explain the clumping we observe, so long as these episodes are neither too short nor too long, and dN/dS is at least ≈4 in the course of an episode (SI Fig. 12). This mechanism has been assumed by Guindon et al. (8) in their analysis of selection acting on env.
Alternatively, a pair of successive nonsynonymous substitutions can be triggered by a single instantaneous change of the fitness landscape. This is unavoidable if reaching the new optimal amino acid requires two (or even three) nonsynonymous substitutions in the codon that encoded the old optimal amino acid. Then, clumping of successive substitutions occurs because at least the last of them, which results in fixation of the new optimal amino acid, is favored by positive selection.
Gillespie (11) argued that a change in the fitness landscape should often lead to a burst of selection-driven amino acid replacements within a protein, which strives to reach a new fitness peak. His arguments, based on the properties of the extreme value distribution, can be applied equally to nonsynonymous substitutions both at the same and at different codons. However, data on mammals (see supporting information figure 1 in ref. 12) and on HIV-1 (not reported) show that clumping is much stronger within than among codons. Thus, within-codon clumping, if caused by isolated changes of the fitness landscape, must be primarily due to the structure of the genetic code.
In fact, reaching the new optimal amino acid can involve multiple nonsynonymous substitution even when the genetic code does not dictate this, so long as the evolving lineage does not follow the shortest path to the new fitness maximum. Redundant multiple substitutions could be common in HIV-1 because of its low effective population size (21, 22), so that the best substitution may not be the first one to get fixed, because of the temporary unavailability of the corresponding mutation. Clumping in pairs of successive nonsynonymous substitutions is approximately the same when the final amino acid can and cannot be reached through a single nonsynonymous substitution (Table 3).
Distinguishing between repeated and isolated changes of the fitness landscape as the cause of bursts of nonsynonymous substitutions would require phylogenies that contain numerous long chains of successive nonsynonymous substitutions. Perhaps the simpler assumption of isolated changes (11) should be regarded as more parsimonious. In any case, it appears that positive selection must be operating at least on the second substitution in the majority of clumped pairs.
Clumping of nonsynonymous substitutions is strongest at the most conservative codon sites, where the total number of nonsynonymous substitutions is the lowest. Thus, the relative role of positive selection in the evolution of conservative amino acids appear to be high and may approach ≈10%. This is not surprising; replacements of a conservative amino acid can seldom be selectively neutral, and a large fraction of replacements that become fixed must be beneficial. Although conventional approaches consistently (14) detect positive selection only at codon sites with high rates of nonsynonymous evolution (Fig. 4 A and B), isolated clumps of nonsynonymous substitutions reveal a previously undescribed class of generally conservative sites in HIV-1 proteins that occasionally evolve under positive selection (Fig. 4C; SI Tables 4–7).
Materials and Methods
Sequences and Phylogenies.
Alignments of nucleotide sequences of all full-length env, gag, pol, and nef protein-coding regions from HIV-1 genomes of subtypes A–H were taken from the 2003 Los Alamos National Laboratory HIV-1 sequence database (23). Sequences known to be recombinant, known contaminants, and sequences containing premature stop codons and ambiguities were removed. For each gene, a maximally parsimonious tree was constructed by PAUP by using whole-length sequences of coding regions. The obtained trees were rooted by using the consensus of consensus sequences for each subtype (24). Trees were then rescaled, and the length of each edge was taken to be the per-codon number of synonymous substitutions within the edge. The resulting trees are available in NEXUS format as SI Data Sets 1–4. Regions of overlapping reading frames were excluded from the subsequent analyses. The analysis was performed by using a set of Bioperl-based (25) scripts, which are available upon request.
Analysis of Nucleotide Substitutions.
We used maximum parsimony to reconstruct the states of the codons at all internal nodes within each tree. Then, for each codon, we inferred the edges of the tree at which each single-nucleotide substitution occurred, as follows. If a pair of successive nodes within a tree differed at one nucleotide site, we assumed that exactly one substitution occurred on the edge connecting these nodes. If the codons at successive nodes differed at more than one nucleotide site, the numbers of synonymous and nonsynonymous substitutions were averaged over all possible orders of substitution events. For each codon, we estimated the number of nonsynonymous substitutions and the number of sy-nonymous substitutions on the whole tree.
We treated synonymous and nonsynonymous substitutions separately, so that synonymous substitutions were ignored when nonsynonymous substitutions were considered and vice versa. If successive substitutions occurred along the path from the tree root to a leaf, for each substitution A, except the first one, the preceding substitution A′ at the same codon site can be uniquely determined. We assume that two substitutions at a codon site form a pair if there is a path from the root of the tree to at least one of the leaves, such that both the substitutions belong to this path, and there are no other substitutions between them. In particular, two substitutions that occurred at the same codon site on the same edge, revealed by the codons at two successive nodes differing from each other at two nucleotide sites, always constitute a pair. In such an ordered pair (A′, A) of successive substitutions, substitution A′ is “ancestral,” and substitution A is “descendant.” Substitutions A′ and A can occur either at the same nucleotide site or at different nucleotide sites of the codon site.
Successive nonsynonymous substitutions at the same nucleotide site constitute a reversal if the second substitution A restores the amino acid encoded before the first substitution A′. For example, a pair of substitutions AAA (Lys) → AAC (Asn) → AAG (Lys) is a reversal. A pair of nonsynonymous substitutions at different nucleotide sites of the codon site is “progressive” if the amino acid encoded after the second substitution cannot be encoded by any single-nucleotide modification of the original codon. For example, a pair of substitutions AAC (Asn) → AAG (Lys) → AGG (Arg) is progressive. The directedness index of the pair of substitutions is defined as dAC/dAB, where A, B, and C are the initial, intermediate, and final amino acids, respectively, and d is the Miyata biochemical distance between the two amino acids (26).
We estimate the distance l between A′ and A as the sum of the lengths of edges between them, assuming that substitutions occur at the middles of edges. If A′ and A occurred within the same edge, l = 0 for them. For each codon site, we calculated the distances within the pairs of successive synonymous and nonsynonymous substitutions and compared them with the distances obtained in simulated evolution for the same total number of substitutions on the phylogeny (see below). We considered substitutions within a codon to be “strongly clumped” if the average distance within their pairs was more than two times smaller than the distance obtained in simulation.
Simulations of Sequence Evolution.
To calculate the expected numbers of pairs of successive substitutions at a codon site and the distance between substitutions in such pairs, we simulated independent occurrence of the same number of substitutions at a codon site. Each edge of the tree got a weight corresponding to the product of its length and the gene-specific a priori probability of substitution at corresponding distance from the tree root, obtained from the observed distribution of substitutions at different distances from the tree root (SI Fig. 9). Simulations were performed separately for each codon site in each gene. First, we counted the total number of substitutions of particular type (synonymous or nonsynonymous) at the given codon on the phylogenetic tree. Next, in each of the 1,000 trials, we distributed the same number of events randomly over the edges of the actual phylogenetic tree with the calculated edge-specific weights. We then counted the numbers of pairs and distances within pairs of successive substitutions in simulation.
Alternating Episodes of Negative Selection and Neutrality.
As in ref. 12, we assumed that negative selection at a codon site switches off and on at random moments. The expected waiting times (in the units of tree height) for off-to-on and on-to-off switches are T and bT, respectively. Thus, negative selection is present with probability of b/(1+b). Assuming that the synonymous substitutions are neutral, which is consistent with the lack of their clumping (Figs. 2 and 3), the fraction of the tree evolving without negative selection equals the observed ratio of nonsynonymous to synonymous substitution rates per site r, and b = 1/r−1. We performed 10,000 Monte Carlo runs for each combination of parameters by using the actual phylogenetic tree and the site-specific r values (only sites with r < 1 were simulated). The negative selection was off at the root of the phylogenetic tree with probability 1/(1+b) and on with probability b/(1+b). Switches of negative selection then occurred in different branches of the tree independently. Substitutions occurred only during the periods of absent negative selection, with the instantaneous per-site rate corresponding to the observed mean rate of synonymous substitutions, c. We then estimated the average distance between the successive substitutions as described above.
Alternating Episodes of Negative and Positive Selection.
We assumed that the only observable impact of positive selection is the increase in the rate of nonsynonymous substitutions. Thus, this situation was modeled analogously to alternating episodes of negative selection and neutrality, except that during the periods of absent negative selection, substitutions occurred with the rate ω≥c, so that b = ω/r−1.
Supplementary Material
Acknowledgments
We thank Mikhail Gelfand, Sergey Kryazhimskiy, and Joshua Plotkin for useful discussions. G.A.B. gratefully acknowledges fellowships from the Pew Charitable Trusts award 2000-002558 and the Burroughs Wellcome Fund award 1001782, both to Princeton University. J.D. and S.A.L. acknowledge support from the National Institutes of Health (award P50 GM071508) and Defense Advanced Research Projects Agency (award HR0011-05-1-0057).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information (SI) online at www.pnas.org/cgi/content/full/0609484103/DC1.
References
- 1.Fisher RA. The Genetical Theory of Natural Selection. Oxford, UK: Clarendon; 1930. [Google Scholar]
- 2.Williams GC. Adaptation and Natural Selection; A Critique of Some Current Evolutionary Thought. Princeton, NJ: Princeton Univ Press; 1966. [Google Scholar]
- 3.Nielsen R. Annu Rev Genet. 2005;39:197–218. doi: 10.1146/annurev.genet.39.073003.112420. [DOI] [PubMed] [Google Scholar]
- 4.Kimura M. The Neutral Theory of Molecular Evolution. Cambridge, UK: Cambridge Univ Press; 1983. [Google Scholar]
- 5.Yang Z, Nielsen R, Goldman N, Pedersen AM. Genetics. 2000;155:431–449. doi: 10.1093/genetics/155.1.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang Z, Nielsen R. Mol Biol Evol. 2002;19:908–917. doi: 10.1093/oxfordjournals.molbev.a004148. [DOI] [PubMed] [Google Scholar]
- 7.Travers SA, O'Connell MJ, McCormack GP, McInerney JO. J Virol. 2005;79:1836–1841. doi: 10.1128/JVI.79.3.1836-1841.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guindon S, Rodrigo AG, Dyer KA, Huelsenbeck JP. Proc Natl Acad Sci USA. 2004;101:12957–12962. doi: 10.1073/pnas.0402177101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McDonald JH, Kreitman M. Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- 10.Smith NG, Eyre-Walker A. Nature. 2002;415:1022–1024. doi: 10.1038/4151022a. [DOI] [PubMed] [Google Scholar]
- 11.Gillespie J. Evolution (Lawrence, Kans) 1984;38:1116–1129. doi: 10.1111/j.1558-5646.1984.tb00380.x. [DOI] [PubMed] [Google Scholar]
- 12.Bazykin GA, Kondrashov FA, Ogurtsov AY, Sunyaev S, Kondrashov AS. Nature. 2004;429:558–562. doi: 10.1038/nature02601. [DOI] [PubMed] [Google Scholar]
- 13.Yang Z. Pac Symp Biocomput. 2001:226–37. [PubMed] [Google Scholar]
- 14.Kosakovsky Pond SL, Frost SD. Mol Biol Evol. 2005;22:1208–1222. doi: 10.1093/molbev/msi105. [DOI] [PubMed] [Google Scholar]
- 15.Yang W, Bielawski JP, Yang Z. J Mol Evol. 2003;57:212–221. doi: 10.1007/s00239-003-2467-9. [DOI] [PubMed] [Google Scholar]
- 16.Choisy M, Woelk CH, Guegan JF, Robertson DL. J Virol. 2004;78:1962–1970. doi: 10.1128/JVI.78.4.1962-1970.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen L, Perlina A, Lee CJ. J Virol. 2004;78:3722–3732. doi: 10.1128/JVI.78.7.3722-3732.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Golding GB. Genet Res. 1987;49:71–82. doi: 10.1017/s0016672300026768. [DOI] [PubMed] [Google Scholar]
- 19.Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ritola K, Pilcher CD, Fiscus SA, Hoffman NG, Nelson JA, Kitrinos KM, Hicks CB, Eron JJ, Jr, Swanstrom R. J Virol. 2004;78:11208–11218. doi: 10.1128/JVI.78.20.11208-11218.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Edwards CT, Holmes EC, Wilson DJ, Viscidi RP, Abrams EJ, Phillips RE, Drummond AJ. BMC Evol Biol. 2006;6:28. doi: 10.1186/1471-2148-6-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Achaz G, Palmer S, Kearney M, Maldarelli F, Mellors JW, Coffin JM, Wakeley J. Mol Biol Evol. 2004;21:1902–1912. doi: 10.1093/molbev/msh196. [DOI] [PubMed] [Google Scholar]
- 23.Korber BT, Brander C, Haynes B, Koup R, Moore JP, Kuiken C, Walker BD, Watkins D. HIV Immunology and Sequence Databases. Los Alamos, NM: Los Alamos National Laboratory; 2000. [Google Scholar]
- 24.Korber B, Muldoon M, Theiler J, Gao F, Gupta R, Lapedes A, Hahn BH, Wolinsky S, Bhattacharya T. Science. 2000;288:1789–1796. doi: 10.1126/science.288.5472.1789. [DOI] [PubMed] [Google Scholar]
- 25.Stajich JE, Block D, Boulez K, Brenner SE, Chervitz SA, Dagdigian C, Fuellen G, Gilbert JG, Korf I, et al. Genome Res. 2002;12:1611–1618. doi: 10.1101/gr.361602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miyata T, Miyazawa S, Yasunaga T. J Mol Evol. 1979;12:219–236. doi: 10.1007/BF01732340. [DOI] [PubMed] [Google Scholar]
- 27.Yamaguchi-Kabata Y, Gojobori T. J Virol. 2000;74:4335–4350. doi: 10.1128/jvi.74.9.4335-4350.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Humphrey W, Dalke A, Schulten K. J Mol Graphics. 1996;14:33–38. 27–28. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.