Abstract
Muscle wasting (cachexia) is an incurable complication associated with chronic infection and cancers that leads to an overall poor prognosis for recovery. Tumor necrosis factor-α (TNFα) is a key inflammatory cytokine associated with cachexia. TNFα inhibits myogenic differentiation and skeletal muscle regeneration through downstream effectors of the p53 cell death pathway including PW1/Peg3, bax, and caspases. We report that p53 is required for the TNFα-mediated inhibition of myogenesis in vitro and contributes to muscle wasting in response to tumor load in vivo. We further demonstrate that PW1 and p53 participate in a positive feedback regulatory loop in vitro. Consistent with this observation, we find that the number of PW1-expressing stem cells in skeletal muscle declines significantly in p53 nullizygous mice. Furthermore, gene transfer of a dominant-negative form of PW1 into muscle tissue in vivo blocks myofiber atrophy in response to tumor load. Taken together, these results show a novel role for p53 in mediating muscle stem cell behavior and muscle atrophy, and point to new targets for the therapeutic treatment of muscle wasting.
Keywords: Skeletal muscle, muscle differentiation, cancer cachexia, p53, PW1/Peg3, Pax7
Severe muscle wasting (cachexia) is a frequent complication associated with a wide variety of chronic diseases and accounts for ∼40%–50% of cancer patient morbidity (Tisdale 2002). It is therefore crucial to identify regulatory proteins that may serve as pharmacological/gene therapy targets (Muscaritoli et al. 2004). Cachexia is characterized by a drastic loss of body weight due to a complete loss of fat and profound loss of skeletal muscle mass (Tisdale 2002). Cachexia is accompanied by elevated circulating levels of inflammatory cytokines, including tumor necrosis factor-α (TNFα), interleukin-1 (IL-1) and IL-6, as well as tumor-derived factors such as proteolysis-inducing factor (PIF) (Todorov et al. 1999; Reid and Li 2001). TNFα (originally referred to as cachectin) plays a pivotal role in mediating muscle wasting by enhancing sarcomere dismantling and protein degradation (Tisdale 2001). The increased muscle protein breakdown observed in cachexia is mediated by a proteasome-dependent pathway. Recently, two muscle-specific E3 ubiquitin–protein ligases, atrogin-1/MAFbx and Murf, have been reported to be involved in skeletal muscle atrophy (Bodine et al. 2001; Gomes et al. 2001). The induction of atrogin-1 in atrophic muscle is dependent on activation of Foxo transcription factors (Sandri et al. 2004).
We and others have demonstrated that TNFα inhibits myogenesis in vitro (Miller et al. 1988; Szalay et al. 1997; Guttridge et al. 2000; Coletti et al. 2002), and recently we extended these observations to regenerating muscle in vivo (Coletti et al. 2005). It is well established that TNFα exposure leads to the activation of NFκB (Israel 2000). Several reports suggest that TNFα exposure leads to a down-regulation of the myogenic factors MyoD and myogenin (Szalay et al. 1997) through an unusual mechanism in which NFκB activation leads to the degradation of MyoD transcripts (Guttridge et al. 2000). In addition, NFκB activation has been reported to inhibit muscle differentiation by induction of cyclin D1 as well as induce cachexia when constitutively activated in transgenic mice (Guttridge et al. 1999; Cai et al. 2004). The functional link between the effects of TNFα on muscle stem cells and subsequent myofiber atrophy remains unresolved. One plausible model is that a prolonged block of muscle regeneration and/or block of stem cell recruitment into mature myofibers contributes to muscle wasting. Indeed, it has been reported that muscle atrophy involves a loss of muscle precursor cells (Mitchell and Pavlath 2004).
After birth, muscle stem cells appear as quiescent, mononuclear cells immediately adjacent to the myofibers under the basal lamina (“satellite cells”) (Mauro 1961). Satellite cells play a pivotal role in postnatal growth and regeneration (for review, see Charge and Rudnicki 2004). Deregulation of satellite cell function or number leads to a failure in postnatal growth and muscle atrophy (Garry et al. 2000; Seale et al. 2000; Nicolas et al. 2005). The mechanisms underlying muscle stem cell specification and maintenance are beginning to be unraveled. Pax7 is a member of the paired-homeobox domain family of transcription factors that plays a crucial role in establishing and/or maintaining satellite cells (Seale et al. 2000; Olguin and Olwin 2004; Oustanina et al. 2004; Zammit et al. 2004). Mice deficient for Pax7 show severe defects in postnatal muscle growth coupled with an inability to sustain a population of satellite cells (Seale et al. 2000). We recently reported that a p53 gene target, PW1, is constitutively expressed in muscle satellite cells as well as a subset of muscle interstitial cells (Nicolas et al. 2005). Transgenic mice expressing a truncated dominant-negative form of PW1 (ΔPW1) under the myogenin promoter show a failure in postnatal muscle growth reminiscent of the Pax7 mutant phenotype (Nicolas et al. 2005). ΔPW1 transgenic mice show increased levels of atrogin-1 and a deregulation of satellite cells, which accumulate as clusters under the basal lamina (Nicolas et al. 2005). These observations support the hypothesis that muscle stem cells are critical for muscle homeostasis, although the mechanism of how perturbation of stem cells trigger fiber atrophy remains unresolved.
The p53 tumor suppressor protein responds to cellular stress by inducing cell cycle arrest or apoptosis (Vousden 2000; Vousden and Woude 2000). p53 activation is directly linked to cellular senescence (Itahana et al. 2001), and several mouse models have shown that p53 is a mediator of organismal senescence (Chin et al. 1999; Rudolph et al. 1999; Vogel et al. 1999; Lim et al. 2000; Maier et al. 2004). p53 nullizygous (p53−/−) mice are highly susceptible to early onset of cancer but otherwise show an overtly normal phenotype. Any resistance to aging of the organism is presumably masked by early death associated with a high rate of lethal tumors. Many studies support the hypothesis that p53 plays a role during differentiation and development of specific lineages including skeletal myogenesis (Soddu et al. 1996; Tamir and Bengal 1998; Porrello et al. 2000). In the case of skeletal muscle, a well-understood cascade of molecular events directs the reprogramming of myogenic stem cells from proliferation-associated gene expression to myofiber-specific gene expression (Walsh and Perlman 1997). The up-regulation of cyclin-dependent kinase inhibitor p21 and inactivation of Cdk activity results in hypophosphorylation of retinoblastoma protein (pRb), which is critical for cell cycle arrest (Walsh and Perlman 1997), whereas p21 is induced by the myogenic factor MyoD (Halevy et al. 1995). p53 is activated during myogenic differentiation and participates with MyoD to promote myogenesis (Weintraub et al. 1991; Tamir and Bengal 1998) by up-regulating pRb (Porrello et al. 2000). Nonetheless, p53 expression is not essential for muscle development since p53−/− mice develop overtly normal skeletal muscle (Donehower et al. 1992) and show normal muscle regenerative capacity (White et al. 2002; our unpublished observations). We demonstrated previously that TNFα inhibits myogenesis by activating the bax–caspase 9 pathway through PW1 (Coletti et al. 2002). Bax and caspases are key targets in the p53-mediated cell death pathway (for review, see Schuler and Green 2001). We showed that PW1 is induced by p53 in the apoptotic pathway in murine fibroblasts (Relaix et al. 2000). In addition, PW1 associates with at least two other p53 gene targets, SIAH-1 and bax, which induce cell death when coexpressed with PW1 (Deng and Wu 2000; Relaix et al. 2000). Forced expression of PW1 alone is sufficient to trigger bax translocation to the mitochondria, which is a key event in the p53-mediated cell death pathway (Deng and Wu 2000).
In addition to a role in the p53 pathway, PW1 participates in the TNFα–NFκB activation pathway via physical interaction with TRAF2 (TNFα receptor-associated factor 2) (Relaix et al. 1998). ΔPW1 blocks TNFα-mediated NFκB activation as well as p53-mediated apoptosis, implicating a pivotal role in mediating both the p53 and TNFα pathways (Relaix et al. 1998, 2000). We originally identified PW1 in a screen for genes involved in the mesodermal to skeletal muscle commitment prior to the overt expression of the MyoD family of muscle transcription factors (Relaix et al. 1996). PW1 expression initiates in the early embryonic mesoderm and is down-regulated in all mesodermal tissues as they differentiate; however, PW1 expression is maintained in a subset of postnatal muscle interstitial and satellite cells (Relaix et al. 1996; Nicolas et al. 2005). While expression of PW1 is not restricted to satellite cells, we note that all primary myoblasts and all myogenic cell lines examined to date—with the exception of the F3 line—express high levels of PW1, whereas fibroblasts express PW1 uniquely upon induction of p53-mediated cell death (Relaix et al. 1996, 2000; Coletti et al. 2002).
The observation that PW1 is expressed in muscle stem cells and mediates both intrinsic and extrinsic cell stress pathways led us to investigate whether p53 plays a role in the TNFα response in skeletal muscle. Mice carrying mutations that lead to a chronic activation of intrinsic cell stress responses such as defects in DNA repair mechanisms show severe muscle atrophy similar to what occurs in the presence of chronically elevated TNFα levels (Nussenzweig et al. 1996). Expression of ΔPW1 in myogenic stem cells in vivo leads to severe muscle atrophy bearing the molecular hallmarks of cachexia (Nicolas et al. 2005). Lastly, muscle wasting is observed after radiotherapy in rodents, and the p53 target gene, p21, is expressed at elevated levels in aged human muscle (Persons et al. 2001; Welle et al. 2004). Taken together, these data suggest a mechanistic link between p53-mediated cell stress and muscle atrophy. Here we demonstrate that the TNFα-mediated inhibition of myogenic differentiation is p53 dependent through a PW1-mediated pathway. p53 is expressed in muscle stem cells and loss of p53 function alters muscle stem cell number. p53−/− mice are less susceptible to TNFα-mediated cachexia, highlighting the importance of p53 for muscle homeostasis. Our data reveal a novel role for p53 and its effector PW1 in mediating stem cell balance and muscle atrophy.
Results
p53 is expressed in myogenic cells and is required for TNFα-mediated inhibition of muscle differentiation
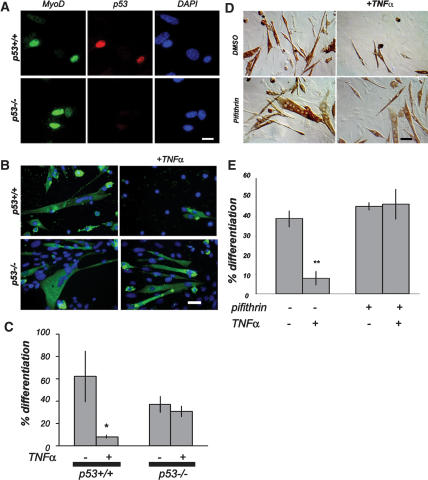
We reported previously that TNFα blocks myogenic differentiation and that this block requires the p53 target gene product, PW1. We further observed that the TNFα-mediated block of myogenesis requires additional p53 effectors, including bax and caspase 9 (Coletti et al. 2002). To test whether p53 directly participates in the TNFα-mediated myogenic block, we first derived primary muscle cultures from p53+/+ and p53−/− mice to characterize p53 expression in normal myogenic cells. Primary cultures consist of a mixture of myoblasts and nonmuscle cells (“fibroblasts”); however, myoblasts can be identified by the expression of the myogenic factor, MyoD1, which is not expressed in fibroblasts (Megeney et al. 1996; Seale et al. 2004). Immunostaining for both MyoD1 and p53 revealed coexpression of MyoD and p53 in primary cultures obtained from p53+/+ skeletal muscle, demonstrating that p53 is specifically expressed in cells committed to the myogenic fate, whereas p53 is not expressed in cells that do not express MyoD (Fig. 1A). We note that p53−/− myoblasts express MyoD, demonstrating that p53 function is not required for myogenic potential in vitro (Fig. 1A). Indeed, p53−/− myoblasts efficiently form multinucleated myotubes expressing myosin heavy chain (MHC) (Fig. 1B). We conclude that p53 serves as a reliable marker for myogenic cells but is not required for myogenic stem cell identity. Moreover, p53−/− myoblasts showed robust differentiation in the presence of TNFα, whereas wild-type myoblasts were markedly inhibited in the presence of TNFα (Fig. 1B,C). Not unexpectedly, p53−/− myoblasts exhibited higher rates of proliferation as compared with p53+/+ myoblasts. In addition, p53−/− myoblasts expand immediately upon plating, whereas p53+/+ myoblasts typically take more than a week before they reach appreciable rates of proliferation, presumably due to “culture shock,” which may reflect a p53-dependent process. These observations raised the possibility that standard primary culture conditions select for qualitatively different lineages of myogenic cells or cells with selected proliferation capacities. To address this concern, we used C2C12 myogenic cells that also express p53 (Latella et al. 2004; data not shown) and assayed differentiation in the presence or absence of the p53 inhibitor Pifithrin (Komarov et al. 1999) in combination with TNFα. As previously observed, TNFα treatment blocks myogenesis (Fig. 1D,E). Treatment with Pifithrin alone does not interfere with differentiation (Fig. 1D,E). In contrast, treatment with Pifithrin rescues muscle differentiation in the presence of TNFα. We conclude that p53 activity is required in the TNFα-mediated block of myogenesis, as demonstrated with both genetic and pharmacological approaches.
Figure 1.
p53 is expressed in myogenic cells and is required for TNFα-mediated inhibition of differentiation. (A) Myogenic cells were identified in primary muscle culture from p53+/+ and p53−/− mice by immunostaining for MyoD (green). Immunostaining for p53 (red) in p53+/+ cells reveals colocalization with MyoD-positive cells. p53−/− cell cultures were tested to verify the specificity of p53 detection. Nuclei were visualized by DAPI staining (blue). Bar, 5 μm. (B) Primary muscle cultures from p53+/+ and p53−/− mice were differentiated in the presence or absence of TNFα as indicated. MHC (green) was used to determine biochemical differentiation. TNFα blocks differentiation of p53+/+ myogenic cells, whereas p53−/− myogenic cells differentiate regardless of treatment. Nuclei were visualized by DAPI staining (blue). Bar, 10 μm. (C) Quantitative analysis of myogenic differentiation (% differentiation) of cells treated as described for B. TNFα treatment does not block differentiation of p53−/− myogenic cells. The values are presented as mean ± SD of at least three independent experiments. (*) p < 0.01 by Student’s t-test. (D) Confluent C2C12 cultured in DM with or without TNFα and/or the p53 inhibitor Pifithrin, as indicated. Immunolocalization of myosin was used as a marker of myogenic differentiation. C2C12 cells do not differentiate in the presence of TNFα. Differentiation is restored in the presence of Pifithrin. The final density of cells shown in D are close to confluence; however, only myotubes are stained. Bar, 30 μm. (E) Quantitative analysis of myogenic differentiation (% differentiation) of cells treated as described for D. Inhibition of p53 by Pifithrin rescues differentiation of C2 cells in the presence of TNFα. The values are presented as mean ± SD from at least three independent experiments. (**) p < 0.01 by Student’s t-test
p53 activation blocks myogenesis in a PW1-dependent manner
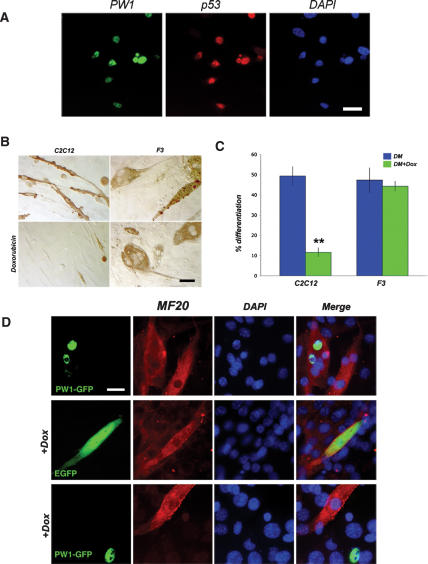
The observation that p53 and a p53 target gene (PW1) are required for the effects of TNFα on myogenesis suggests direct interaction and consequently coexpression of these two gene products in myoblasts. Immunostaining reveals that p53 and PW1 colocalize in the nuclei of primary myoblasts (Fig. 2A). To test whether p53 can mediate myogenesis through p53-dependent cell stress response pathways, we treated C2C12 cells with the DNA-damaging agent Doxorubicin and evaluated subsequent myogenic differentiation. As reported by others (Kurabayashi et al. 1993, 1994), we found that Doxorubicin treatment was sufficient to inhibit muscle differentiation (Fig. 2B,C). Since our previous studies revealed that PW1 acts downstream in the p53 cell stress response, we examined F3 myoblasts that do not express PW1 under any conditions (Coletti et al. 2002). In contrast to C2C12 cells, we note that F3 differentiation is not inhibited in the presence of Doxorubicin (Fig. 2B,C). Identical results were obtained using another p53 activator Etoposide (see Puri et al. 2002; data not shown). Forced expression of a GFP-PW1 fusion construct in F3 cells has no effect on F3 differentiation (Fig. 2D, top panel); however, GFP-PW1-expressing cells poorly differentiate in the presence of Doxorubicin as compared with cells that were transfected with GFP alone (3% vs. 36% of differentiation) (Fig. 2D). These data indicate that PW1 expression blocks myogenesis in response to p53-mediated cell stress.
Figure 2.
PW1 confers p53 sensitivity in myogenic cells. (A) Immunolocalization of PW1 (green) and p53 (red) in proliferating primary myoblasts showing coexpression in vitro. Nuclei were visualized by DAPI (blue). Bar, 20 μm. (B) Myogenic cells cultured in DM in the presence or absence of Doxorubicin. Immunohistochemistry of myosin shows that C2C12 (PW1-expressing) cells do not differentiate in the presence of Doxorubicin. In contrast, F3 cells (PW1 negative) differentiate regardless of treatment. Bar, 30 μm. (C) The myogenic potentials of C2C12 and F3 cells cultured as shown in B were evaluated by quantitative analysis of myogenic differentiation (% differentiation). F3 cells are resistant to genotoxic stress-mediated inhibition of differentiation. Values are expressed as the mean ± SD of at least three independent experiments. (**) p < 0.01 by Student’s t-test. (D) F3 cells were transfected with either EGFP or PW1-GFP expression vector and induced to differentiate in the presence or absence of Doxorubicin. Myosin (red) was immunostained to assess differentiation in cells expressing either EGFP (green) or PW1-GFP (green). Nuclei were visualized by DAPI staining (blue). (Top panels) Ectopic PW1 expression in F3 cells does not affect differentiation. (Middle panels) F3 cells transfected with EGFP differentiate in the presence of Doxorubicin. (Bottom panels) In contrast, virtually all F3 cells expressing PW1-GFP fail to differentiate in the presence of Doxorubicin. Only PW1-negative F3 cells are myosin positive upon Doxorubicin treatment as shown in the middle and bottom panels. Bar, 15 μm.
PW1 regulates p53 expression and activity
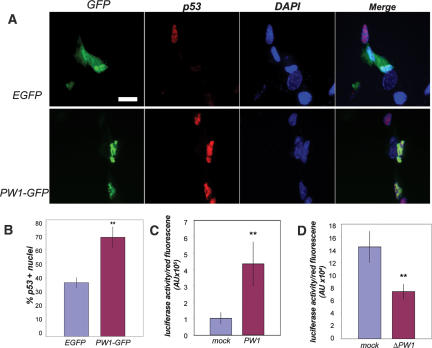
While we previously demonstrated that p53 can induce PW1 expression de novo in fibroblasts, the situation in myoblasts appears different. The observation that p53 and PW1 colocalize in muscle cells, and that PW1 is necessary for p53-mediated cell stress responses in myoblasts, suggests that PW1 positively regulates p53 expression and activity. We therefore examined p53 expression following forced expression of GFP or GFP-PW1 in fibroblasts that do not normally express PW1 or exhibit elevated levels of p53 except under cell stress conditions (Relaix et al. 2000). We detected a significant increase in nuclear p53 localization in cells overexpressing PW1 as compared with cells transfected with EGFP alone (Fig. 3A,B). Using the luciferase gene reporter under control of the p21 promoter containing p53-binding sites, we show that PW1 expression leads to bona fide p53 activation (transcriptional activity) (Fig. 3C). To investigate whether endogenous PW1 regulates endogenous p53 activity in myogenic cells, we cotransfected C2C12 cells with the dominant-negative ΔPW1 construct and the p21–luciferase reporter construct. ΔPW1 expression results in a down-regulation of the p21–luciferase promoter activity (Fig. 3D). We conclude that PW1 activates p53, and that p53 is constitutively active in myogenic cells.
Figure 3.
PW1 up-regulates p53 expression and activity. (A) Immunofluorescence analysis of p53 expression in 10T1/2 fibroblasts. 10T1/2 cells were transfected with either EGFP or PW1-GFP expression vector (PW1-GFP) followed by immunolocalization of p53 (red). Nuclei were visualized by DAPI staining (blue). The results were subjected to quantitative analyses as shown below in B. Bar, 5 μm. (B) Quantification of p53-labeled 10T1/2 cells reveals a significant increase in p53-positive cells following PW1-GFP transfection. The values are the mean ± SD of 100–200 transfected cells in three independent experiments. (**) p < 0.01 by Student’s t-test. (C) Quantification of p53 activity using the p21–luciferase reporter construct. 10T1/2 cells transfected with PW1 show an increase in p53 transcriptional activity as compared with control (mock)-transfected 10T1/2 cells. Luciferase activity was normalized for transfection efficiency with a cotransfected RFP expression construct (see Materials and Methods) (**) p < 0.05 by Student’s t-test. (D) C2C12 cells transfected with the dominant-negative PW1 (ΔPW1) show a decrease in p53 transcriptional activity as compared with C2C12 cells transfected with control vector (mock). Luciferase activity was normalized for transfection efficiency as described above and in Materials and Methods. (**) p < 0.05 by Student’s t-test.
p53 mediates skeletal muscle atrophy
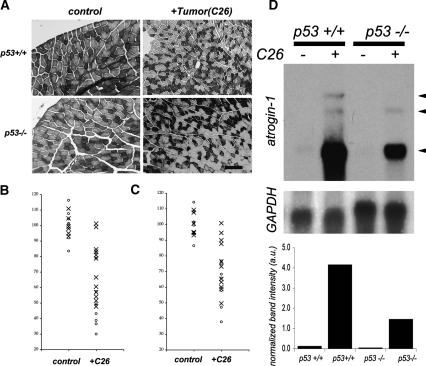
Our data reveal regulatory cross-talk between the TNFα pathway, which is known to mediate muscle atrophy, and a p53 pathway in myogenic cells in vitro. To determine if this relationship exists in vivo, we tested whether p53+/+ and p53−/− mice show a difference in muscle atrophy in response to tumor load. We used the established C26 tumor graft model of cancer-associated cachexia that induces a pronounced elevation of circulating inflammatory cytokines (Tanaka et al. 1990; Sturlan et al. 2002). Since p53−/− mice develop tumors between 3 and 6 mo of age (Donehower et al. 1992; Jacks et al. 1994; Tyner et al. 2002), we used young (7 wk) mice that were analyzed at 10 wk of age—well before the onset of spontaneous tumor occurrence. We nonetheless performed biopsies on all of the mice at the end of the experiment to monitor for spontaneous tumor formation. We first observed that weight loss in response to tumor load was more pronounced in p53+/+ as compared with p53−/− mice (data not shown). However, loss of body weight that occurs in cachectic subjects has multiple causes, stemming from diminished appetite and fat loss, as well as skeletal muscle wasting. We therefore restricted our analyses to skeletal muscle responses to tumor load. Previous studies have shown a difference in response to atrophic stimuli between fiber types (Desplanches et al. 1987). Transverse cryosections at the mid-belly of Tibialis anterior (TA) muscle were collected and stained for NADH to distinguish between fast and slow fibers. As shown in Figure 4A, different fiber types can be readily distinguished. A fiber cross-section area was subjected to a two-way ANOVA (analysis of variance) to assess the effects of genotype and tumor load individually as well as their potential interaction. These analyses reveal that tumor load has a highly significant effect on fiber size (p < 0.0001, F value = 81.62) regardless of phenotype. We also note a weaker effect of genotype on fiber size in that p53−/− fibers are slightly smaller at 10 wk of age as compared with p53+/+ mice, regardless of tumor load (p = 0.02, F value = 5.74). Importantly, we find that the p53 genotype is a very significant factor in muscle fiber size in response to tumor load (p = 0.0003, F value = 16.85). In order to eliminate the variation due to genotype, we normalized changes in fiber size as a function of percentage change following tumor load compared with control values and subjected these data to a Student’s t-test used for post-hoc analysis (see Materials and Methods). As shown in Figure 4B, analyses of fast fiber sizes from both TA and gastrocnemius (GA) reveal a significant decrease in p53+/+ tumor-bearing mice as compared with p53+/+ controls, whereas p53−/− mice show a markedly reduced response to tumor load. We note that the tumor-induced atrophy is significantly higher in p53+/+ mice when directly compared with p53−/− mice. p53+/+ mice undergo an average loss of 52% in fiber size in response to tumor load as compared with p53−/− mice, which show only an average loss of 26% (see Fig. 4B). We also compared changes in slow fibers as shown in Figure 4C. ANOVA was similarly performed for these data, revealing that tumor load causes a decrease in slow fiber size (p < 0.0001, F value = 54.85). In contrast to our findings with fast fibers, genotype has no significant effect on slow fiber size (p > 0.05, F value = 3.95). We note that tumor load shows a significant interaction with genotype, affecting the final slow fiber size outcome (p < 0.05, F value = 4.82). In p53+/+ mice, there is an average reduction of 38% in slow fiber size due to tumor load in contrast to an average reduction of 25% in p53−/− mice (Fig. 4C). We note that loss of p53 has a less pronounced effect on changes in slow fiber size due to tumor load (p = 0.08) as compared with fast fibers (p < 0.005; see Fig. 4B).
Figure 4.
p53 regulates cancer-associated cachexia. (A) Representative photomicrographs of cross-sections from TA removed 3 wk following tumor grafting (+C26) or sham surgery (control). Sections were stained for NADH activity to identify fast (light staining) and slow (dark staining) fibers. Bar, 60 μm. (B) Changes in fast fiber sizes resulting from tumor graft (+C26) in p53+/+ (O) and p53−/− (X) mice. Data are represented as percentage of change from the mean value of controls as compared with values obtained from tumor-bearing mice within genotypic groups (p53+/+ and p53−/−). In tumor-bearing p53+/+ mice, the average fiber size decreases to 48% of control values (SD = 15%; p < 0.000005 by Student’s t-test). In contrast, fast fiber size in tumor-bearing p53−/− mice shows a less marked decrease to an average value of 74% of control values (SD = 17%; p < 0.001 by Student’s t-test). The decrease in fiber size due to tumor load is significantly more pronounced in p53+/+ mice as compared with p53−/− mice (p < 0.005 by Student’s t-test). (C) Changes in slow fiber size resulting from tumor implant (+C26) in p53+/+ (O) and p53−/− (X) mice. All comparisons were performed as described for B. In tumor-bearing p53+/+ mice, fiber size declines to an average value of 62% of control values (SD = 13%; p < 0.00002 by Student’s t-test). In contrast, fiber size in p53−/− tumor-bearing (+C26) mice declines to an average value of 75% of control values (SD 17%; p < 0.0005 by Student’s t-test). We note that loss of p53 has a less pronounced effect on changes in slow fiber size due to tumor load (p = 0.08) as compared with fast fibers (p < 0.005, see B). (D) Northern blot analysis for atrogin-1 transcripts (arrows) in p53+/+ and p53−/− muscles following (sham surgery, —) or tumor load (+C26). (Top and bottom panels) Mice subjected to sham surgery do not express detectable levels of atrogin-1. (Top and bottom panels) Following tumor implantation, p53+/+ mice show a marked up-regulation of atrogin-1, whereas p53−/− mice show only a modest increase in transcript levels. (Middle panel) GAPDH was used as a loading control. (Bottom panel) Densitometric analysis of total atrogin transcript levels are presented normalized against GAPDH, revealing a diminished increase in atrogin expression in p53−/− muscle as compared with p53+/+ muscle following tumor load.
The molecular signature of cachexia-associated muscle fiber atrophy is elevated expression of atrogin-1 (Fig. 4D). As expected, control p53+/+ and p53−/− muscle showed nearly undetectable levels of atrogin-1 (Fig. 4D). In response to tumor load, atrogin-1 levels are dramatically up-regulated in p53+/+ muscle, whereas this up-regulation is attenuated in p53−/− muscle (Fig. 4D). Densitometric measurements of atrogin-1 levels normalized to GAPDH levels show a >75% reduction in atrogin-1 levels in p53−/− mice as compared with p53+/+ mice following tumor load (Fig. 4D). Taken together, these data reveal that p53 function mediates muscle atrophy in response to tumor load in vivo.
p53 levels affect muscle stem cell populations
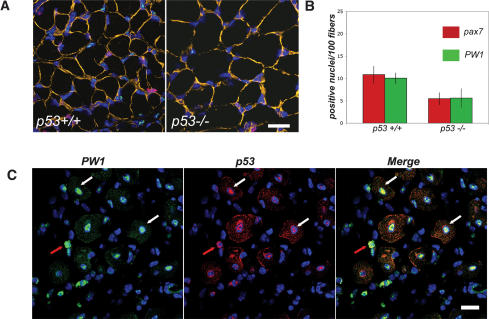
Previous studies by our laboratory and others reveal a strong correlation between postnatal muscle growth and maintenance of muscle stem cell number (Garry et al. 2000; Seale et al. 2000; Mitchell and Pavlath 2004; Coletti et al. 2005; Nicolas et al. 2005). Given that Pax7 and PW1 identify muscle stem cells, we examined their expression in 2-wk-old muscle from p53+/+ and p53−/− mouse hindlimbs. Tissue sections were also stained for laminin and the nuclei were stained with 4′,6-Diamidino-2-phenylindole (DAPI) to allow for the normalization of labeled cells (nuclei) to muscle fiber number (Fig. 5A,B). While we observed no change in the overall distribution pattern of PW1 and Pax7 in p53+/+ as compared with p53−/− hindlimb muscles (Fig. 5A), we observed a twofold decrease in the number of PW1- and Pax7-positive cells in p53−/− muscle as compared with wild type (Fig. 5A,B). We note a potential trend in larger fiber sizes in 2-wk-old p53−/− mice as compared with p53+/+ mice; however, careful analysis of this fails to reveal a statistically significant difference (data not shown).
Figure 5.
p53 regulates myogenic stem cell number and colocalizes with PW1 in vivo. (A) Photomicrographs of representative cross-sections of 14-d-old hindlimb muscles from p53+/+ and p53−/− mice immunostained for PW1 (green), Pax7 (red), and laminin (orange). Nuclei were counterstained with DAPI (blue). We note that very few cells are positive for PW1 and Pax7 in the p53−/− muscles as compared with the wild type (p53+/+). Bar, 15 μm. (B) Quantification of PW1- and Pax7-positive cells per 100 fibers from sections as shown in A. p53−/− muscles show a statistically significant decrease in PW1- and Pax7-positive nuclei as compared with p53+/+. At least 100 fibers per randomly chosen field for each individual were counted and the mean value ± SD was calculated from at least three animals per genotype. (*) p < 0.05 by Student’s t-test. (C) Representative cross-sections of adult muscle 5 d after focal injury immunostained for PW1 (green) and p53 (red) and processed with DAPI (blue) to visualize nuclei. We note that both single cells (red arrow) and myonuclei (centrally located; indicated by white arrow) show expression of p53 during the regenerative myogenic response. We further note coexpression of p53 and PW1 in centrally located nuclei (white arrows) as well as in cells outside the muscle fibers (red arrows). Bar, 15 μm.
We have shown here that p53 is readily detected in primary myoblasts (Fig. 1A); however, we do not detect significant levels of p53 expression in postnatal muscle in vivo (data not shown). If p53 plays a role in myogenic cells in vivo, we would expect to observe p53 expression in myogenic cells in muscle tissue. Since PW1 as well as Pax7 are difficult to detect in adult muscle in the absence of stress or injury, we extended our analysis to regenerating skeletal muscle. Five days after focal injury of adult muscle, we observed strong expression of PW1 and p53 in the regenerating regions (Fig. 5C). Furthermore, as observed in primary myoblasts in vitro, p53 colocalizes with PW1 in undifferentiated cells infiltrating the regenerating area (Fig. 5C) and in centrally located nuclei that are typical of newly regenerated fibers (Fig. 5C).
PW1 mediates muscle atrophy
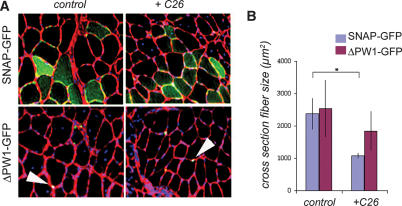
Our data reveal a role for PW1 and p53 in mediating the effects of TNFα on myogenesis in vitro as well as a role for p53 in mediating muscle atrophy in vivo. To test whether PW1 directly participates in muscle atrophy in vivo, we delivered the GFP-ΔPW1 dominant-negative expression vector to adult TA skeletal muscle using electroporation-mediated gene transfer in control and tumor-implanted mice. Gene delivery was performed as previously described (Coletti et al. 2005), and transfected fibers were compared with fibers transfected with the GFP expression construct as an internal control. Expression of the GFP construct in electroporated muscle was visualized by fluorescent microscopy in combination with immunostaining for laminin to visualize fibers and DAPI to identify nuclei (Fig. 6A). In the absence of tumor load, the nuclear expression of ΔPW1 or GFP alone did not significantly affect fiber size (Fig. 6A,B). The atrophic response to the tumor presence was greatly attenuated in ΔPW1-expressing fibers, whereas SNAP-GFP-expressing fibers showed significant fiber size reduction in response to tumor load (Fig. 6A,B). We conclude that PW1 is required for the p53-dependent cachectic response to occur in skeletal muscle.
Figure 6.
PW1 modulates fiber atrophy in response to tumor load. (A) Representative sections of TA electroporated with SNAP-GFP (green) or ΔPW1-GFP (green) from tumor-bearing (+C26) or control (sham-operated) mice. The basal lamina was visualized by laminin immunofluorescence (red), and nuclei were counterstained with DAPI (blue). SNAP-GFP appears throughout the myofiber, whereas ΔPW1-GFP fusion protein is restricted to nuclei (white arrowheads). Bar, 30 μm. (B) Quantification of transfected fiber cross-section area from muscles treated as shown in A. As expected, SNAP-GFP electroporated muscle fiber area decreases in response to tumor load (+C26). In contrast, ΔPW1-GFP-electroporated fibers do not show a significant size change in response to tumor load (+C26) as compared with controls. The values are presented as the mean ± SD of six animals. (*) p < 0.05 by Student’s t-test.
Discussion
It is widely recognized that cachexia diminishes the quality of life as well as the prognosis for recovery from cancer and other chronic disease (Inui 2002). Cellular mechanisms underlying cachexia include increased muscle protein breakdown and impaired satellite cell function (Glass 2003a, b; Dasarathy et al. 2004). TNFα is one of the major cytokines implicated in triggering cachexia and has been demonstrated to provoke an up-regulation of the ubiquitin–proteasome pathway via atrogin-1 in vitro (Li et al. 2005) and in vivo (Coletti et al. 2005). Additionally, TNFα has been shown to inhibit skeletal muscle differentiation of myogenic cells in vitro and in vivo (Miller et al. 1988; Szalay et al. 1997; Guttridge et al. 2000; Langen et al. 2004). We recently demonstrated that elevated levels of TNFα result in both cachexia and inhibition of muscle regeneration following experimentally induced injury in vivo, raising the possibility that TNFα-induced muscle wasting involves impaired myogenic stem cell function (Coletti et al. 2005). This proposed mechanism is consistent with results demonstrating that the inhibition of TNFα activity slows the breakdown of dystrophic muscle and improves regeneration and recovery in dystrophic (mdx) mice (Grounds and Torrisi 2004). Indeed, a recent study has provided a direct functional link between the process of muscle atrophy and the maintenance of muscle integrity through dystrophin function in mdx mice (Acharyya et al. 2005).
TNFα inhibits muscle differentiation through activation of components of the p53-dependent cell death pathway, including bax, caspase 9, and PW1/Peg3 (Coletti et al. 2002). In nonmuscle cells (fibroblasts) in which PW1 is not constitutively expressed, p53 activates PW1, bax translocation to mitochondria (Deng and Wu 2000), and caspase activation (Relaix et al. 2000). Therefore, PW1 provides a mechanistic link between the TNFα signaling and the p53 apoptotic pathway. These observations suggest that cell death effectors play a role in the regulation of the myogenic program (Coletti et al. 2002).
In this study we show that p53, in concert with PW1, plays a role in cachexia by blocking muscle differentiation in the presence of TNFα and regulating muscle stem cell populations in vivo. We further demonstrate that TNFα acts through a signaling pathway that requires p53 function to inhibit myogenic differentiation. In support of this hypothesis, we found that Pifithrin, a p53 inhibitor shown to protect from the side effects of cancer therapy such as weight loss and muscle atrophy (Komarov et al. 1999), can reverse the inhibitory effect of TNFα on muscle differentiation. Doxorubicin treatment of myoblasts leads to p53 activation via phosphorylation (Latella et al. 2004), and it has been shown that Doxorubicin/genotoxic stress inhibits myogenic differentiation by down-regulating MyoD and myogenin (Kurabayashi et al. 1993, 1994; Puri et al. 2002). We show here that this effect requires PW1. Taken together, these results imply a fundamental role for p53 in the TNFα signaling pathway.
Several studies suggest a role for p53 in regulating muscle homeostasis. For example, increased p53 levels are found in skeletal muscle during unloading-induced muscle atrophy (Siu and Alway 2005) and in immobilized rat muscle (Machida and Booth 2005), as well as in aging skeletal muscle (Chung and Ng 2006). Under these conditions, p53 can be localized in muscle stem cells (satellite cells) (Machida and Booth 2004). Several in vivo models have been developed that show that p53 plays a crucial role in the onset of early aging phenotypes including profound loss of muscle mass (Vogel et al. 1999; Tyner et al. 2002; Maier et al. 2004; Varela et al. 2005). In addition, the procachectic factor myostatin (Zimmers et al. 2002) inhibits muscle differentiation via p21 and p53 (Joulia et al. 2003). We and others (Yarasheski et al. 2002) have observed an increased level of myostatin with aging. Radiation therapy also provokes muscle wasting (Persons et al. 2001), which can be blocked by chemical inhibition of p53 (Komarov et al. 1999). Mice deficient for a protein important for the repair of DNA double-strand (Ku80−/−) undergo premature senescence. The senescent phenotype has been previously demonstrated to be dependent on p53 activity in cells obtained from Ku80 mutant mice (Vogel et al. 1999). Here we show that hallmarks of cachexia, including a decline in muscle fiber size as well as an increase in atrogin-1 transcription, are attenuated in p53-deficient mice. Our observation that p53-null mice are resistant to cachexia provides genetic evidence that p53 is a key mediator of muscle wasting and homeostasis in vivo.
PW1 was identified in a screen for novel regulatory factors in myogenic stem cell determination (Relaix et al. 1996) as well as in a screen for novel effectors in p53-mediated cell death (Relaix et al. 2000). Whereas PW1 is normally not expressed in nonmuscle cells, it is expressed following the activation or forced expression of p53 leading to cell death (Relaix et al. 2000). In this context, it is curious that PW1 is constitutively expressed in myoblasts and most myogenic cell lines in vitro even though myogenic cells appear healthy and capable of high levels of mitosis (Relaix et al. 1996; Coletti et al. 2002). We observe that satellite cells as well as a subset of interstitial cells express PW1 in postnatal muscle (Nicolas et al. 2005). This expression in interstitial cells is not seen in other tissue types, suggesting that PW1 also identifies another population of cells with potential stem cell properties. Unpublished results indicate that pluripotent stem cells (mesoangioblasts) derived from embryonic vascular tissue with pronounced myogenic capacity (De Angelis et al. 1999) are also positive for PW1 (G. Marazzi and D. Sassoon, unpubl.; G. Cossu, pers. comm.). These data lend further support to a role for PW1 in regulating postnatal stem cells in muscle tissue. We observed that p53 colocalizes with PW1 as well as other myogenic markers including MyoD in primary myoblasts. We note that p53 expression is as reliable an indicator of myogenic cells as Pax7 and MyoD in primary cultures derived from wild-type postnatal muscle even though the cells do not undergo cell death. These observations may explain a long-accepted methodology in preparing “enriched” populations of primary myoblasts in which single-cell preparations obtained from muscle are first preplated for several hours to allow fibroblasts to settle, whereas nonadherent cells (myoblasts) are transferred to new culture plates as they appear to undergo stress-induced growth arrest. The observation that p53 is constitutively active in myoblasts and not in fibroblasts may underlie the “slower” kinetics of re-entry into the cell cycle of primary myoblasts in culture (i.e., “culture shock”).
We find that the number of both PW1- and Pax7-expressing cells is decreased in p53−/− mice. In addition, we observe that PW1 overexpression induces p53 expression and transcriptional activation in fibroblasts, while the inhibition of PW1 pathways results in repressed p53 transcriptional activity in myoblasts. Taken together, these data suggest that PW1 and p53 participate in a positive feedback loop whereby elevated PW1 expression in muscle stem cells inhibits subsequent stem cell expansion and/or maintenance. This is consistent with the observation that impaired satellite cell functions result in atrophy (Dasarathy et al. 2004). Disrupting normal muscle stem cell populations can lead to compromised postnatal growth or directly to muscle atrophy, as seen in the Pax7−/− mice and in mice without MMF (forkhead/winged helix protein), which is also selectively expressed in satellite cells in adult animals (Garry et al. 2000; Seale et al. 2000). Transgenic mice overexpressing ΔPW1, which has been shown to inhibit p53-mediated pathways, show a deregulation of postnatal muscle stem cells (Nicolas et al. 2005) consistent with a role for p53 regulation of muscle stem cells. Taken together, the results presented here reveal a new role for p53 in concert with the p53 target PW1 in skeletal muscle pathology and stem cell behavior. This study demonstrates that in addition to acting as a tumor suppressor, p53 also plays a more global role in the organism, contributing to muscle wasting in response to tumor load. The identification of key effectors in this pathway may lead to the identification of new targets for therapeutic approach in the treatment of cachexia.
Materials and methods
Cells culture and transfection procedures
Primary cultures of satellite cells were obtained from 8-d-old mouse hindlimbs and maintained in culture in high-serum medium (GM) as previously described (Megeney et al. 1996; Coletti et al. 2002). C2 and F3 myoblasts and 10T1/2 fibroblasts were grown in GM as previously described (Coletti et al. 2002). Myogenic differentiation of primary cultures and cell lines was induced by shifting the medium to DMEM supplemented with 2% horse serum (DM). For differentiation experiments, cells were plated on gelatin-coated coverslips and treated for 24 h with 30 μM Pifithrin (Calbiochem) or 65 nM Doxorubicin (MP Biomedicals LLC) as indicated. TNFα (Roche Molecular Biochemicals) was supplemented at 20 ng/mL in GM for 8 h prior to changing to DM supplemented with 20 ng/mL TNFα. DMSO was used for control experiments. The medium was changed daily for all experiments to maintain constant cytokine levels.
Cells plated on gelatin-coated coverslips were transfected with EGFP-PW1 (kindly provided by Dr. Xiangwei Wu, Baylor College of Medicine, Houston, TX) (Deng and Wu 2000) or EGFP using Lipofectamine 2000 reagents according to the manufacturer’s instructions (Invitrogen). Cells were treated with DMSO or 65 nmol Doxorubicin in DMSO 24 h after transfection. Cells were allowed to grow for 2 d following transfection and then were differentiated for 3 d. The medium was changed daily to keep levels of Doxorubicin constant.
The human p53 reporter construct E16TATAp215′-Luc (courtesy of Dr. James Manfredi, Mount Sinai Medical Center, New York, NY) and a red fluorescent protein (RFP) construct (dsRED-C1, BD Bioscience) were cotransfected in suspension with either pcDNA, PW1, or ΔPW1-GFP in a 1:1:1 ratio, by mixing 2 μg of each construct using Fugene transfection reagent (Roche Molecular Biochemicals). The ΔPW1-GFP construct was generated from the ΔPW1 fragment previously identified in our laboratory (Relaix et al. 1998) that was subcloned into the EGFP-C1 backbone obtained from Clontech. Following transfection, cells were seeded in replicate dishes and were collected for luciferase assay and fluorometric analysis or fixed for immunofluorescence after 24 h.
Immunohistochemistry, immunofluorescence, and differentiation assays
Cells were grown on gelatin-coated coverslips and fixed in 4% paraformaldehyde. Myosin was detected using the MF20 antibody (Developmental Hybridoma Bank of the University of Iowa) followed by goat anti-mouse Ab and streptavidin-conjugated horseradish peroxidase (Jackson Laboratories) as previously described (Coletti et al. 2002). Additional assays were carried out in which cells were stained with antibodies against MHC (MF20), MyoD (Santa Cruz Biotechnology, Inc.), PW1, or p53 (Pab 242, kindly provided by Dr. David Lane, University of Dundee, Dundee, Scotland, UK). AlexaFluor568-conjugated anti-mouse, AlexaFluor488-conjugated anti-rabbit, and AlexaFluor488-conjugated anti-mouse (Molecular Probes) were used as secondary antibodies. Nuclei were counterstained with DAPI. Photomicrographs were obtained using a Zeiss Axiophot microscope fitted with a SPOT RT Slider camera (Diagnostic Instruments).
Quantitative analysis of differentiation was performed by determining the number of nuclei in MF20-positive cells and expressing this as a percentage of the total number of nuclei in multiple microscopic fields (% differentiation). At least 150–300 cells from randomly chosen fields in three independent experiments were analyzed.
Luciferase and fluorometric assays
Luciferase activity was assayed in duplicate by mixing 5 μL of cell extracts with 100 μL of luciferase assay reagent (Promega). Reporter gene expression was expressed as arbitrary units of RFP expression normalized for transfection efficiency. Expression of RFP was assayed on the same lysates used for the luciferase assay. The fluorescence of 10 μL of cell lysates (diluted in 2 mL of water) was measured in duplicate with a 558/583 Ex/Em pair by using a luminescence spectrometer (Perkin Elmer)
Mice and cachexia mouse models
The animal models used were p53+/+ and p53−/− BALB/c mice (The Jackson Laboratory, strain name C.129S2(b6)-Trp53tmaTyj/J) (Jacks et al. 1994) and p53−/− C57BL/6 mice (Taconic, strain name C57BL/6 TSG-p53) (Donehower et al. 1992).
Cachexia was induced by subcutaneous grafting of a 1-mm3 solid fragment of colon carcinoma (C26, obtained from the National Cancer Institute) (Tanaka et al. 1990) in the back of 7-wk-old p53−/− and p53+/+ BALB/c mice and compared with sham-operated controls. Siblings derived from matings of heterozygous couples were used for all experiments. The animals were sacrificed 3 wk after tumor implantation, at which time the tumor typically grew to a weight of ∼1 g. At the end of the experiment, mice were 10 wk old, prior to the time when p53−/− mice develop tumors (3–6 mo of age) (Jacks et al. 1994). Nonetheless, heart, lung, kidney, liver, spleen, and intestine were surveyed for the presence of metastasis and spontaneous primary tumors. No tumor was ever observed at this stage in mice used for these experiments.
Northern blot analysis
Northern blot analysis of RNA extracted from skeletal muscle was performed as previously described (Gomes et al. 2001; Coletti et al. 2005). Membranes were probed with a KpnI/PstI fragment from the atrogin-1 plasmid (kind gift of Dr. Alfred Goldberg, Beth Israel Deaconess Medical Center, Boston, MA) and with GAPDH probe as a loading control. Band intensity was quantified using an Aida analysis system (Reytest). The sum of the intensity of the three atrogin-1 bands was normalized to GAPDH band intensity.
Muscle tissue analyses
To independently evaluate both fast and slow fiber size in the TA and the GA, NADH transferase staining (NADH and NBT from Roche Molecular Biochemicals) was performed on 8-μm-thick cryosections (cross-sections in the mid-belly region of the muscle) as described previously (Degenhardt and Sassoon 2001). Morphometric analysis was performed on type IIb (no NADH transferase activity–fast) and type I (strong NADH transferase activity–slow) fibers separately, as described previously (Coletti et al. 2005). For each muscle, average fiber size (cross-section area) was calculated on 200–600 fibers. For each experimental group (p53+/+ and p53−/− with and without tumor), six to 12 animals were analyzed. Both the genotype and tumor load were considered as variables using two-way ANOVA. ANOVA was performed using the VassarStats calculation pages (http://faculty.vassar.edu/lowry/VassarStats.htm). Based on the ANOVA results, we then performed Student’s t-test as a post-hoc analysis, comparing normalized fiber size changes between control and tumor-bearing mice within genotypic groups (p53+/+ and p53−/−). We further compared the effect of tumor bearing between the two genotypic groups.
For immunofluorescence experiments, hindlimb muscles from 14-d-old postnatal pups were snap frozen in isopentane-cooled liquid nitrogen. Cryosections were post-fixed with 4% paraformaldehyde and immunostained for PW1, Pax7 (Developmental Hybridoma Bank of the University of Iowa), and laminin (Sigma) as previously described (Nicolas et al. 2005).
Antibody binding was visualized by using Alexa488-conjugated goat anti-rabbit IgG (Molecular Probes), biotin-conjugated goat anti-mouse IgG1 specific followed by Cy3-conjugated streptavidin (Jackson Immunoresearch), and Cy5-conjugated goat anti-rabbit IgG (Jackson Immunoresearch). Nuclei were visualized by DAPI staining. Photomicrographs were captured using a Leica TCS-SP(UV) confocal microscope (MSSM-Microscopy Shared Resource Facility).
Regeneration of skeletal muscle was induced by a single freeze–crush injury of the TA as previously described (McGeachie and Grounds 1987). For all regeneration experiments, 2-mo-old mice were used. Five days after injury, tissues were fixed overnight in 4% paraformaldehyde, and frozen in OCT. Cryosections were immunostained for PW1 and p53 (CM-5 Ab kindly provided by Dr. David Lane) as described above.
In vivo gene transfer
To induce exogenous expression of DNA in skeletal muscle, we injected 25 μg of EGFP-ΔPW1 into the TA of BALB/c mice, or 25 μg of pCMV-Snap25-GFP (kindly provided by Dr. Tullio Pozzan, University of Padova, Padova, Italy) into the contra-lateral muscle. DNA injection was immediately followed by electric pulses delivered using 3 × 5-mm Genepaddle electrodes (BTX) as described previously (Dona et al. 2003; Coletti et al. 2005).
Acknowledgments
We gratefully acknowledge the advice and use of reagents from Drs. L. Donehower and S. Aaronson. Confocal laser scanning microscopy was performed at the MSSM-Microscopy Shared Resource Facility, supported with funding from NIH-NCI shared resources grant (5R24 CA095823-04), NSF Major Research Instrumentation grant (DBI-9724504), and NIH shared instrumentation grant (1 S10 RR0 9145-01). This work was supported by NIH PO1 CA80058-05 (project 3) and a grant from the Muscular Dystrophy Association (USA) to D.S. and a grant from the MIUR-Rientro dei cervelli to D.C.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.412606
References
- Acharyya, S., Butchbach, M.E., Sahenk, Z., Wang, H., Saji, M., Carathers, M., Ringel, M.D., Skipworth, R.J., Fearon, K.C., Hollingsworth, M.A., et al. Dystrophin glycoprotein complex dysfunction: A regulatory link between muscular dystrophy and cancer cachexia. Cancer Cell. 2005;8:421–432. doi: 10.1016/j.ccr.2005.10.004. [DOI] [PubMed] [Google Scholar]
- Bodine, S.C., Stitt, T.N., Gonzalez, M., Kline, W.O., Stover, G.L., Bauerlein, R., Zlotchenko, E., Scrimgeour, A., Lawrence, J.C., Glass, D.J., et al. Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nat. Cell Biol. 2001;3:1014–1019. doi: 10.1038/ncb1101-1014. [DOI] [PubMed] [Google Scholar]
- Cai, D., Frantz, J.D., Tawa, N.E., Jr., Melendez, P.A., Oh, B.C., Lidov, H.G., Hasselgren, P.O., Frontera, W.R., Lee, J., Glass, D.J., et al. IKKβ/NF-κB activation causes severe muscle wasting in mice. Cell. 2004;119:285–298. doi: 10.1016/j.cell.2004.09.027. [DOI] [PubMed] [Google Scholar]
- Charge, S.B., Rudnicki, M.A. Cellular and molecular regulation of muscle regeneration. Physiol. Rev. 2004;84:209–238. doi: 10.1152/physrev.00019.2003. [DOI] [PubMed] [Google Scholar]
- Chin, L., Artandi, S.E., Shen, Q., Tam, A., Lee, S.L., Gottlieb, G.J., Greider, C.W., DePinho, R.A. p53 deficiency rescues the adverse effects of telomere loss and cooperates with telomere dysfunction to accelerate carcinogenesis. Cell. 1999;97:527–538. doi: 10.1016/s0092-8674(00)80762-x. [DOI] [PubMed] [Google Scholar]
- Chung, L., Ng, Y.C. Age-related alterations in expression of apoptosis regulatory proteins and heat shock proteins in rat skeletal muscle. Biochim. Biophys. Acta. 2006;1762:103–109. doi: 10.1016/j.bbadis.2005.08.003. [DOI] [PubMed] [Google Scholar]
- Coletti, D., Yang, E., Marazzi, G., Sassoon, D. TNFα inhibits skeletal myogenesis through a PW1-dependent pathway by recruitment of caspase pathways. EMBO J. 2002;21:631–642. doi: 10.1093/emboj/21.4.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coletti, D., Moresi, V., Adamo, S., Molinaro, M., Sassoon, D. Tumor necrosis factor-α gene transfer induces cachexia and inhibits muscle regeneration. Genesis. 2005;43:119–127. doi: 10.1002/gene.20160. [DOI] [PubMed] [Google Scholar]
- Dasarathy, S., Dodig, M., Muc, S.M., Kalhan, S.C., McCullough, A.J. Skeletal muscle atrophy is associated with an increased expression of myostatin and impaired satellite cell function in the portacaval anastamosis rat. Am. J. Physiol. Gastrointest. Liver Physiol. 2004;287:G1124–G1130. doi: 10.1152/ajpgi.00202.2004. [DOI] [PubMed] [Google Scholar]
- De Angelis, L., Berghella, L., Coletta, M., Lattanzi, L., Zanchi, M., Cusella-De Angelis, M.G., Ponzetto, C., Cossu, G. Skeletal myogenic progenitors originating from embryonic dorsal aorta coexpress endothelial and myogenic markers and contribute to postnatal muscle growth and regeneration. J. Cell Biol. 1999;147:869–878. doi: 10.1083/jcb.147.4.869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degenhardt, K., Sassoon, D.A. A role for Engrailed-2 in determination of skeletal muscle physiologic properties. Dev. Biol. 2001;231:175–189. doi: 10.1006/dbio.2000.0131. [DOI] [PubMed] [Google Scholar]
- Deng, Y., Wu, X. Peg3/Pw1 promotes p53-mediated apoptosis by inducing Bax translocation from cytosol to mitochondria. Proc. Natl. Acad. Sci. 2000;97:12050–12055. doi: 10.1073/pnas.97.22.12050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desplanches, D., Mayet, M.H., Sempore, B., Flandrois, R. Structural and functional responses to prolonged hindlimb suspension in rat muscle. J. Appl. Physiol. 1987;63:558–563. doi: 10.1152/jappl.1987.63.2.558. [DOI] [PubMed] [Google Scholar]
- Dona, M., Sandri, M., Rossini, K., Dell’Aica, I., Podhorska-Okolow, M., Carraro, U. Functional in vivo gene transfer into the myofibers of adult skeletal muscle. Biochem. Biophys. Res. Commun. 2003;312:1132–1138. doi: 10.1016/j.bbrc.2003.11.032. [DOI] [PubMed] [Google Scholar]
- Donehower, L.A., Harvey, M., Slagle, B.L., McArthur, M.J., Montgomery, C.A., Jr., Butel, J.S., Bradley, A. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- Garry, D.J., Meeson, A., Elterman, J., Zhao, Y., Yang, P., Bassel-Duby, R., Williams, R.S. Myogenic stem cell function is impaired in mice lacking the forkhead/winged helix protein MNF. Proc. Natl. Acad. Sci. 2000;97:5416–5421. doi: 10.1073/pnas.100501197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass, D.J. Molecular mechanisms modulating muscle mass. Trends Mol. Med. 2003a;9:344–350. doi: 10.1016/s1471-4914(03)00138-2. [DOI] [PubMed] [Google Scholar]
- Glass, D.J. Signalling pathways that mediate skeletal muscle hypertrophy and atrophy. Nat. Cell Biol. 2003b;5:87–90. doi: 10.1038/ncb0203-87. [DOI] [PubMed] [Google Scholar]
- Gomes, M.D., Lecker, S.H., Jagoe, R.T., Navon, A., Goldberg, A.L. Atrogin-1, a muscle-specific F-box protein highly expressed during muscle atrophy. Proc. Natl. Acad. Sci. 2001;98:14440–14445. doi: 10.1073/pnas.251541198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grounds, M.D., Torrisi, J. Anti-TNFα (Remicade) therapy protects dystrophic skeletal muscle from necrosis. FASEB J. 2004;18:676–682. doi: 10.1096/fj.03-1024com. [DOI] [PubMed] [Google Scholar]
- Guttridge, D.C., Albanese, C., Reuther, J.Y., Pestell, R.G., Baldwin, A.S., Jr. NF-κB controls cell growth and differentiation through transcriptional regulation of cyclin D1. Mol. Cell. Biol. 1999;19:5785–5799. doi: 10.1128/mcb.19.8.5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttridge, D.C., Mayo, M.W., Madrid, L.V., Wang, C.Y., Baldwin, A.S., Jr. NF-κB-induced loss of MyoD messenger RNA: Possible role in muscle decay and cachexia. Science. 2000;289:2363–2366. doi: 10.1126/science.289.5488.2363. [DOI] [PubMed] [Google Scholar]
- Halevy, O., Novitch, B.G., Spicer, D.B., Skapek, S.X., Rhee, J., Hannon, G.J., Beach, D., Lassar, A.B. Correlation of terminal cell cycle arrest of skeletal muscle with induction of p21 by MyoD. Science. 1995;267:1018–1021. doi: 10.1126/science.7863327. [DOI] [PubMed] [Google Scholar]
- Inui, A. Cancer anorexia–cachexia syndrome: Current issues in research and management. CA Cancer J. Clin. 2002;52:72–91. doi: 10.3322/canjclin.52.2.72. [DOI] [PubMed] [Google Scholar]
- Israel, A. The IKK complex: An integrator of all signals that activate NF-κB? Trends Cell Biol. 2000;10:129–133. doi: 10.1016/s0962-8924(00)01729-3. [DOI] [PubMed] [Google Scholar]
- Itahana, K., Dimri, G., Campisi, J. Regulation of cellular senescence by p53. Eur. J. Biochem. 2001;268:2784–2791. doi: 10.1046/j.1432-1327.2001.02228.x. [DOI] [PubMed] [Google Scholar]
- Jacks, T., Remington, L., Williams, B.O., Schmitt, E.M., Halachmi, S., Bronson, R.T., Weinberg, R.A. Tumor spectrum analysis in p53-mutant mice. Curr. Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- Joulia, D., Bernardi, H., Garandel, V., Rabenoelina, F., Vernus, B., Cabello, G. Mechanisms involved in the inhibition of myoblast proliferation and differentiation by myostatin. Exp. Cell Res. 2003;286:263–275. doi: 10.1016/s0014-4827(03)00074-0. [DOI] [PubMed] [Google Scholar]
- Komarov, P.G., Komarova, E.A., Kondratov, R.V., Christov-Tselkov, K., Coon, J.S., Chernov, M.V., Gudkov, A.V. A chemical inhibitor of p53 that protects mice from the side effects of cancer therapy. Science. 1999;285:1733–1737. doi: 10.1126/science.285.5434.1733. [DOI] [PubMed] [Google Scholar]
- Kurabayashi, M., Jeyaseelan, R., Kedes, L. Antineoplastic agent doxorubicin inhibits myogenic differentiation of C2 myoblasts. J. Biol. Chem. 1993;268:5524–5529. [PubMed] [Google Scholar]
- Kurabayashi, M., Jeyaseelan, R., Kedes, L. Doxorubicin represses the function of the myogenic helix–loop–helix transcription factor MyoD. Involvement of Id gene induction. J. Biol. Chem. 1994;269:6031–6039. [PubMed] [Google Scholar]
- Langen, R.C., Van Der Velden, J.L., Schols, A.M., Kelders, M.C., Wouters, E.F., Janssen-Heininger, Y.M. Tumor necrosis factor-α inhibits myogenic differentiation through MyoD protein destabilization. FASEB J. 2004;18:227–237. doi: 10.1096/fj.03-0251com. [DOI] [PubMed] [Google Scholar]
- Latella, L., Lukas, J., Simone, C., Puri, P.L., Bartek, J. Differentiation-induced radioresistance in muscle cells. Mol. Cell. Biol. 2004;24:6350–6361. doi: 10.1128/MCB.24.14.6350-6361.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y.P., Chen, Y., John, J., Moylan, J., Jin, B., Mann, D.L., Reid, M.B. TNF-α acts via p38 MAPK to stimulate expression of the ubiquitin ligase atrogin1/MAFbx in skeletal muscle. FASEB J. 2005;19:362–370. doi: 10.1096/fj.04-2364com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, D.S., Vogel, H., Willerford, D.M., Sands, A.T., Platt, K.A., Hasty, P. Analysis of ku80-mutant mice and cells with deficient levels of p53. Mol. Cell. Biol. 2000;20:3772–3780. doi: 10.1128/mcb.20.11.3772-3780.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida, S., Booth, F.W. Increased nuclear proteins in muscle satellite cells in aged animals as compared to young growing animals. Exp. Gerontol. 2004;39:1521–1525. doi: 10.1016/j.exger.2004.08.009. [DOI] [PubMed] [Google Scholar]
- Machida, S., Booth, F.W. Changes in signalling molecule levels in 10-day hindlimb immobilized rat muscles. Acta Physiol. Scand. 2005;183:171–179. doi: 10.1111/j.1365-201X.2004.01395.x. [DOI] [PubMed] [Google Scholar]
- Maier, B., Gluba, W., Bernier, B., Turner, T., Mohammad, K., Guise, T., Sutherland, A., Thorner, M., Scrable, H. Modulation of mammalian life span by the short isoform of p53. Genes & Dev. 2004;18:306–319. doi: 10.1101/gad.1162404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauro, A. Satellite cell of skeletal muscle fibers. J. Biophys. Biochem. Cytol. 1961;9:493–495. doi: 10.1083/jcb.9.2.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGeachie, J.K., Grounds, M.D. Initiation and duration of muscle precursor replication after mild and severe injury to skeletal muscle of mice. An autoradiographic study. Cell Tissue Res. 1987;248:125–130. doi: 10.1007/BF01239972. [DOI] [PubMed] [Google Scholar]
- Megeney, L.A., Kablar, B., Garrett, K., Anderson, J.E., Rudnicki, M.A. MyoD is required for myogenic stem cell function in adult skeletal muscle. Genes & Dev. 1996;10:1173–1183. doi: 10.1101/gad.10.10.1173. [DOI] [PubMed] [Google Scholar]
- Miller, S.C., Ito, H., Blau, H.M., Torti, F.M. Tumor necrosis factor inhibits human myogenesis in vitro. Mol. Cell. Biol. 1988;8:2295–2301. doi: 10.1128/mcb.8.6.2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell, P.O., Pavlath, G.K. Skeletal muscle atrophy leads to loss and dysfunction of muscle precursor cells. Am. J. Physiol. Cell Physiol. 2004;287:C1753–C1762. doi: 10.1152/ajpcell.00292.2004. [DOI] [PubMed] [Google Scholar]
- Muscaritoli, M., Bossola, M., Bellantone, R., Rossi Fanelli, F. Therapy of muscle wasting in cancer: What is the future? Curr. Opin. Clin. Nutr. Metab. Care. 2004;7:459–466. doi: 10.1097/01.mco.0000134366.07148.2e. [DOI] [PubMed] [Google Scholar]
- Nicolas, N., Marazzi, G., Kelley, K., Sassoon, D. Embryonic deregulation of muscle stress signaling pathways leads to altered postnatal stem cell behavior and a failure in postnatal muscle growth. Dev. Biol. 2005;281:171–183. doi: 10.1016/j.ydbio.2005.02.022. [DOI] [PubMed] [Google Scholar]
- Nussenzweig, A., Chen, C., da Costa Soares, V., Sanchez, M., Sokol, K., Nussenzweig, M.C., Li, G.C. Requirement for Ku80 in growth and immunoglobulin V(D)J recombination. Nature. 1996;382:551–555. doi: 10.1038/382551a0. [DOI] [PubMed] [Google Scholar]
- Olguin, H.C., Olwin, B.B. Pax-7 up-regulation inhibits myogenesis and cell cycle progression in satellite cells: A potential mechanism for self-renewal. Dev. Biol. 2004;275:375–388. doi: 10.1016/j.ydbio.2004.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oustanina, S., Hause, G., Braun, T. Pax7 directs postnatal renewal and propagation of myogenic satellite cells but not their specification. EMBO J. 2004;23:3430–3439. doi: 10.1038/sj.emboj.7600346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persons, C.C., Hermens, A.F., Franken, N.A., Wondergem, J. Muscle wasting after radiotherapy in young and adult rats. Oncol. Rep. 2001;8:1117–1122. doi: 10.3892/or.8.5.1117. [DOI] [PubMed] [Google Scholar]
- Porrello, A., Cerone, M.A., Coen, S., Gurtner, A., Fontemaggi, G., Cimino, L., Piaggio, G., Sacchi, A., Soddu, S. p53 regulates myogenesis by triggering the differentiation activity of pRb. J. Cell Biol. 2000;151:1295–1304. doi: 10.1083/jcb.151.6.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri, P.L., Bhakta, K., Wood, L.D., Costanzo, A., Zhu, J., Wang, J.Y. A myogenic differentiation checkpoint activated by genotoxic stress. Nat. Genet. 2002;32:585–593. doi: 10.1038/ng1023. [DOI] [PubMed] [Google Scholar]
- Reid, M.B., Li, Y.P. Tumor necrosis factor-α and muscle wasting: A cellular perspective. Respir. Res. 2001;2:269–272. doi: 10.1186/rr67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relaix, F., Weng, X., Marazzi, G., Yang, E., Copeland, N., Jenkins, N., Spence, S.E., Sassoon, D. Pw1, a novel zinc finger gene implicated in the myogenic and neuronal lineages. Dev. Biol. 1996;177:383–396. doi: 10.1006/dbio.1996.0172. [DOI] [PubMed] [Google Scholar]
- Relaix, F., Wei, X.J., Wu, X., Sassoon, D.A. Peg3/Pw1 is an imprinted gene involved in the TNF–NFκB signal transduction pathway. Nat. Genet. 1998;18:287–291. doi: 10.1038/ng0398-287. [DOI] [PubMed] [Google Scholar]
- Relaix, F., Wei, X., Li, W., Pan, J., Lin, Y., Bowtell, D.D., Sassoon, D.A., Wu, X. Pw1/Peg3 is a potential cell death mediator and cooperates with Siah1a in p53-mediated apoptosis. Proc. Natl. Acad. Sci. 2000;97:2105–2110. doi: 10.1073/pnas.040378897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph, K.L., Chang, S., Lee, H.W., Blasco, M., Gottlieb, G.J., Greider, C., DePinho, R.A. Longevity, stress response, and cancer in aging telomerase-deficient mice. Cell. 1999;96:701–712. doi: 10.1016/s0092-8674(00)80580-2. [DOI] [PubMed] [Google Scholar]
- Sandri, M., Sandri, C., Gilbert, A., Skurk, C., Calabria, E., Picard, A., Walsh, K., Schiaffino, S., Lecker, S.H., Goldberg, A.L. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell. 2004;117:399–412. doi: 10.1016/s0092-8674(04)00400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler, M., Green, D.R. Mechanisms of p53-dependent apoptosis. Biochem. Soc. Trans. 2001;29:684–688. doi: 10.1042/0300-5127:0290684. [DOI] [PubMed] [Google Scholar]
- Seale, P., Sabourin, L.A., Girgis-Gabardo, A., Mansouri, A., Gruss, P., Rudnicki, M.A. Pax7 is required for the specification of myogenic satellite cells. Cell. 2000;102:777–786. doi: 10.1016/s0092-8674(00)00066-0. [DOI] [PubMed] [Google Scholar]
- Seale, P., Ishibashi, J., Holterman, C., Rudnicki, M.A. Muscle satellite cell-specific genes identified by genetic profiling of MyoD-deficient myogenic cell. Dev. Biol. 2004;275:287–300. doi: 10.1016/j.ydbio.2004.07.034. [DOI] [PubMed] [Google Scholar]
- Siu, P.M., Alway, S.E. Id2 and p53 participate in apoptosis during unloading-induced muscle atrophy. Am. J. Physiol. Cell Physiol. 2005;288:C1058–C1073. doi: 10.1152/ajpcell.00495.2004. [DOI] [PubMed] [Google Scholar]
- Soddu, S., Blandino, G., Scardigli, R., Coen, S., Marchetti, A., Rizzo, M.G., Bossi, G., Cimino, L., Crescenzi, M., Sacchi, A. Interference with p53 protein inhibits hematopoietic and muscle differentiation. J. Cell Biol. 1996;134:193–204. doi: 10.1083/jcb.134.1.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturlan, S., Beinhauer, B.G., Oberhuber, G., Huang, L., Aasen, A.O., Rogy, M.A. In vivo gene transfer of murine interleukin-4 inhibits colon-26-mediated cancer cachexia in mice. Anticancer Res. 2002;22:2547–2554. [PubMed] [Google Scholar]
- Szalay, K., Razga, Z., Duda, E. TNF inhibits myogenesis and downregulates the expression of myogenic regulatory factors myoD and myogenin. Eur. J. Cell Biol. 1997;74:391–398. [PubMed] [Google Scholar]
- Tamir, Y., Bengal, E. p53 protein is activated during muscle differentiation and participates with MyoD in the transcription of muscle creatine kinase gene. Oncogene. 1998;17:347–356. doi: 10.1038/sj.onc.1201929. [DOI] [PubMed] [Google Scholar]
- Tanaka, Y., Eda, H., Tanaka, T., Udagawa, T., Ishikawa, T., Horii, I., Ishitsuka, H., Kataoka, T., Taguchi, T. Experimental cancer cachexia induced by transplantable colon 26 adenocarcinoma in mice. Cancer Res. 1990;50:2290–2295. [PubMed] [Google Scholar]
- Tisdale, M.J. Loss of skeletal muscle in cancer: Biochemical mechanisms. Front. Biosci. 2001;6:D164–D174. doi: 10.2741/tisdale. [DOI] [PubMed] [Google Scholar]
- Tisdale, M.J. Cachexia in cancer patients. Nat. Rev. Cancer. 2002;2:862–871. doi: 10.1038/nrc927. [DOI] [PubMed] [Google Scholar]
- Todorov, P.T., Field, W.N., Tisdale, M.J. Role of a proteolysis-inducing factor (PIF) in cachexia induced by a human melanoma (G361) Br. J. Cancer. 1999;80:1734–1737. doi: 10.1038/sj.bjc.6690590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyner, S.D., Venkatachalam, S., Choi, J., Jones, S., Ghebranious, N., Igelmann, H., Lu, X., Soron, G., Cooper, B., Brayton, C., et al. p53 mutant mice that display early ageing-associated phenotypes. Nature. 2002;415:45–53. doi: 10.1038/415045a. [DOI] [PubMed] [Google Scholar]
- Varela, I., Cadinanos, J., Pendas, A.M., Gutierrez-Fernandez, A., Folgueras, A.R., Sanchez, L.M., Zhou, Z., Rodriguez, F.J., Stewart, C.L., Vega, J.A., et al. Accelerated ageing in mice deficient in Zmpste24 protease is linked to p53 signalling activation. Nature. 2005;437:564–568. doi: 10.1038/nature04019. [DOI] [PubMed] [Google Scholar]
- Vogel, H., Lim, D.S., Karsenty, G., Finegold, M., Hasty, P. Deletion of Ku86 causes early onset of senescence in mice. Proc. Natl. Acad. Sci. 1999;96:10770–10775. doi: 10.1073/pnas.96.19.10770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vousden, K.H. p53: Death star. Cell. 2000;103:691–694. doi: 10.1016/s0092-8674(00)00171-9. [DOI] [PubMed] [Google Scholar]
- Vousden, K.H., Woude, G.F. The ins and outs of p53. Nat. Cell Biol. 2000;2:E178–E180. doi: 10.1038/35036427. [DOI] [PubMed] [Google Scholar]
- Walsh, K., Perlman, H. Cell cycle exit upon myogenic differentiation. Curr. Opin. Genet. Dev. 1997;7:597–602. doi: 10.1016/s0959-437x(97)80005-6. [DOI] [PubMed] [Google Scholar]
- Weintraub, H., Hauschka, S., Tapscott, S.J. The MCK enhancer contains a p53 responsive element. Proc. Natl. Acad. Sci. 1991;88:4570–4571. doi: 10.1073/pnas.88.11.4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welle, S., Brooks, A.I., Delehanty, J.M., Needler, N., Bhatt, K., Shah, B., Thornton, C.A. Skeletal muscle gene expression profiles in 20–29 year old and 65–71 year old women. Exp. Gerontol. 2004;39:369–377. doi: 10.1016/j.exger.2003.11.011. [DOI] [PubMed] [Google Scholar]
- White, J.D., Rachel, C., Vermeulen, R., Davies, M., Grounds, M.D. The role of p53 in vivo during skeletal muscle post-natal development and regeneration: Studies in p53 knockout mice. Int. J. Dev. Biol. 2002;46:577–582. [PubMed] [Google Scholar]
- Yarasheski, K.E., Bhasin, S., Sinha-Hikim, I., Pak-Loduca, J., Gonzalez-Cadavid, N.F. Serum myostatin-immunoreactive protein is increased in 60–92 year old women and men with muscle wasting. J. Nutr. Health Aging. 2002;6:343–348. [PubMed] [Google Scholar]
- Zammit, P.S., Golding, J.P., Nagata, Y., Hudon, V., Partridge, T.A., Beauchamp, J.R. Muscle satellite cells adopt divergent fates: A mechanism for self-renewal? J. Cell Biol. 2004;166:347–357. doi: 10.1083/jcb.200312007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmers, T.A., Davies, M.V., Koniaris, L.G., Haynes, P., Esquela, A.F., Tomkinson, K.N., McPherron, A.C., Wolfman, N.M., Lee, S.J. Induction of cachexia in mice by systemically administered myostatin. Science. 2002;296:1486–1488. doi: 10.1126/science.1069525. [DOI] [PubMed] [Google Scholar]