Abstract
The choice of self-renewal versus differentiation is a fundamental issue in stem cell and cancer biology. Neural progenitors of the Drosophila post-embryonic brain, larval neuroblasts (NBs), divide asymmetrically in a stem cell-like fashion to generate a self-renewing NB and a Ganglion Mother Cell (GMC), which divides terminally to produce two differentiating neuronal/glial daughters. Here we show that Aurora-A (AurA) acts as a tumor suppressor by suppressing NB self-renewal and promoting neuronal differentiation. In aurA loss-of-function mutants, supernumerary NBs are produced at the expense of neurons. AurA suppresses tumor formation by asymmetrically localizing atypical protein kinase C (aPKC), an NB proliferation factor. Numb, which also acts as a tumor suppressor in larval brains, is a major downstream target of AurA and aPKC. Notch activity is up-regulated in aurA and numb larval brains, and Notch signaling is necessary and sufficient to promote NB self-renewal and suppress differentiation in larval brains. Our data suggest that AurA, aPKC, Numb, and Notch function in a pathway that involved a series of negative genetic interactions. We have identified a novel mechanism for controlling the balance between self-renewal and neuronal differentiation during the asymmetric division of Drosophila larval NBs.
Keywords: Neuroblast, stem cells, asymmetric division, tumor suppressor, self-renewal
Drosophila larval NBs divide asymmetrically to produce a larger NB daughter that is capable of self-renewal, and a smaller Ganglion Mother Cell (GMC) daughter that divides once to produce progeny, which undergo terminal differentiation into two post-mitotic neurons/glia. Studies primarily in the embryonic ventral cord have revealed the mechanism of NB asymmetric divisions, which appears to be conserved between embryonic and larval NBs (for reviews, see Jan and Jan 2001; Betschinger and Knoblich 2004; Wodarz 2005; Yu et al. 2006). The asymmetric division of NBs involves the asymmetric localization and segregation of cell fate determinants Numb, Prospero (Pros), Brat, and their adaptor proteins Partner of Numb (Pon) and Miranda (Mira) into the basal GMC (Rhyu et al. 1994; Knoblich et al. 1995; Ikeshima-Kataoka et al. 1997; Shen et al. 1997; Lu et al. 1998). During NB asymmetric divisions, the mitotic spindle is oriented along an axis perpendicular to the epithelial layer and an asymmetric spindle is generated to give rise to two unequal-sized daughter cells with distinct cell fates. NB asymmetric divisions are controlled by an apically localized complex of proteins that include the Drosophila homologs of the conserved Par3 (Bazooka, Baz)/Par6 (DmPar6)/atypical protein kinase C(DaPKC) proteins (Kuchinke et al. 1998; Wodarz et al. 2000; Petronczki and Knoblich 2001), Inscuteable (Insc) (Kraut et al. 1996), and heterotrimeric G proteins Gαi (Schaefer et al. 2001; Yu et al. 2003) and their regulators Partner of Insc (Pins) (Yu et al. 2000), Locomotion defects (Loco) (Yu et al. 2005), and a Pins-interacting protein mushroom body defective (Mud) (Bowman et al. 2006; Izumi et al. 2006; Siller et al. 2006). The asymmetric localization of Gαi requires Gβ (Schaefer et al. 2001; Yu et al. 2003) and Gγ (Fuse et al. 2003) and its membrane localization requires Ric-8 (Hampoelz et al. 2005; Wang et al. 2005). Basal protein localization and segregation are mediated by apical proteins through cortically localized tumor suppressors, Discs large (Dlg) and Lethal (2) giant larvae (Lgl) (Ohshiro et al. 2000; Peng et al. 2000). Phosphorylation of Lgl by apically localized aPKC leads to Lgl inactivation, while nonphosphorylated Lgl at the basal cortex is active and allows the localization of basal proteins (Betschinger et al. 2003).
Drosophila larval NBs have emerged as a model system for studying stem cell self-renewal as larval NBs, unlike embryonic NBs, share many features of stem cells as they can undergo growth and self-renewal for extended periods and produce a large number of progeny (see reviews Jan and Jan 2001; Betschinger and Knoblich 2004; Wodarz 2005; Yu et al. 2006). There is increasing support for the idea that tumors can arise from tumor stem cells in which the normal control of self-renewal versus differentiation is disturbed (Passegue 2006). The regulation of self-renewal versus differentiation, associated with asymmetric cell divisions, is tightly controlled during Drosophila larval brain development. Perturbation of the asymmetric division process can lead to uncontrolled proliferation and aberrant terminal differentiation. When transplanted into wild-type adults, mutant larval brain tissue from pins, mira, numb, or pros can form malignant tumors that rapidly kill the host (Caussinus and Gonzalez 2005). Recent studies have also suggested that NBs utilize the asymmetric localization/segregation machinery to distribute “proliferation factors” to the NB daughter and “differentiation factors” to the GMC daughter during asymmetric cell division. aPKC, which is inherited by NBs, is necessary to promote NB self-renewal, and when a membrane targeted version is overexpressed and localized to the entire cortex, leads to NB overgrowth (Lee et al. 2006a). In contrast, “differentiation factors” such as tumor suppressors Brain tumor (Brat) and Prospero (Pros), which are asymmetrically segregated into the GMC daughter act to suppress self-renewal and promote differentiation in GMC. Loss of Brat or Pros results in massive production of NBs at the expense of neurons (Bello et al. 2006; Betschinger et al. 2006; Lee et al. 2006b). Delocalization of aPKC and Brat/Pros in lethal giant larvae (lgl) mutants can lead to tumor formation in larval brains (Betschinger et al. 2006; Lee et al. 2006a, b). However, the mechanism of self-renewal versus differentiation is still largely unknown. For example, aPKC is not required for proliferation in brat mutant and the downstream target(s) of aPKC for the regulation of NB proliferation remains unclear. It appears that additional novel mechanisms might act to suppress excessive NB self-renewal.
Here we show that Aurora-A (AurA) acts as a tumor suppressor by suppressing NB self-renewal and promoting neuronal differentiation during larval brain development. We show that AurA acts upstream of aPKC to regulate NB self-renewal. AurA is required for the asymmetric localization of aPKC and preventing it from localizing to the basal cortex. We also suggest that Numb is one major target of AurA and aPKC in regulating NB self-renewal. The overproliferation phenotype of aurA or numb mutants is due to the up-regulation of Notch activity, but appears to be largely independent of Brat and Pros. Thus, our data provides evidence for a novel pathway that involves AurA, aPKC, Numb, and Notch for the regulation of neuroblast (NB) self-renewal versus neuronal differentiation.
Results
AurA acts as a novel tumor suppressor in Drosophila larval brains
We screened a collection of pupal lethal mutants (L. Wang, J. Evans, H. Andrews, R. Beckstead, C.S. Thummel, and A. Bashirullah, in prep.) to isolate mutants that are defective in asymmetric cell division or self-renewal of larval NBs. Three recessive EMS-induced pupal lethal mutants (Fig. 1A; data not shown) belonging to a single complementation group were identified. In these mutants, supernumerary NBs that express both Mira and Inscuteable (Insc) {Fig. 1A [wild type], A′ [l(3)LL-8839 (aurA8839)]; data not shown} were produced. Genetic analyses and sequencing of these mutations, aurA8839 (Lys 377 to a stop codon), aurA14641 (Val 302 to Glutamate) and aurA17961 (Asp 344 to Asparagine), revealed that they each harbor either a single missense or a nonsense mutation in the conserved C-terminal kinase domain of aurora-A (aurA) (Fig. 1B). An available strong hypomorphic allele, aur87Ac-5, also shows a similar tumorous larval brain phenotype (data not shown). These alleles of aurA represent an allelic series, with aurA8839 showing the strongest phenotype. The aurA8839 tumor phenotype and lethality are fully rescued by an aurA-GFP transgene expressed with a NB-specific driver, worniu-Gal4 (wor-Gal4) (data not shown). In addition, AurA protein, which normally localizes on both centrosomes in wild-type metaphase NBs (Fig. 1C), is undetectable on the centrosomes of aurA8839 larval NBs (Fig. 1C′). These data indicate that loss of AurA function is responsible for the overproliferation phenotype, and that aurA8839 is either a strong hypomorphic or null allele.
Figure 1.
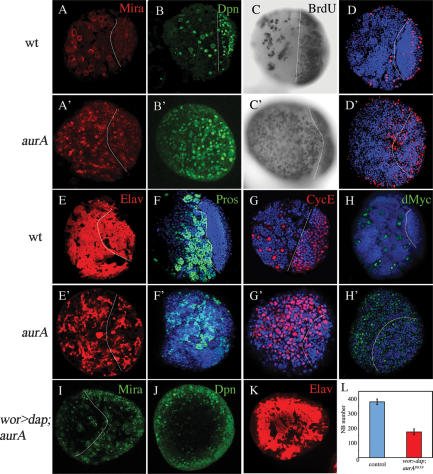
AurA is a novel tumor suppressor gene. (A,A′) Wild-type (A) and aurA8839 (A′) larval central brain regions at 96 h ALH were stained with the NB markers Mira (red) and Insc (green); DNA staining is in blue in all panels. (B) Schematic representation of AurA domains and mutated sites for aurA8839, aurA14641, and aurA17961. (C,C′) AurA is undetectable in aurA8839 mutant larval NBs. AurA in wild-type metaphase NBs (C) is localized to both centrosomes, but is undetectable in aurA8839 metaphase NBs (C′). (D) Quantification of wild-type and aurA8839 central brain NB numbers from 24- to 96-h-ALH. n = 20 per time point per genotype. (E,E′) aur8839 larval brain (E′) can grow to a massive size compared with wild-type brains (E). Arrows point to the brain lobes.
Loss of AurA leads to larval NB overgrowth
We quantified central brain NB numbers in aurA8839 mutant larvae from 24 h to 96 h after larval hatching (ALH). Larval NBs can be identified unambiguously by the expression of the markers Worniu, Deadpan (Dpn) and Mira (which is also transiently present in newborn GMCs) and the absence of the neuronal marker, Elav (Betschinger et al. 2006; Lee et al. 2006a, b). Wild-type larval central brains have 31 ± 6 NBs at 24 h ALH and the NB number increases to 89 ± 16 by 96 h ALH just prior to metamorphosis (Figs. 1D, 2A,B; Lee et al. 2006a). In aurA8839 mutant larvae, NBs start with a similar number at 24 h ALH. However, NB number increases dramatically to 371 ± 85 at 96 h ALH (Figs. 1D, 2A′,B′) and to more than a thousand at 120 h ALH during an extended larval life (data not shown). Before puparium formation, the aurA8839 larval brains (Fig. 1E′, arrow) can grow up to 10 times the wild-type size (Fig. 1E, arrow). BrdU incorporation experiments indicate a large increase in the number of S-phase cells in aurA mutant (Fig. 2C′) compared with wild-type brains (Fig. 2C). A significant increase of mitotic cells labeled by phospho-Histone H3 was also observed in aurA brains (Fig. 2D [wild type], D′). There is also a significant decrease in cells expressing the neuronal markers, Elav (Fig. 2E′) and nuclear Pros (Fig. 2F′), in aurA compared with wild-type brains (Fig. 2E,F), suggesting that the increase in NB-like cells occurs at the expense of differentiated neurons.
Figure 2.
AurA acts to suppress self-renewal and promote neuronal differentiation. (A–H′), Confocal single-scanning images of wild-type (A–G) and aurA mutant (A′–G′) larval brains at 96 h ALH stained with NB markers Mira (A,A′, in red), Dpn (B,B′, in green), BrdU incorporation and labeling (C,C′, in black), phospho-Histone H3 (D,D′, in red), neuronal marker Elav (E,E′, in red), nuclear Pros (F,F′, in green), and CycE (G,G′, in red). (H,H′), The number of dMyc-expressing cells is increased in aurA mutant (H′, in green) compared with wild type (H, in green). (I–L) NB overgrowth is dependent on CDK2/CycE activity. UAS-dap; aurA8839 (data not shown) and wor-Gal4, UAS-dap (wor > dap); aurA8839 brains at 96 h ALH were stained with Mira (I, in green), Dpn (J, in green), and Elav (K, in red). (L) Quantification of NB numbers in UAS-dap; aurA8839 (control) and wor > dap; aurA8839 brains at 96 h ALH. n = 20 for each genotype. In panels that contain both central brain and optic lobe, the approximate margin between the two regions is marked by a dashed line with the central brain region to the left and optic lobe to the right (optic lobe NBs are smaller than central brain NBs). Note that in aurA mutants, both regions are enlarged compared with wild type. Posterior brain regions are imaged in all panels.
Mammalian AurA is thought to be oncogenic, as overexpression of mammalian AurA causes cell transformation (Zhou et al. 1998; Meraldi et al. 2002). Human AurA phosphorylates P53, a tumor suppressor that regulates apoptosis, leading to its degradation and thus facilitating the oncogenic transformation of cells (Katayama et al. 2004). However, overexpression of AurA with wor-Gal4 did not affect NB number in the larval brains (data not shown). Apoptosis also appears not to be affected in aurA8839 mutant larval brains, as the numbers of cells labeled by Caspase-3 (Supplementary Fig. 1A′) or DIAP-1 (data not shown) in aurA8839 are similar to those of wild-type brains (Supplementary Fig. 1A; data not shown). These data suggest that AurA acts via a novel mechanism in regulating Drosophila larval NB proliferation.
CycE/CDK2 is an important cell cycle regulator and CycE levels were shown to be up-regulated in imaginal discs of tumor suppressor mutants including warts, salvador, and hippo (Vidal and Cagan 2006). We also observed a dramatic increase in the number of CycE-positive cells in aurA larval brains (Fig. 2G′) compared with wild type (Fig. 2G). To investigate whether this is a causal effect, we overexpressed Dacapo, a member of the P21/P27 family of CDK inhibitors (Lane et al. 1996), by wor-Gal4 in the aurA8839 mutant. The tumor phenotype of aurA8839 is largely suppressed (Fig. 2I–L) by overexpressing Dacapo. However, overexpression of CycE alone does not cause overgrowth of larval NBs (Betschinger et al. 2006; data not shown), suggesting that elevated levels of CycE are necessary but not sufficient to induce tumor formation. In addition to CycE, dMyc, another important growth factor, is also up-regulated in aurA8839 mutant (Fig. 2H′) compared with wild-type larval brains (Fig. 2H). aurA mutant also generate weak NB overproliferation phenotype in mitotic clones by the Mosaic Analysis with a Repressible Cell Marker (MARCM) system (Supplementary Fig. 2; Lee et al. 2000), presumably due to protein perdurance in the clone.
Asymmetric localization of aPKC and Numb requires AurA function
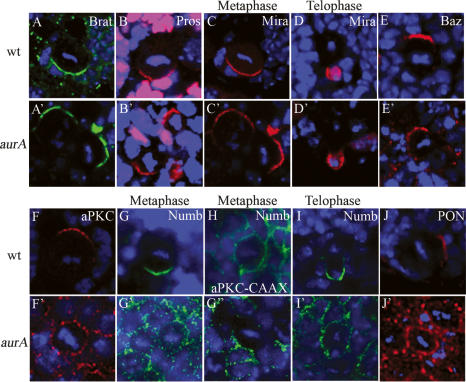
Tumor suppressors Brat (96%, n = 27, Fig. 3A′; wild type, 100%, n = 19, Fig. 3A) and Pros (96%, n = 25, Fig. 3B′; wild type, 100%, n = 16, Fig. 3B) remain asymmetrically localized in aurA8839 mutants. Consistently, asymmetric localization (94%, n = 36, Fig. 3C′; wild type, 100%, n = 20, Fig. 3C) and segregation (100%, n = 18, Fig. 3D′; wild type, 100%, n = 20, Fig. 3D) of Mira, an adaptor protein of Brat and Pros, is also unaffected in aurA mutants. Localization of Inscuteable (Insc) (97%, n = 32, Fig. 1A′; wild type, 100%, n = 21, Fig. 1A) and Bazooka (97%, n = 34, Fig. 3E′; wild type, 100%, n = 15, Fig. 3E), remain asymmetric in aurA mutants.
Figure 3.
AurA is required for asymmetric localization of aPKC and Numb/Pon and regulates spindle alignment. Brat (A,A′), Pros (B,B′), and Mira (C,C′), remain asymmetrically localized in both wild-type (A–C) and aurA8839 (A′–C ′) mutant metaphase NBs; DNA staining is in blue in all panels. Mira is exclusively segregated to the GMC at telophase in both wild-type (D) and aurA mutant (D′) NBs. (E,E′) Baz has a slight expansion to the cortex but remains asymmetrically localized in aurA8839 (E′) compared with wild-type (E) NBs. (F,F′) aPKC is delocalized to the entire cortex in aurA8839 NBs (F′) in contrast to a crescent seen in a wild-type NB (F). (G–G″) During metaphase, Numb is either seen as weakly scattered to the entire cortex (G′) or a strongly reduced crescent (G″) in NBs from 120-h-ALH larvae. Numb is delocalized to the entire cortex in 24.7% of NBs and Numb crescent is reduced in 28.8% NBs from 96-h-ALH larvae (n = 73). (H) Ectopic cortical localization of aPKC leads to delocalization of Numb. When aPKC-CAAX is overexpressed, Numb is delocalized in metaphase NBs. At telophase, Numb asymmetric segregation is defective in aurA8839 larval NBs (I′) compared with that in wild-type larval NBs (I). Pon (J′) is also often delocalized to the entire cortex in aurA8839 larval NBs, while they are seen as crescents in wild-type NBs (J).
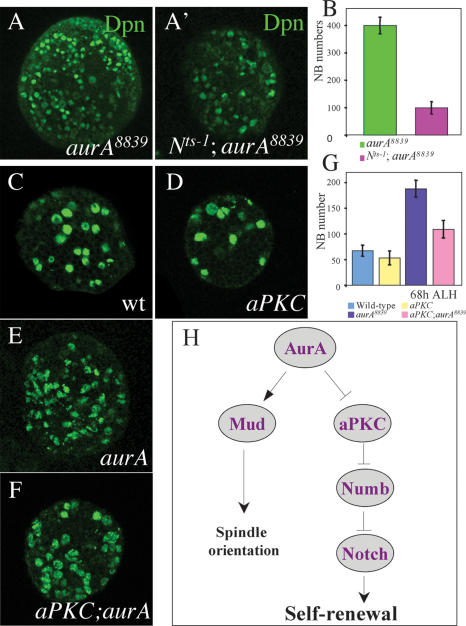
Apically localized aPKC is a NB proliferation factor that seems to act independently of Brat (Lee et al. 2006a, b). We tested whether AurA regulates NB self-renewal by modulating aPKC function. Indeed, in aurA8839 larval metaphase NBs, asymmetric localization of aPKC is strongly affected and it is delocalized to the entire cortex with punctuate staining (58%, n = 33 at 96 h ALH; Fig. 3F′). This phenotype is first observed at 48 h ALH (31.8%, n = 22) and is more severe in NBs from 120-h-ALH larvae (88.4% of delocalization, n = 69) in which most of maternal AurA had been presumably depleted. In this context, Mira remains largely asymmetrically localized (76.1% normal crescent, 15.2% reduced crescent, and 8.6% missing or cytoplsmic; n = 46), presumably because Baz and Insc remain asymmetrically localized. Overexpressed aPKC-CAAX (a membrane-targeted version of aPKC) causes ectopic localization of aPKC to both the apical and basal cortex and results in tumor formation (Lee et al. 2006a). Delocalization of aPKC in aurA mutants raises the interesting possibility that overproliferation of aurA may result from ectopic localization of aPKC on the cortex. To examine whether this is the case, we generated an aPKC;aurA double mutant. We found that aPKC significantly suppresses the aurA overproliferation phenotype at 68 h ALH (number of NB per brain lobe for each genotype: wild type, 67.2 ± 10.6; aPKC, 53.2 ± 13.5; aur8839, 187.9 ± 16.4; aPKC;aur8839 double mutant, 109 ± 17.1) (Fig. 7C–G, below). Thus, AurA may suppress self-renewal by preventing aPKC from localizing to the basal cortex.
Figure 7.
Genetic interactions of AurA with Notch and aPKC. (A,A′) Notchts-1 suppresses the aurA overproliferation phenotype. Brains double mutant for Notchts-1; aurA8839 do not display overgrowth of NBs (A′) as indicated by the Dpn staining compared with aurA8839 (A) at 68 h ALH at 29°C. (B) Quantification of NB numbers per brain lobe for Notchts-1; aurA8839 and aurA8839 alone. (C–G) aPKC significantly suppresses NB overgrowth phenotype in aurA. Larval brains of wild-type (C), aPKC (D), aurA8839 (E), and aPKC; aurA8839 double mutant (F) were stained with Dpn and number of NBs per brain lobe were quantified at 68 h ALH (G). n = 20. (H) A model showing a pathway composed of AurA, aPKC, Numb, and Notch in the regulation of larval NB self-renewal. AurA is also involved in spindle orientation through regulating Mud asymmetric localization. However, in our model the proposed role for aPKC is formal and our genetic analysis does not imply a direct molecular role of AurA on aPKC.
We also found that in aurA8839 larval metaphase NBs (at 96 h ALH), Numb levels are strongly reduced, and its asymmetric localization is also compromised (24.7% weakly cortical localization; 28.8% strongly reduced crescent, n = 73) (data not shown) compared with wild-type NBs (100% crescents, n = 16, Fig. 3G,H). Delocalization of Numb has been observed at 48 h ALH (32% weakly cortical or strongly reduced crescent, n = 25) in aurA8839, and these defects were more prominent (66.7% weakly cortical with punctate staining, Fig. 3G′; 29.8% strongly reduced crescent, Fig. 3G″; n = 57) in aurA8839 metaphase NBs from 120-h-ALH larvae. During anaphase/telophase, Numb is often missegregated to both daughter cells (57.9%, n = 19, Fig. 3I′) in aurA larval NBs, whereas segregation is exclusively to one daughter in wild type (100%, n = 12, Fig. 3I). These are consistent with the previous finding that AurA is required for the asymmetric localization of Numb in sensory organ precursors (SOPs) of the Drosophila peripheral nervous system (Berdnik and Knoblich 2002). Pon, an adaptor protein required for the asymmetric localization of Numb (Lu et al. 1998), was also often delocalized to the entire cortex of aurA metaphase NBs (34.1%, n = 41, 96 h ALH, Fig. 3J′; 58.6%, n = 29, 120 h ALH) in contrast to wild-type metaphase NBs (100% crescents, n = 11, Fig. 3J). We then tested to see whether delocalization of aPKC to the entire NB cortex might affect asymmetric localization of Numb. Under conditions in which aPKC-CAAX is overexpressed, where it localizes uniformly to the NB cortex and causes overproliferation (Lee et al. 2006a), Numb also delocalized to the entire cortex (86.2%, n = 29, Fig. 3H). These data further suggest that AurA acts upstream of aPKC in controlling NB self-renewal, probably by regulating Numb function.
AurA is required for proper spindle orientation
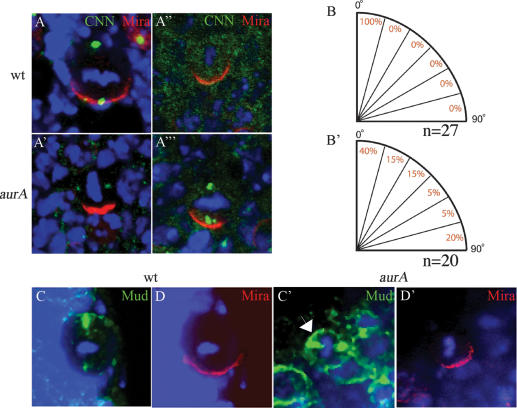
aurA has a function in centrosome separation and chromosome segregation (Glover et al. 1995; Giet et al. 2002). We investigated whether AurA might be required for proper orientation of the mitotic spindle. In wild-type metaphase NBs, the mitotic spindle, as judged by the positions of centrosomes, orients perpendicular to the Mira crescent (Fig. 4A,B). In 60% of aurA metaphase NBs, the spindle appears to be misaligned relative to the Mira crescent (Fig. 4A′,B′). Centrosomin (CNN) is absent in the majority of metaphase NBs and appears cytoplasmic (Fig. 4A″). The centrosome number in aurA mutants is also abnormal, and NBs with one (data not shown) to three (Fig. 4A′″) centrosomes can be observed. mud NBs also exhibit spindle orientation and centrosome organization defects (Bowman et al. 2006; Izumi et al. 2006; Siller et al. 2006) similar to those seen for aurA. Mud is required for proper spindle orientation in NBs and it is normally localized to both centrosomes and forms an apical crescent in wild-type NBs (Fig. 4C). Given that both AurA and Mud localize to the centrosomes and exhibit similar defects in spindle orientation, they may act together to regulate spindle orientation. Indeed, in aurA8839 larval NBs Mud cortical localization was no longer asymmetric, and was uniformly cortical (Fig. 4C′, arrow), while its centrosomal localization was unaffected (Fig. 4C′). These observations suggest that AurA may act through Mud to regulate spindle orientation in larval NBs.
Figure 4.
AurA controls mitotic spindle orientation through regulating asymmetric localization of Mud. (A,A′) The mitotic spindle is often misoriented relative to the Mira crescent in aurA17961 mutants (A′), in contrast to wild type (A) in which the spindle orientation inferred by two CNN-labeled centrosomes is always aligned perpendicular to the Mira crescent. CNN staining is often lost (A″) or strongly reduced (A′) in aurA8839 mutant NBs. In aurA mutants, one (data not shown) or three (A′″) centrosomes can also be observed. The quantification of the distribution of spindle orientation in wild-type (B) and aurA17961 mutant (B′) NBs based on NBs that have two CNN+ centrosomes. (C–D′) AurA is required for Mud asymmetric cortical localization but not its centrosomal localization. (C) In wild-type metaphase NBs, Mud is asymmetrically localized as a cortical crescent at the apical cortex as well as to both centrosomes. (C′) In contrast, in aurA metaphase NBs, Mud is delocalized to almost the entire cortex but remains localized to both centrosomes (arrow). (D,D′) Mira is asymmetrically localized in both wild-type NBs (D) and aurA mutant (D′).
Numb acts as a tumor suppressor that functions downstream from AurA
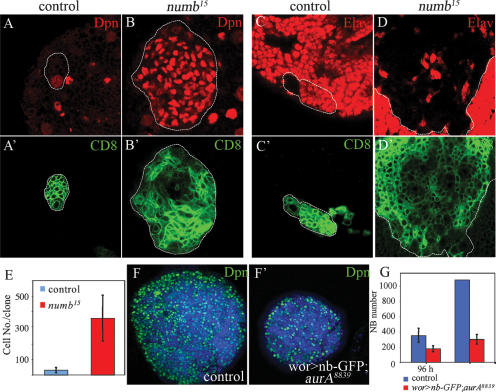
To examine whether Numb might be a downstream target of AurA and aPKC in regulating NB self-renewal, we generated numb15 NB MARCM clones in larval brains. numb15 clones contained a greater number of cells compared with wild-type clones (Fig. 5E, numb15, 342 ± 82; wild type, 38 ± 18) and most of them express Dpn (Fig. 5B) and Mira (Supplementary Fig. 3B) in contrast to wild-type NB clones in which only a single cell expresses Dpn (Fig. 5A) and Mira (Supplementary Fig. 3A). numb15 clones contain very few neurons expressing Elav (Fig. 5C [wild type], D) and nuclear Pros (Supplementary Fig. 3C,D [wild type]). Thus, similar to AurA, Numb acts as a tumor suppressor to suppress self-renewal and promote differentiation during larval brain development. Ectopic expression of Numb significantly suppresses the aurA overproliferation phenotype (178 ± 40 NBs per brain lobe at 96 h ALH and 302 ± 63 NBs per brain lobe at 120 h ALH, Fig. 5F′,G) compared with control aurA8839 mutant (352 ± 94 NBs per brain lobe at 96 h ALH, and >1000 at 120 h ALH, Fig. 5F,G), suggesting that Numb acts downstream from AurA to regulate NB self-renewal. In this case, Numb expressed at high levels remains mislocalized on the cortex in a significant proportion of aurA mutant NBs (25%, n = 24 at 96 h ALH) (data not shown), indicating that the level of Numb has probably exceeded the threshold necessary to suppress proliferation in a proportion of these cells.
Figure 5.
AurA acts upstream of Numb to suppress tumor formation. (A–E) Numb acts as a tumor suppressor during larval brain development. (A′,B′) NB MARCM clones marked by CD8-GFP in third instar larval brains. Most cells in numb15 clones express Dpn (B) but very few cells express Elav (D), whereas in wild-type clones usually only a single cell (the NB) expresses Dpn (A) and most cells express Elav (C). (E) Quantification of the number of cells per clone in wild-type and numb15 clones. (F–G) Overexpression of Numb (nb)-GFP suppresses the NB overproliferation phenotype in aurA8839 brains. Overexpression of Numb-GFP using wor-Gal4 in aurA8839 mutant background (wor > nb-GFP;aurA8839) reduces brain size (F′) compared with the control wor-Gal4, aurA8839 (F) at 120 h ALH, and the NB number is also significantly reduced (G; n = 20 per genotype per time point).
The role of Notch in NB proliferation
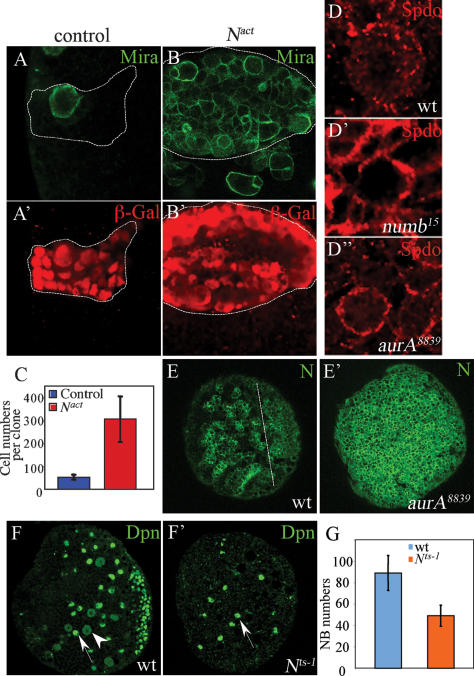
There is increasing evidence that Notch—a transmembrane receptor that is cleaved to release its intracellular domain, which directly participates in transcriptional regulation (Louvi and Artavanis-Tsakonas 2006)—is involved in the control of proliferation versus differentiation (Go et al. 1998; Androutsellis-Theotokis et al. 2006; Ferres-Marco et al. 2006; Micchelli and Perrimon 2006; Ohlstein and Spradling 2006). Numb can antagonize Notch signaling to specify distinct sibling cell fates during asymmetric divisions of various progenitor cells (Rhyu et al. 1994; Spana and Doe 1996; Skeath and Doe 1998). Overexpression of a Notch dominant-active form (Nact, the intracellular domain of Notch) in NB clones also leads to the overproliferation of larval NBs (Fig. 6B,B′). These mutant clones (O/E Nact, 306 ± 99 cells per clone) (Fig. 6C) were of much larger size than control clones (53 ± 11 cells per clone) (Fig. 6C) and contained mostly NBs that expressed Mira (Fig. 6A [control], B) and Dpn (Supplementary Fig. 4A [control], B), and were essentially devoid of cells expressing Elav (Supplementary Fig. 4C [control], D). A previous study (Almeida and Bray 2005) suggested that Notch does not play a role in controlling NB proliferation in larval ventral nerve cord (VNC). The discrepancies in our findings may be due to differences in which proliferation of VNC and central brain NBs are regulated. To test whether Notch might be hyperactivated in aurA mutants, we examined the membrane localization of Sanpodo (Spdo), which is required to activate Notch but is inhibited by Numb during GMC and SOP asymmetric divisions (O’Connor-Giles and Skeath 2003; Le Borgne et al. 2005). In wild-type interphase larval NBs, Spdo displayed a weak cortical staining and in punctate structures throughout the cytoplasm (100%, n = 38; Fig. 6D). In both numb15 (93%, n = 15; Fig. 6D′) and aurA8839 (57%, n = 21; Fig. 6D″) larval NBs, Spdo was localized strongly to the cortex. In addition, aurA larval NBs expressed elevated levels of Notch (Fig. 6E′) compared with wild type (Fig. 6E) using antibodies against Notch intracellular domain or extracellular domain (data not shown). The number of cells expressing CycE and dMyc are drastically increased when Nact is overexpressed (Supplementary Fig. 4E–G′ and legend).
Figure 6.
Notch activity is up-regulated in aurA mutants and Notch is necessary and sufficient for NB self-renewal. (A–B′) Overexpression of activated Notch (the intracellular domain of Notch, Nact) induced NB overgrowth and tumor formation. (A′,B′) Mitotic clones were marked by β-galactosidase (β-Gal). In Nact clones, most cells express Mira (B), whereas only one cell (NB) expresses Mira in control clone (A). (C) Quantification of the number of cells per clone in control and activated Notch clones. (D,D″) Spdo localization in wild-type (D), numb15 (D′), and aurA8839 (D″) mutant larval NBs. (D) In wild-type NBs, Spdo is mostly observed as weak punctate structures on the cortex as well as throughout the cytoplasm. In numb15 (D′) and aurA8839 (D″) mutants, Spdo is almost exclusively localized at the cortex. (E,E′) Notch levels indicated by an antibody against its intracellular domain are up-regulated in aurA mutant brains (E′) compared with wild type (E). (F,F′) Notch is a critical proliferation factor for larval NBs. Notchts-1 third instar larval brains (68 h ALH, 29°C from larval hatching) contain fewer NBs, marked by Dpn in green (F′) compared with wild type (F). Note that wild-type larval NBs exhibit either nuclear (arrow) or cytosolic (arrowhead) Dpn signals, whereas most of the Notchts-1 larval NBs exhibit nuclear (arrow) but not cytosolic Dpn. (G) Quantification of NB numbers per brain lobe for wild type and Notchts-1. Genotypes are hs-FLP; actin-FRT-y+-FRT-Gal4, UAS-nlsLacZ(A,A′), and hs-FLP; actin-FRT-y+-FRT-Gal4, UAS-nlsLacZ/UAS-Nact (B,B′).
A Notch temperature-sensitive (ts) mutant (Nts-1), when shifted to the restrictive temperature from the first instar larval stage onward, produced far fewer NBs in third instar larval brains (49 ± 9.8, n = 27; Fig. 6F′,G) compared with wild type (89 ± 16.3, n = 10; Fig. 6F,G). Moreover, only 1.1% of the NBs are dividing in the mutant brains (n = 276; Fig. 6F′), as evidenced by cytoplasmic Dpn, compared with 18% in wild type (n = 293; Fig. 6F, arrowhead). The number of CycE-expressing NBs was also drastically decreased in the Notchts-1 mutant (Supplementary Fig. 4H′) compared with wild-type brains (Supplementary Fig. 4H). Thus, Notch is necessary and sufficient to promote NB self-renewal during larval brain development. To further investigate whether AurA acts through regulation of Notch, we analyzed whether loss-of-Notch could suppress the aurA overgrowth phenotype. In double-mutant Nts-1;aurA8839 larval brains, the NB overgrowth phenotype is largely suppressed (Fig. 7A′,B, 99 ± 22.9 NBs per brain lobe) compared with aurA8839 (Fig. 5A,B; 400 ± 30.5 NBs per brain lobe), suggesting that AurA acts upstream of Notch to control NB self-renewal.
Discussion
Here we described a novel function of AurA as a tumor suppressor involved in the regulation of larval NB self-renewal. When aurA function is compromised, mutant NBs acquire some features of cancer stem cells. They divide to generate a large number of daughter cells capable of self-renewal. This excessive self-renewal occurs at the expense of neuronal differentiation, suggesting that the normally asymmetric NB divisions have been altered such that the mutant NBs can divide symmetrically to generate two NB-like daughters. Cell cycle regulator CycE and cell growth factor dMyc are expressed in most of these tumor-like cells. We show that up-regulation of CycE is required for aurA overgrowth phenotype. AurA also regulates proper orientation of the mitotic spindle probably by controlling asymmetric localization of Mud. Both proteins are localized to centrosomes and are required for centrosome function. Centrosome abnormality and chromosome segregation defects in aurA could lead to aneuploidy, and many cancer cells exhibit centrosome defects and chromosome instability (Badano et al. 2005). Mammalian AurA when overexpressed can be oncogenic. However, future studies on its possible role as a tumor suppressor will be particularly interesting.
Our data suggest that aurA negatively regulates aPKC function to regulate NB self-renewal. aPKC appears to act as a NB proliferation factor since overexpression of a modified membrane-targeted version, aPKC-CAAX, which exhibits ectopic cortical localization throughout the NB cortex, leads to overproliferation and tumor formation (Lee et al. 2006a), similar to loss of aurA. AurA is required for the asymmetric localization of aPKC and restrict aPKC to the cortical region associated with the future NB daughter and loss of aurA results in delocalization of aPKC to the entire cortex. Consistent with and supporting this notion, we have shown that loss of aPKC can suppress, albeit partially, the aurA mutant overgrowth phenotype.
In contrast to the well-studied role of Numb as a cell fate determinant during asymmetric divisions of embryonic GMCs, SOPs, or muscle progenitors (Rhyu et al. 1994; Spana and Doe 1996; Carmena et al. 1998), a role for Numb during NB asymmetric divisions has not been described. We show here that Numb also acts as a tumor suppressor in Drosophila larval brains, and that Numb is a key downstream target of AurA and aPKC in the regulation of NB self-renewal. In both aurA mutant NBs or NBs overexpressing aPKC-CAAX, the asymmetric localization of Numb is compromised and the resultant overgrowth phenotype is consistent with that of numb loss-of-function. numb and aurA mutant NBs also share several common features including excessive self-renewal at the expense of neuronal differentiation as well as the membrane enrichment of Spdo, a positive regulator of Notch signaling. Our data suggest that AurA positively regulates Numb function. Genetic analysis is consistent with the notion that this is achieved through the negative regulation of aPKC that in turn negatively regulates Numb.
Numb is known to be a negative regulator of Notch signaling (Rhyu et al. 1994; Spana and Doe 1996; Carmena et al. 1998). Our findings indicate that Notch is necessary and sufficient for promoting larval NB proliferation and suppressing neuronal differentiation. Our genetic epistasis studies suggest that an AurA–aPKC–Numb–Notch genetic hierarchy acts to regulate self-renewal of Drosophila neural progenitor cells (Fig. 7H). During a wild-type larval NB asymmetric division, aurA acts to negatively regulate aPKC and restrict its localization to the cortical region associated with the future NB daughter; aPKC negatively regulates Numb and ensures that its localization/activity is restricted to the future GMC where Numb acts to antagonize Notch. The net effect is that Notch is asymmetrically activated in the NB daughter where it acts to promote self-renewal and suppress differentiation. Although our data suggest that aurA acts through the aPKC/Numb/Notch pathway, given the partial suppression seen in the double mutants aPKC;aurA and Notchts-1;aurA, we cannot exclude the possibility that additional mechanisms may be involved.
Materials and methods
Fly strains
Fly strains used were aurA8839 [l(3)LL8839], aurA14641 [l(3)LL14641], aurA17961 [l(3)LL17961], worniu-Gal4, and brat11 (C.Q. Doe); aPKC (A. Wodarz and D. Bilder); UAS-aurA-GFP (J. Knoblich); UAS-CycE (P. O’Farrells); UAS-Dacapo (P. O’Farrells); UAS-aPKC-CAAX (C.Q. Doe); numb15 (J. Knoblich); Notchts-1, UAS-Numb-GFP (Y.N. Jan); UAS-actNotch (S. Artavanis-Tsakonas); yw, hs-Flp; act-FRT-y+-FRT-Gal4, UAS-LacZ(nls) (J. Knoblich); and Tub-Gal80ts and UAS-Sanpodo (J. Skeath).
Clonal analysis
To generate NB clones, 24-h-ALH larvae were heat-shocked at 37°C for 90 min and further aged for 3 d at 25°C.
Immunohistochemistry
For larval brain immunofluorescence stainings, larvae were dissected in PBS and fixed for 20 min in 4% formaldehyde, and processed similar to embryo stainings as described (Yu et al. 2000). Antibodies used were rabbit anti-Insc (1:1000), mouse anti-Mira (1:50, F. Matsuzaki), guinea pig anti-Dpn (1:1000, J. Skeath), mouse anti-Worniu (1:500), rabbit anti-phospho-Histone H3 (1:1000, Sigma), rat anti-Elav (1:10, DSHB), mouse anti-CycE (1:10, H. Richadson), mouse anti-dMyc (1:5, B. Edgar), rabbit anti-AurA (1:100, D. Glover), mouse anti-Pros (1:10, DSHB), rabbit anti-Brat (1:100, J. Knoblich), rat anti-Brat (1:100, R.P. Wharton), guinea pig anti-Numb (1:1000, J. Skeath), rabbit anti-Pon (1:500, Y.N. Jan), rabbit anti-Mud (1:200, F. Matusuzaki), rabbit anti-GFP (1:500, Molecular Probes), guinea pig anti-Spdo (1:1000, J. Skeath), rabbit anti-cleaved-Caspase-3 (1:75, Cell Signaling Technology), mouse anti-BrdU (1:20, Roche), mouse anti-Diap-1 (1:200, B. Hay), rabbit anti-CNN (1:5000, E. Schejter), and rabbit and mouse anti-β-Gal (1:1000, Promega).
BrdU labeling
Proliferating cells within whole brains were detected as previously described (Ceron et al. 2001). Dissected larval tissue was given a 40-min pulse of 37.5 μg/mL BrdU in Shields and Sang 3M insect medium. Tissue was then fixed for 15 min in 3.7% formaldehyde, and DNA denatured with 2N HCl for 40 min, before washing in PBS and incubating with anti-BrdU.
Acknowledgments
We thank S. Artavanis-Tsakonas, D. Bilder, C.Q. Doe, B. Edgar, P. O’Farrells, D. Glover, B. Hay, Y.N. Jan, T. Kaufman, J.A. Knoblich, F. Matsuzaki, H. Richadson, E. Schejter, J. Skeath, G. Struhl, Developmental Studies Hybridoma Bank (University of Iowa), and the Bloomington Stock Center for generously providing antibodies and fly stocks. We thank A. Gould for generously providing the MARCM driver lines, and H.W. thanks D. Stainier and Y.N. Jan for providing confocal microscopes, S. Younger for discussion, and M. Eddison for critical reading and comments on the manuscript. F.Y. and H.W. especially thank Y.N. Jan’s generous support. This work was supported by an NIH grant (to U.H.), Singapore Millennium Foundation (F.Y.), and Temasek Life Sciences Laboratory (F.Y. and W.C.).
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1487506
References
- Almeida, M.S., Bray, S.J. Regulation of post-embryonic neuroblasts by Drosophila Grainyhead. Mech. Dev. 2005;122:1282–1293. doi: 10.1016/j.mod.2005.08.004. [DOI] [PubMed] [Google Scholar]
- Androutsellis-Theotokis, A., Leker, R.R., Soldner, F., Hoeppner, D.J., Ravin, R., Poser, S.W., Rueger, M.A., Bae, S.K., Kittappa, R., McKay, R.D. Notch signalling regulates stem cell numbers in vitro and in vivo. Nature. 2006;442:823–826. doi: 10.1038/nature04940. [DOI] [PubMed] [Google Scholar]
- Badano, J.L., Teslovich, T.M., Katsanis, N. The centrosome in human genetic disease. Nat. Rev. Genet. 2005;6:194–205. doi: 10.1038/nrg1557. [DOI] [PubMed] [Google Scholar]
- Bello, B., Reichert, H., Hirth, F. The brain tumor gene negatively regulates neural progenitor cell proliferation in the larval central brain of Drosophila. Development. 2006;133:2639–2648. doi: 10.1242/dev.02429. [DOI] [PubMed] [Google Scholar]
- Berdnik, D., Knoblich, J.A. Drosophila Aurora-A is required for centrosome maturation and actin-dependent asymmetric protein localization during mitosis. Curr. Biol. 2002;12:640–647. doi: 10.1016/s0960-9822(02)00766-2. [DOI] [PubMed] [Google Scholar]
- Betschinger, J., Knoblich, J.A. Dare to be different: Asymmetric cell division in Drosophila, C. elegans and vertebrates. Curr. Biol. 2004;14:R674–R685. doi: 10.1016/j.cub.2004.08.017. [DOI] [PubMed] [Google Scholar]
- Betschinger, J., Mechtler, K., Knoblich, J.A. The Par complex directs asymmetric cell division by phosphorylating the cytoskeletal protein Lgl. Nature. 2003;422:326–330. doi: 10.1038/nature01486. [DOI] [PubMed] [Google Scholar]
- Betschinger, J., Mechtler, K., Knoblich, J.A. Asymmetric segregation of the tumor suppressor brat regulates self-renewal in Drosophila neural stem cells. Cell. 2006;124:1241–1253. doi: 10.1016/j.cell.2006.01.038. [DOI] [PubMed] [Google Scholar]
- Bowman, S.K., Neumuller, R.A., Novatchkova, M., Du, Q., Knoblich, J.A. The Drosophila NuMA Homolog Mud regulates spindle orientation in asymmetric cell division. Dev. Cell. 2006;10:731–742. doi: 10.1016/j.devcel.2006.05.005. [DOI] [PubMed] [Google Scholar]
- Carmena, A., Murugasu-Oei, B., Menon, D., Jimenez, F., Chia, W. Inscuteable and numb mediate asymmetric muscle progenitor cell divisions during Drosophila myogenesis. Genes & Dev. 1998;12:304–315. doi: 10.1101/gad.12.3.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caussinus, E., Gonzalez, C. Induction of tumor growth by altered stem-cell asymmetric division in Drosophila melanogaster. Nat. Genet. 2005;37:1125–1129. doi: 10.1038/ng1632. [DOI] [PubMed] [Google Scholar]
- Ceron, J., Gonzalez, C., Tejedor, F.J. Patterns of cell division and expression of asymmetric cell fate determinants in postembryonic neuroblast lineages of Drosophila. Dev. Biol. 2001;230:125–138. doi: 10.1006/dbio.2000.0110. [DOI] [PubMed] [Google Scholar]
- Ferres-Marco, D., Gutierrez-Garcia, I., Vallejo, D.M., Bolivar, J., Gutierrez-Avino, F.J., Dominguez, M. Epigenetic silencers and Notch collaborate to promote malignant tumours by Rb silencing. Nature. 2006;439:430–436. doi: 10.1038/nature04376. [DOI] [PubMed] [Google Scholar]
- Fuse, N., Hisata, K., Katzen, A.L., Matsuzaki, F. Heterotrimeric G proteins regulate daughter cell size asymmetry in Drosophila neuroblast divisions. Curr. Biol. 2003;13:947–954. doi: 10.1016/s0960-9822(03)00334-8. [DOI] [PubMed] [Google Scholar]
- Giet, R., McLean, D., Descamps, S., Lee, M.J., Raff, J.W., Prigent, C., Glover, D.M. DrosophilaAurora A kinase is required to localize D-TACC to centrosomes and to regulate astral microtubules. J. Cell Biol. 2002;156:437–451. doi: 10.1083/jcb.200108135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover, D.M., Leibowitz, M.H., McLean, D.A., Parry, H. Mutations in aurora prevent centrosome separation leading to the formation of monopolar spindles. Cell. 1995;81:95–105. doi: 10.1016/0092-8674(95)90374-7. [DOI] [PubMed] [Google Scholar]
- Go, M.J., Eastman, D.S., Artavanis-Tsakonas, S. Cell proliferation control by Notch signaling in Drosophila development. Development. 1998;125:2031–2040. doi: 10.1242/dev.125.11.2031. [DOI] [PubMed] [Google Scholar]
- Hampoelz, B., Hoeller, O., Bowman, S.K., Dunican, D., Knoblich, J.A. Drosophila Ric-8 is essential for plasma-membrane localization of heterotrimeric G proteins. Nat. Cell Biol. 2005;7:1099–1105. doi: 10.1038/ncb1318. [DOI] [PubMed] [Google Scholar]
- Ikeshima-Kataoka, H., Skeath, J.B., Nabeshima, Y., Doe, C.Q., Matsuzaki, F. Miranda directs Prospero to a daughter cell during Drosophila asymmetric divisions. Nature. 1997;390:625–629. doi: 10.1038/37641. [DOI] [PubMed] [Google Scholar]
- Izumi, Y., Ohta, N., Hisata, K., Raabe, T., Matsuzaki, F. Drosophila Pins-binding protein Mud regulates spindle-polarity coupling and centrosome organization. Nat. Cell Biol. 2006;8:586–593. doi: 10.1038/ncb1409. [DOI] [PubMed] [Google Scholar]
- Jan, Y.N., Jan, L.Y. Asymmetric cell division in the Drosophila nervous system. Nat. Rev. Neurosci. 2001;2:772–779. doi: 10.1038/35097516. [DOI] [PubMed] [Google Scholar]
- Katayama, H., Sasai, K., Kawai, H., Yuan, Z.M., Bondaruk, J., Suzuki, F., Fujii, S., Arlinghaus, R.B., Czerniak, B.A., Sen, S. Phosphorylation by aurora kinase A induces Mdm2-mediated destabilization and inhibition of p53. Nat. Genet. 2004;36:55–62. doi: 10.1038/ng1279. [DOI] [PubMed] [Google Scholar]
- Knoblich, J.A., Jan, L.Y., Jan, Y.N. Asymmetric segregation of Numb and Prospero during cell division. Nature. 1995;377:624–627. doi: 10.1038/377624a0. [DOI] [PubMed] [Google Scholar]
- Kraut, R., Chia, W., Jan, L.Y., Jan, Y.N., Knoblich, J.A. Role of inscuteable in orienting asymmetric cell divisions in Drosophila. Nature. 1996;383:50–55. doi: 10.1038/383050a0. [DOI] [PubMed] [Google Scholar]
- Kuchinke, U., Grawe, F., Knust, E. Control of spindle orientation in Drosophila by the Par-3-related PDZ-domain protein Bazooka. Curr. Biol. 1998;8:1357–1365. doi: 10.1016/s0960-9822(98)00016-5. [DOI] [PubMed] [Google Scholar]
- Lane, M.E., Sauer, K., Wallace, K., Jan, Y.N., Lehner, C.F., Vaessin, H. Dacapo, a cyclin-dependent kinase inhibitor, stops cell proliferation during Drosophila development. Cell. 1996;87:1225–1235. doi: 10.1016/s0092-8674(00)81818-8. [DOI] [PubMed] [Google Scholar]
- Le Borgne, R., Bardin, A., Schweisguth, F. The roles of receptor and ligand endocytosis in regulating Notch signaling. Development. 2005;132:1751–1762. doi: 10.1242/dev.01789. [DOI] [PubMed] [Google Scholar]
- Lee, T., Winter, C., Marticke, S.S., Lee, A., Luo, L. Essential roles of Drosophila RhoA in the regulation of neuroblast proliferation and dendritic but not axonal morphogenesis. Neuron. 2000;25:307–316. doi: 10.1016/s0896-6273(00)80896-x. [DOI] [PubMed] [Google Scholar]
- Lee, C.Y., Robinson, K.J., Doe, C.Q. Lgl, Pins and aPKC regulate neuroblast self-renewal versus differentiation. Nature. 2006a;439:594–598. doi: 10.1038/nature04299. [DOI] [PubMed] [Google Scholar]
- Lee, C.Y., Wilkinson, B.D., Siegrist, S.E., Wharton, R.P., Doe, C.Q. Brat is a miranda cargo protein that promotes neuronal differentiation and inhibits neuroblast self-renewal. Dev. Cell. 2006b;10:441–449. doi: 10.1016/j.devcel.2006.01.017. [DOI] [PubMed] [Google Scholar]
- Louvi, A., Artavanis-Tsakonas, S. Notch signalling in vertebrate neural development. Nat. Rev. Neurosci. 2006;7:93–102. doi: 10.1038/nrn1847. [DOI] [PubMed] [Google Scholar]
- Lu, B., Rothenberg, M., Jan, L.Y., Jan, Y.N. Partner of Numb colocalizes with Numb during mitosis and directs Numb asymmetric localization in Drosophila neural and muscle progenitors. Cell. 1998;95:225–235. doi: 10.1016/s0092-8674(00)81753-5. [DOI] [PubMed] [Google Scholar]
- Meraldi, P., Honda, R., Nigg, E.A. Aurora-A overexpression reveals tetraploidization as a major route to centrosome amplification in p53−/− cells. EMBO J. 2002;21:483–492. doi: 10.1093/emboj/21.4.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micchelli, C.A., Perrimon, N. Evidence that stem cells reside in the adult Drosophila midgut epithelium. Nature. 2006;439:475–479. doi: 10.1038/nature04371. [DOI] [PubMed] [Google Scholar]
- O’Connor-Giles, K.M., Skeath, J.B. Numb inhibits membrane localization of Sanpodo, a four-pass transmembrane protein, to promote asymmetric divisions in Drosophila. Dev. Cell. 2003;5:231–243. doi: 10.1016/s1534-5807(03)00226-0. [DOI] [PubMed] [Google Scholar]
- Ohlstein, B., Spradling, A. The adult Drosophila posterior midgut is maintained by pluripotent stem cells. Nature. 2006;439:470–474. doi: 10.1038/nature04333. [DOI] [PubMed] [Google Scholar]
- Ohshiro, T., Yagami, T., Zhang, C., Matsuzaki, F. Role of cortical tumour-suppressor proteins in asymmetric division of Drosophila neuroblast. Nature. 2000;408:593–596. doi: 10.1038/35046087. [DOI] [PubMed] [Google Scholar]
- Passegue, E. Cancer biology: A game of subversion. Nature. 2006;442:754–755. doi: 10.1038/442754a. [DOI] [PubMed] [Google Scholar]
- Peng, C.Y., Manning, L., Albertson, R., Doe, C.Q. The tumour-suppressor genes lgl and dlg regulate basal protein targeting in Drosophila neuroblasts. Nature. 2000;408:596–600. doi: 10.1038/35046094. [DOI] [PubMed] [Google Scholar]
- Petronczki, M., Knoblich, J.A. DmPAR-6 directs epithelial polarity and asymmetric cell division of neuroblasts in Drosophila. Nat. Cell Biol. 2001;3:43–49. doi: 10.1038/35050550. [DOI] [PubMed] [Google Scholar]
- Rhyu, M.S., Jan, L.Y., Jan, Y.N. Asymmetric distribution of numb protein during division of the sensory organ precursor cell confers distinct fates to daughter cells. Cell. 1994;76:477–491. doi: 10.1016/0092-8674(94)90112-0. [DOI] [PubMed] [Google Scholar]
- Schaefer, M., Petronczki, M., Dorner, D., Forte, M., Knoblich, J.A. Heterotrimeric G proteins direct two modes of asymmetric cell division in the Drosophila nervous system. Cell. 2001;107:183–194. doi: 10.1016/s0092-8674(01)00521-9. [DOI] [PubMed] [Google Scholar]
- Shen, C.P., Jan, L.Y., Jan, Y.N. Miranda is required for the asymmetric localization of Prospero during mitosis in Drosophila. Cell. 1997;90:449–458. doi: 10.1016/s0092-8674(00)80505-x. [DOI] [PubMed] [Google Scholar]
- Siller, K.H., Cabernard, C., Doe, C.Q. The NuMA-related Mud protein binds Pins and regulates spindle orientation in Drosophila neuroblasts. Nat. Cell Biol. 2006;8:594–600. doi: 10.1038/ncb1412. [DOI] [PubMed] [Google Scholar]
- Skeath, J.B., Doe, C.Q. Sanpodo and Notch act in opposition to Numb to distinguish sibling neuron fates in the Drosophila CNS. Development. 1998;125:1857–1865. doi: 10.1242/dev.125.10.1857. [DOI] [PubMed] [Google Scholar]
- Spana, E.P., Doe, C.Q. Numb antagonizes Notch signaling to specify sibling neuron cell fates. Neuron. 1996;17:21–26. doi: 10.1016/s0896-6273(00)80277-9. [DOI] [PubMed] [Google Scholar]
- Vidal, M., Cagan, R.L. Drosophila models for cancer research. Curr. Opin. Genet. Dev. 2006;16:10–16. doi: 10.1016/j.gde.2005.12.004. [DOI] [PubMed] [Google Scholar]
- Wang, H., Ng, K.H., Qian, H., Siderovski, D.P., Chia, W., Yu, F. Ric-8 controls Drosophila neural progenitor asymmetric division by regulating heterotrimeric G proteins. Nat. Cell Biol. 2005;7:1091–1098. doi: 10.1038/ncb1317. [DOI] [PubMed] [Google Scholar]
- Wodarz, A. Molecular control of cell polarity and asymmetric cell division in Drosophila neuroblasts. Curr. Opin. Cell Biol. 2005;17:475–481. doi: 10.1016/j.ceb.2005.08.005. [DOI] [PubMed] [Google Scholar]
- Wodarz, A., Ramrath, A., Grimm, A., Knust, E. Drosophila atypical protein kinase C associates with Bazooka and controls polarity of epithelia and neuroblasts. J. Cell Biol. 2000;150:1361–1374. doi: 10.1083/jcb.150.6.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, F., Morin, X., Cai, Y., Yang, X., Chia, W. Analysis of partner of inscuteable, a novel player of Drosophila asymmetric divisions, reveals two distinct steps in inscuteable apical localization. Cell. 2000;100:399–409. doi: 10.1016/s0092-8674(00)80676-5. [DOI] [PubMed] [Google Scholar]
- Yu, F., Cai, Y., Kaushik, R., Yang, X., Chia, W. Distinct roles of Gαi and Gβ13F subunits of the heterotrimeric G protein complex in the mediation of Drosophila neuroblast asymmetric divisions. J. Cell Biol. 2003;162:623–633. doi: 10.1083/jcb.200303174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, F., Wang, H., Qian, H., Kaushik, R., Bownes, M., Yang, X., Chia, W. Locomotion defects, together with Pins, regulates heterotrimeric G-protein signaling during Drosophila neuroblast asymmetric divisions. Genes & Dev. 2005;19:1341–1353. doi: 10.1101/gad.1295505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, F., Kuo, C.T., Jan, Y.N. Drosophila neuroblast asymmetric cell division: Recent advances and implications for stem cell biology. Neuron. 2006;51:13–20. doi: 10.1016/j.neuron.2006.06.016. [DOI] [PubMed] [Google Scholar]
- Zhou, H., Kuang, J., Zhong, L., Kuo, W.L., Gray, J.W., Sahin, A., Brinkley, B.R., Sen, S. Tumour amplified kinase STK15/BTAK induces centrosome amplification, aneuploidy and transformation. Nat. Genet. 1998;20:189–193. doi: 10.1038/2496. [DOI] [PubMed] [Google Scholar]