Abstract
The progression of neurogenesis relies on proneural basic helix–loop–helix (bHLH) transcription factors. These factors operate in undifferentiated neural stem cells and induce cell cycle exit and the initiation of a neurogenic program. However, the transient expression of proneural bHLH proteins in neural progenitors indicates that expression of neuronal traits must rely on previously unexplored mechanisms operating downstream from proneural bHLH proteins. Here we show that the HMG-box transcription factors Sox4 and Sox11 are of critical importance, downstream from proneural bHLH proteins, for the establishment of pan-neuronal protein expression. Examination of a neuronal gene promoter reveals that Sox4 and Sox11 exert their functions as transcriptional activators. Interestingly, the capacity of Sox4 and Sox11 to induce the expression of neuronal traits is independent of mechanisms regulating the exit of neural progenitors from the cell cycle. The transcriptional repressor protein REST/NRSF has been demonstrated to block neuronal gene expression in undifferentiated neural cells. We now show that REST/NRSF restricts the expression of Sox4 and Sox11, explaining how REST/NRSF can prevent precocious expression of neuronal proteins. Together, these findings demonstrate a central regulatory role of Sox4 and Sox11 during neuronal maturation and mechanistically separate cell cycle withdrawal from the establishment of neuronal properties.
Keywords: Sox proteins, proneural bHLH proteins, REST/NRSF, neurogenesis, development, CNS
The progression of vertebrate neurogenesis relies on mechanisms that in an orderly fashion direct precursor cells to exit the cell cycle, down-regulate progenitor cell identities, and to subsequently initiate the expression of neuronal properties. Despite advances in defining mechanisms that control the initiation of neurogenesis, the genetic program that drives the acquisition of the neuronal phenotype of post-mitotic neurons remains to be characterized.
Insights into the mechanisms that regulate pan-neuronal gene expression have been derived from studies of the zinc finger repressor protein REST (RE1 silencing transcription factor, also known as NRSF) (Chong et al. 1995; Schoenherr and Anderson 1995), which is ubiquitously expressed in neural precursors and has the capacity to bind and repress a large number of genes encoding neuronal proteins. In contrast, the proneural basic helix–loop–helix (bHLH) transcription factors, including Ngn1, Ngn2, and Mash1, function in neural stem cells to initiate the progression of neurogenesis (Bertrand et al. 2002). While proneural bHLH proteins mediate this function by committing stem cells to the neuronal linage and by inducing cell cycle exit, their expression is generally suppressed before progenitor cells exit the proliferative zone and begin to express neuronal properties (Gradwohl et al. 1996; Fode et al. 2000). Thus, the ability of proneural proteins to promote the terminal steps of neurogenesis must rely on downstream regulatory programs that subsequently establish the expression of neuronal properties in post-mitotic neural cells.
The bHLH genes Math3 and NeuroD (Lee et al. 1995; Perron et al. 1999) and the non-basic HLH gene Ebf1 (Garcia-Dominguez et al. 2003) are examples of transcription factors that have been suggested to function downstream from proneural bHLH proteins during the maturation steps of neurogenesis. Despite the fact that these proteins can induce ectopic formation of neurons in Xenopus (Lee et al. 1995; Perron et al. 1999; Garcia-Dominguez et al. 2003), mice deficient for NeuroD, Math3, or Ebf1 display only minor neurogenic defects (Naya et al. 1997; Garel et al. 1999; Tomita et al. 2000), and their role during neurogenesis remains unclear. Furthermore, a substantial number of neurons are generated prior to the induction of NeuroD expression (Lee et al. 1995; Roztocil et al. 1997). Thus, the molecular mechanism that controls the terminal steps of neurogenesis and the expression of neuronal properties has not yet been identified.
The HMG-box transcription factors of the Sox gene family have diverse regulatory functions during the formation of the vertebrate CNS (Pevny and Placzek 2005). Sox1, Sox2, and Sox3, which are expressed by most precursor cells, act to maintain the expression of progenitor identities and thus preserve cells in an undifferentiated state (Bylund et al. 2003; Graham et al. 2003), whereas another HMG-box protein, Sox21, has the opposite activity and allows cells to initiate a differentiation program (Sandberg et al. 2005). Hence, B-group Sox proteins appear to have key regulatory roles in the commitment of progenitors to neurogenesis. In contrast to Sox1–3 and Sox21, Sox4 and Sox11, which constitute the C-group of the Sox gene family (Kamachi et al. 2000), are mainly expressed in neural cells that have already been committed to neuronal differentiation (Uwanogho et al. 1995; Cheung et al. 2000), raising the possibility that these proteins control later aspects of neurogenesis. Mice, in which either the function of Sox4 or Sox11 has been inactivated, do not reveal any significant role of C-group Sox proteins during neurogenesis (Cheung et al. 2000; Sock et al. 2004), but structural similarities and the conserved expression patterns among these proteins indicate that functional redundancy may compensate for the loss of an individual Sox4 or Sox11 gene.
In this study, we have examined the role of Sox4 and Sox11 in the formation of neurons in the vertebrate CNS. We report that Sox4 and Sox11 operate downstream from proneural bHLH proteins and are vital for the establishment of pan-neuronal protein expression. Interestingly, misexpression of Sox4 and Sox11 does not cause progenitor cells to exit the division cycle or commit to a neuronal differentiation program. Instead, Sox4 and Sox11 can induce precocious expression of neuronal markers in self-renewing precursors. Examination of a neuronal gene promoter indicates that Sox4 and Sox11 can mediate their functions as transcriptional activators. Collectively, these findings establish an essential role of Sox4 and Sox11 in neuronal maturation and separate mechanistically cell cycle exit and the induction of pan-neuronal protein expression.
Results
The expression of Sox4 and Sox11 is confined to post-mitotic differentiating neural cells
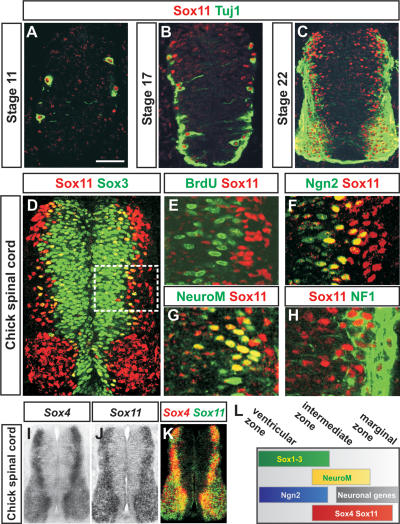
To examine the role of group-C Sox proteins during neurogenesis, we defined the expression of Sox11 in the spinal cord of chick embryos from Hamburger-Hamilton (HH) stages 10–22. At forelimb levels, Sox11 protein could first be detected at stage 11, which coincides with the appearance of differentiated neurons expressing the neuronal marker Tuj1 (Fig. 1A; Moody and Stein 1988). Between stages 11 and 22, the expression of Sox11 increased significantly (Fig. 1B,C), and at stage 22, the expression of Sox11 could be detected both medial to and within the domain of Tuj1+ neurons (Fig. 1C).
Figure 1.
Expression of Sox4 and Sox11 in the developing chick spinal cord. (A–C) Expression of Sox11 and Tuj1 protein in spinal cords from HH stages 11 to 22. (D–H) Expression of Sox11 in comparison with Sox3+ (D), BrdU+ (E), Ngn2+ (F), NeuroM/Math3+ (G), and NF1+ (H) cells. The white box in D indicates the analyzed area in E–H. (I–K) Expression of Sox4 (I) and Sox11 (J) mRNA. (K) The image represents an overlay of images I and J. (L) The summary figure indicates the distribution of Sox1–3, Ngn2, NeuroM/Math3, Sox4, and Sox11 and neuronal proteins in the developing neural tube. Bars: A,B, 40 μm; C,D, 60 μm;, E–H), 15 μm; I–K, 100 μm.
To better characterize how the distribution of Sox11 protein varies during the course of neurogenesis, we next compared the expression of Sox11 with molecular markers defining progenitor cells or differentiating neurons. In the ventricular zone, expression of Sox11 protein was restricted to post-mitotic cells expressing the neural progenitor marker Sox3 (Fig. 1D,E; Pevny et al. 1998; Bylund et al. 2003). Furthermore, most of these cells coexpressed the proneural bHLH transcription factor Ngn2 (Fig. 1F; Fode et al. 1998), whereas Sox11+ cells in the intermediate zone expressed the bHLH protein NeuroM/Math3 (Fig. 1G), a marker for post-mitotic neural cells that are in the process of down-regulating progenitor markers and initiating the expression of neuronal genes (Roztocil et al. 1997). Sox11 protein could also be detected in more differentiated neurons that had up-regulated expression of the pan-neuronal marker NF1 (Fig. 1H; Karlsson et al. 1987). Thus, the expression of Sox11 is initiated as post-mitotic Ngn2+ cells up-regulate NeuroM/Math3 and is maintained in differentiating neurons as these start to express neuronal properties (Fig. 1L). In addition, the other group-C member, Sox4, was expressed in a similar pattern as Sox11 (Fig. 1I,J), and in the stage 24 chick neural tube, their expression could be detected at most positions along the dorsoventral axis (Fig. 1K). However, while the distribution of Sox4 and Sox11 mRNAs overlapped extensively, the expression of Sox4 became progressively weaker in differentiated neurons located in the lateral aspect of the marginal zone (Fig. 1K; Cheung et al. 2000).
Sox4 and Sox11 can direct the expression of neuronal properties
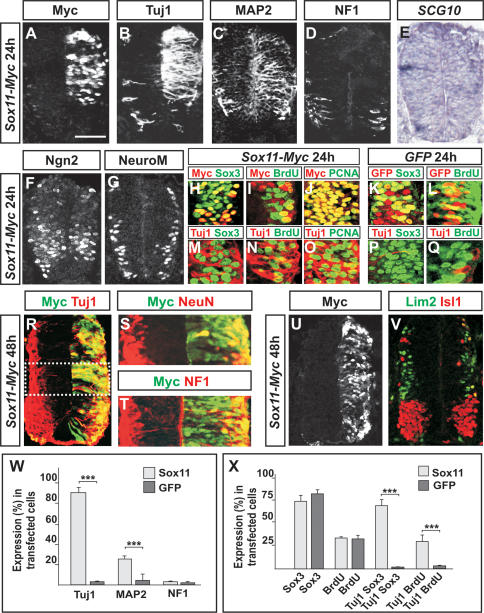
As the expression of Sox4/11 is restricted to post-mitotic differentiating neurons, it is possible that Sox4/11 have regulatory roles during the terminal steps of neurogenesis. To begin to address this issue, we examined the expression of pan-neuronal markers after misexpression of Sox11 in the ventricular zone. Misexpression of Sox11 for 24 h resulted in strong ectopic expression of the neuronal proteins Tuj1 and MAP2 (Fig. 2A–C,W; Riederer and Matus 1985). However, no premature or ectopic expression could be detected of the neuronal markers NF1 and SCG10 (Fig. 2D,E,W; Curmi et al. 1997). Sox4 had similar activity as Sox11 and caused transfected cells to up-regulate the expression of Tuj1 and MAP2 but not that of NF1 or SCG10 (Supplementary Fig. 1; data not shown). Thus, Sox4 and Sox11 have redundant functions and can promote precursor cells to up-regulate the expression of pan-neuronal markers.
Figure 2.
Sox4 and Sox11 can induce the expression of neuronal proteins. (A–E,W) Misexpression of Sox11 for 24 h (A) increased the number of Tuj1+ (B,W) and MAP2+ cells (C,W), whereas no change in the expression levels of NF1 (D,W) or SCG10 (E) could be detected (n ≥ 6 embryos; [*] p < 0.05; [**] p < 0.01; [***] p < 0.001). (F,G) Electroporation of Sox11 did not alter the expression of the proneural bHLH protein Ngn2 (F) or the bHLH protein NeuroM/Math3 (G). (H–L,X) Many Sox11-transfected cells expressed the progenitor marker Sox3 (H,X), and the number of Sox11-transfected cells that incorporated BrdU (I,X) or expressed the cell cycle marker PCNA (J) was comparable with cells electroporated with a GFP control vector (K,L,X) (n ≥ 6 embryos; [***] p < 0.001). (M–Q,X) Most of the ectopic Tuj1+ cells coexpressed Sox3 (M,X), and several of these also incorporated BrdU (N,X) and expressed PCNA (O). No or very few Tuj1+/Sox3+-incorporating cells (P,X) or Tuj1+/BrdU-incorporating cells (Q,X) could be detected in embryos electroporated with a GFP control vector. (R–T) Forty-eight hours after Sox11 transfection (R), many of the electroporated cells were post-mitotic and also expressed NeuN (S) and NF1 (T). (U,V) Misexpression of Sox11 for 48 h (U) did not disrupt the expression pattern of the subtype-specific neuronal markers Lim2 and Isl1 (V). Results are represented as mean ± SD. Bars: A–E,H–Q,R,U,V, 60 μm; F,G, 75 μm; S,T, 40 μm.
The induced expression of neuronal proteins in Sox4/11-electroporated cells could either reflect that these cells have completed a neurogenic program prematurely or that the transfected progenitor cells have bypassed the initial steps of neurogenesis and up-regulated the expression of neuronal markers in an ectopic fashion. To address these different possibilities, we next analyzed Sox11-electroporated cells with markers discriminating between self-renewing progenitors and cells that have initiated a neurogenic program. Transfection of Sox11 for 24 h did not induce any significant alteration in the expression of the bHLH proteins Ngn2 or NeuroM/Math3 (Fig. 2F,G), markers characteristic of cells that have initiated a neurogenic program. Furthermore, many electroporated cells expressed the progenitor marker Sox3 (Fig. 2H,K,X), and the number of transfected cells that were in a self-renewing state was comparable to progenitor cells electroporated with a GFP-expressing control vector (Fig. 2I,J,L,X). Comparable results were obtained with Sox4 (data not shown). Thus, misexpression of Sox4 and Sox11 does not promote progenitor cells to exit the division cycle or initiate a neuronal differentiation program. Instead, a high incidence of the ectopic Tuj1+ cells coexpressed the progenitor marker Sox3 (Fig. 2M,P,X), and several of these cells were also in a self-renewing state (Fig. 2N,O,Q,X). Together, these results indicate that Sox4/11 function during the terminal steps of neurogenesis and can activate the expression of neuronal traits independently of mechanisms directing the exit of progenitors from the cell cycle.
Sox11-electroporated neural tubes, which were allowed to develop for 48 h instead of 24 h, contained many transfected cells that were post-mitotic, located in the marginal zone, and expressed all pan-neuronal markers examined, including NeuN and NF1 (Fig. 2R–T). In line with this, Sox11-transfected cells located in the marginal zone also expressed the interneuron marker Lim2 and the motor neuron marker Isl1/2 (Tsuchida et al. 1994) in a normal dorsoventral pattern (Fig. 2U,V). Thus, although misexpression of Sox4 and Sox11 causes self-renewing progenitor cells to up-regulate premature ectopic expression of neuronal properties, the transfected cells are still capable of undergoing the normal pathway of neurogenesis and establishing a complete neuronal phenotype.
The establishment of pan-neuronal gene expression requires Sox4 and Sox11 function
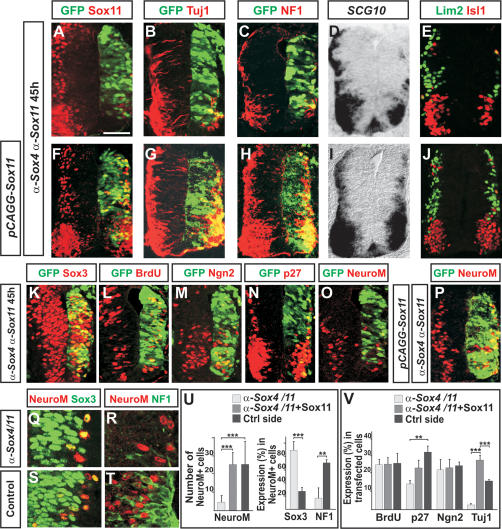
To further examine the role of group-C Sox proteins during the formation of neurons, we blocked the expression of Sox4 and Sox11 using RNA interference (siRNA) (Grishok and Mello 2002). Electroporation of siRNAs (Rao et al. 2004) directed against chick Sox4 or Sox11 (α-Sox4 and α-Sox11) decreased the level of Sox4 and Sox11 mRNA transcripts, whereas scrambled control versions of α-Sox4 and α-Sox11 had no effect on the expression of Sox4 or Sox11 (Supplementary Fig. 2). Cotransfection of α-Sox4 together with α-Sox11 resulted in a simultaneous reduction of both Sox4 and Sox11 mRNA transcripts and decreased the number of cells expressing Sox11 protein (Fig. 3A; Supplementary Fig. 2). Notably, 45 h after α-Sox4 and α-Sox11 coelectroporation, the number of cells expressing the neuronal markers Tuj1, NF1, SCG10, Lim2, and Isl1/2 was significantly reduced (Fig. 3B–E,V), suggesting that Sox4/11 function is required for the formation of cells expressing neuronal properties. In accordance with this, the presence of cotransfected vectors, expressing high levels of Sox11, rescued the generation of neurons in α-Sox4- and α-Sox11-electroporated neural tubes (Fig. 3F–J,V).
Figure 3.
Decreased levels of Sox4 and Sox11 block the establishment of a neuronal phenotype. (A–E,V) Forty-five hours after siRNA transfection, the expression of Sox11 (A), Tuj1 (B,V), NF1 (C), SCG10 (D), and Lim2 and Isl1 (E) was considerably reduced compared with the nontransfected control side (n ≥ 6 embryos; [**] p < 0.01; [***] p < 0.001). (F–J,V) The generation of Tuj1+ (G,V), NF1+ (H), SCG10+ (I), and Lim2+ and Isl1+ (J) neurons could be rescued in α-Sox4- and α-Sox11-electroporated neural tubes by the presence of Sox11 expression vectors (F). (K–M,V) Transfection of α-Sox4 and α-Sox11 siRNAs did not alter the expression of Sox3 (K), the rate of BrdU incorporation (L,V), or the expression of Ngn2 (M,V). (N–P,U,V) α-Sox4 and α-Sox11 siRNAs reduced the number of p27Kip1+ and NeuroM/Math3+ cells (N,O,U,V), a reduction that could be rescued by the presence of cotransfected Sox11 expression vectors (P,U,V) (n ≥ 6 embryos; [**] p < 0.01; [***] p < 0.001). (Q–U) In α-Sox4 and α-Sox11 siRNA-transfected neural tubes, most NeuroM/Math3+ cells coexpressed the progenitor marker Sox3 (Q,U) but had failed to up-regulate NF1 expression (R,U). In contrast, in the nonelectroporated control side, 27% of the NeuroM/Math3+ cells coexpressed Sox3 (S,U) and >70% coexpressed NF1 (T,U). Results are represented as mean ± SEM. Bars: A–J, 60 μm; K–P, 90 μm; Q–T, 120 μm.
Next we determined at which step during neurogenesis a reduction in Sox4 and Sox11 expression obstructs the establishment of neuronal properties. Transfection of α-Sox4 and α-Sox11 did not affect the general expression level of the progenitor marker Sox3 or the proportion of cells incorporating BrdU (Fig. 3K,L,V). Furthermore, the expression of the proneural protein Ngn2 was comparable with the nontransfected control side (Fig. 3M,V). However, although a fraction of the transfected cells expressed the post-mitotic marker p27Kip1 (Fig. 3N; El Wakil et al. 2006) and NeuroM/Math3 (Fig. 3O), their expression was decreased by 61% and 65%, respectively (Fig. 3U,V), a reduction that could be rescued by the presence of cotransfected Sox11 (Fig. 3P,U,V).
NeuroM/Math3 is expressed during the transition period when differentiating cells are in the process of suppressing progenitor specific markers and initiating the expression of neuronal proteins. In the nontransfected control side of the neural tube, 27% of the NeuroM/Math3+ cells coexpressed the progenitor marker Sox3, and 70% of the NeuroM/Math3+ cells coexpressed the neuronal marker NF1 (Fig. 3S–U). In the siRNA-treated side of the neural tube, most of the remaining NeuroM/Math3+ cells coexpressed Sox3 (Fig. 3Q,U), and very few NeuroM/Math3+ cells that expressed NF1 could be detected (Fig. 3R,U). Thus, decreased levels of Sox4/11 expression appear to block the differentiation step at which post-mitotic NeuroM/Math3+ cells up-regulate the expression of neuronal properties. Notably, in the siRNA-treated neural tubes, but not in embryos coelectroporated with siRNAs together with a Sox11 expression vector, we could detect a small increase in the number of apoptotic cells (Supplementary Fig. 3). Thus, one possibility is that the failure of NeuroM/Math3+ cells to up-regulate the expression of neuronal proteins results in cell degeneration. Furthermore, although the expression of Sox4/11 and NeuroM/Math3 appears to be initiated simultaneously (Fig. 1G), we cannot rule out the possibility that the reduction of NeuroM/Math3+ cells is reflecting an additional role of Sox4/11 upstream of NeuroM/Math3 expression.
Repression of Sox11 downstream targets blocks the expression of neuronal properties
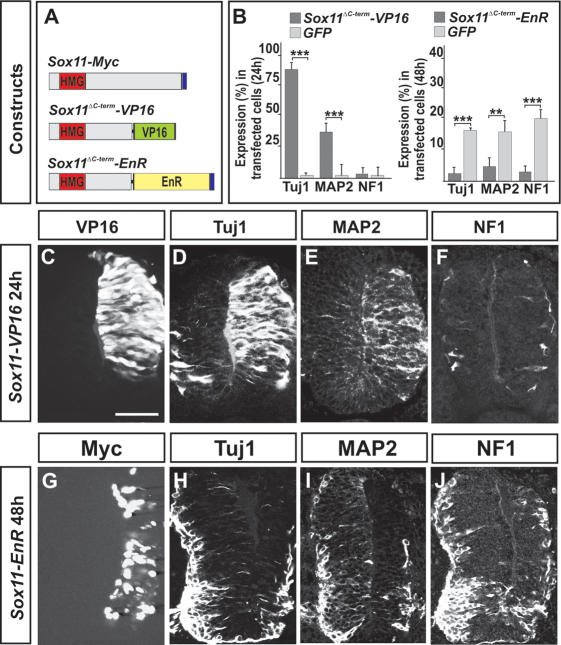
In vitro studies have revealed that group-C Sox proteins can function as transcriptional activators (van de Wetering et al. 1993; Kuhlbrodt et al. 1998; Schmidt et al. 2003; Wiebe et al. 2003), an activity that has been mapped to their C-terminal regions (Hargrave et al. 1997; Kuhlbrodt et al. 1998; Wegner 1999). To further determine the mechanism by which group-C Sox proteins direct the expression of neuronal characters, we generated obligate repressor or activator variants of Sox11. Sox11 cDNA, lacking the C-terminal putative activation domain, was either fused to the transactivation domain of the cDNA encoding the viral protein VP16 (Sox11ΔC-term–VP16) (Berk et al. 1998) or to the transcriptional repressor domain of the Drosophila Engrailed gene (Sox11ΔC-term–EnR) (Fig. 4A; Smith and Jaynes 1996). Misexpression of Sox11ΔC-term–VP16 mimicked the activity of full-length Sox11 and induced ectopic expression of Tuj1 and MAP2 but not the expression of NF1 or SCG10 (Fig. 4B–F; data not shown). In contrast, the repressor form of Sox11, Sox11ΔC-term–EnR, had the opposite activity and suppressed the endogenous expression of both Tuj1 and MAP2 (Fig. 4B, G–I). In addition, Sox11ΔC-term–EnR also blocked the expression of NF1 and SCG10 (Fig. 4B,J; data not shown). Thus, active repression of Sox11 target genes perturbs the induction of neuronal properties. Together, these results indicate that the ability of Sox11 to direct the expression of neuronal traits relies on its function as a transcriptional activator.
Figure 4.
Active repression of Sox11 target genes prevents the expression of pan-neuronal properties. (A) The constructs used were as follows: The C-terminal part of Sox11 was replaced with the transactivation domain of VP16 (Sox11ΔC-term–VP16), or with the repressor domain of the Drosophila Engrailed gene (Sox11ΔC-term–EnR). (B–F) Sox11ΔC-term–VP16 (C) behaved like full-length Sox11 and induced the expression of Tuj1 (B,D) and MAP2 (B,E) but not that of NF1 (B,F). (B,G–J) Forced expression of Sox11ΔC-term–EnR (G) prevented neuronal protein expression, including Tuj1 (B,H), MAP2 (B,I), and NF1 (B,J) (n ≥ 6 embryos; [**] p < 0.01; [***] p < 0.001). Results are represented as mean ± SD. Bars: C–F, 60 μm; G–J, 80 μm.
Sox4 and Sox11 can function as transcriptional activators of a neuronal gene promoter
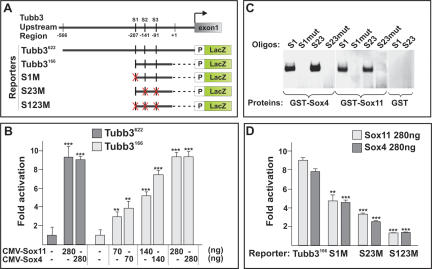
To further examine the mechanism by which group-C Sox proteins control neuronal gene expression, we focused on the upstream 622-base-pair (bp) non-coding region of the mouse Tubb3 gene (Tubb3622; also known as classIII β-tubulin or Tuj1) (Fig. 5A), as this DNA segment is sufficient to drive the expression of a reporter gene in EC cells during neuronal differentiation (Dennis et al. 2002). To determine whether group-C Sox proteins functionally interact with this genomic element, COS1 cells were transfected with expression constructs encoding Sox4 and Sox11 proteins, together with a LacZ reporter containing the isolated Tubb3 genomic fragment and a minimal β-globin promoter (Tubb3622-LacZ) (Fig. 5A). Sox4 and Sox11 displayed similar activities in this system and activated the Tubb3622-LacZ reporter nearly ninefold (Fig. 5B). Notably, examination of the Tubb3622 sequence revealed that three potential binding sites for Sox4 and Sox11 (van Beest et al. 2000) were located within a domain −91 to −207 bp upstream of the Tubb3 transcriptional start site (Tubb3166) (Fig. 5A). In a DNA-binding gel shift assay, both recombinant Sox4 and Sox11 proteins interacted with these sites (Fig. 5C) in a sequence-specific manner (Fig. 5C; Supplementary Fig. 4). In line with these findings, the Tubb3166-LacZ reporter (Fig. 5A) could be activated by Sox4 and Sox11 in a dose-dependent manner (Fig. 5B), and mutations of each individual Sox4/11-binding site reduced the transactivation of the Tubb3166-LacZ reporter more than sixfold (Fig. 5D). Moreover, Ngn2 or Sox3, which have been demonstrated to induce and repress Tuj1 expression, respectively (Novitch et al. 2001; Bylund et al. 2003; Graham et al. 2003), could not regulate the Tubb3166-LacZ reporter through the defined Sox4/11-binding sites (Supplementary Fig. 5). Hence, using Tubb3 as an example, this set of experiments indicates that Sox4 and Sox11 can function as transcriptional activators of a neuronal gene promoter.
Figure 5.
Sox4 and Sox11 interact with promoter elements of the neuronal gene Tubb3. (A) Tubb3 promoter constructs used in transactivation studies. S1–S3 indicate potential Sox4- and Sox11-binding sites, S1 (TTCTATTGTCCCC), S2, and S3 (CCGCATTGTGCGG). X marked in red indicates mutated sites. (B) Transactivation of the Tubb3166-LacZ and Tubb3622-LacZ reporters by Sox4 or Sox11. (C) Recombinant Sox4 and Sox11 proteins were able to bind to S1, S2, and S3 (S23) in a DNA-binding gel shift assay. No binding could be detected when these sites were mutated: S1mut (TTCTCCCGTCCCC) and S23mut (CCGCGGGGTGCGG). (D) The ability of Sox4 and Sox11 to transactivate the Tubb3166-LacZ reporter construct was reduced more than sixfold when S1 and S23 were mutated (S1M and S23M). The error bars in B and D indicate the standard deviation (SD) of three independent transfections. Each experiment was repeated six times. (**) p < 0.01; (***) p < 0.001.
REST and proneural proteins regulate the expression of Sox4 and Sox11
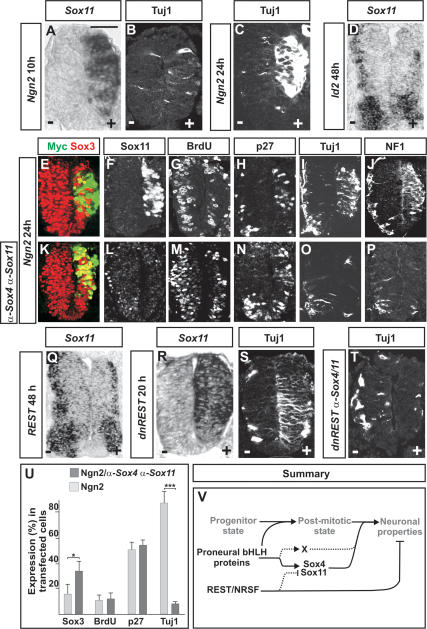
Proneural bHLH proteins can promote the generation of neurons from precursor cells (Sun et al. 2001; Lo et al. 2002). However, as the expression of proneural proteins is suppressed before neural progenitors exit the proliferative zone (Gradwohl et al. 1996; Fode et al. 2000), their ability to induce the expression of pan-neuronal markers must rely on downstream transcriptional programs. Since the expression of Sox11 is initiated in differentiating Ngn2+ cells (Fig. 1F), the expression of group-C Sox genes may be controlled by proneural protein activity. To examine this possibility, we misexpressed either Ngn2 or Id2 in the developing chick neural tube. Indeed, electroporation of Ngn2 for 10 h induced high levels of Sox11 expression (Fig. 6A) before transfected cells had up-regulated high levels of Tuj1 (Fig. 6B,C). Comparable results were obtained with another proneural gene, Ascl1 (also known as Mash1) (Guillemot and Joyner 1993; data not shown). Id2, which functions as a passive repressor of proneural protein activity (Yokota 2001), had the opposite function compared with Ngn2 and decreased the level of Sox11 gene expression 48 h after electroporation (Fig. 6D). In addition, similar to Sox11, the expression of Sox4 was induced by Ngn2 and repressed by Id2 activity (data not shown). Thus, the induction of Sox4/11 expression in differentiating neural cells appears to depend on proneural protein activity.
Figure 6.
The expression of Sox4 and Sox11 is controlled by REST and proneural proteins. (A–C) Electroporation of Ngn2 induced high levels of ectopic Sox11 expression already 10 h after transfection (A), whereas the expression of Tuj1 first could be readily detected 24 h after electroporation (B,C). (D) Electroporation of Id2 for 48 h reduced the expression of Sox11. (E–J,U) Overexpression of Ngn2 for 24 h promoted cells to suppress progenitor characters (E,G,U) and up-regulate the expression of Sox11 (F), p27Kip1 (H,U), and neuronal proteins (I,J,U) (n ≥ 6 embryos; [*] p < 0.05; [***] p < 0.001). (K–P,U) Cells cotransfected with Ngn2 and α-Sox4 and α-Sox11 siRNAs for 24 h expressed reduced levels of Sox3 (K,U) and had exited the cell cycle (M,U) and up-regulated p27Kip1 (N,U). Cotransfected cells did not up-regulate the expression of Sox11 (L) or neuronal markers (O,P,U). (Q–T) Misexpression of REST for 48 h decreased the expression of Sox11 (Q) (48% ± 11% reduction; [***] p < 0.001), whereas a dominant-negative version of REST, dnREST, 20 h after electroporation had induced high levels of ectopic Sox11 (R) and Tuj1 expression (S). (T) dnREST-transfected cells could not up-regulate the expression of Tuj1 when the expression of Sox4 and Sox11 was prevented by siRNAs. Results are represented as mean ± SD. Bars: A–C,R–T, 60 μm; E–P, 80 μm; D,Q, 50 μm. (V) Proposed molecular network regulating the establishment of pan-neuronal properties. Proneural bHLH proteins drive the initial steps of neurogenesis and direct the exit of neural cells from the division cycle. Proneural proteins also induce the expression of Sox4 and Sox11, which in turn activate neuronal gene expression. According to this model, proneural proteins induce the expression of an additional genetic program (designated X) that together with Sox4 and Sox11 activates a complete neuronal phenotype in differentiating neurons. REST/NRSF prevents precocious expression of neuronal proteins in undifferentiated neural cells both by a direct repression of neuronal genes and by restricting the expression of Sox4 and Sox11 to neural cells that have exited the cell cycle.
Since the expression of group-C Sox genes is regulated by proneural proteins, we next examined whether the ability of Ngn2 to direct the establishment of a neuronal phenotype is dependent on the activity of Sox4/11. To address this issue, Ngn2 was misexpressed either alone or together with coelectroporated α-Sox4 and α-Sox11 siRNAs. Twenty-four hours after Ngn2 electroporation, a majority of the transfected cells had down-regulated the expression of Sox3 and up-regulated the expression of Sox11 (Fig. 6E,F,U). At this stage, many of the cells had exited the division cycle and up-regulated the expression of p27Kip1 (Fig. 6G,H,U) and the definitive neuronal markers Tuj1 and NF1 (Fig. 6I,J,U). The presence of α-Sox4 and α-Sox11 siRNAs reduced the ability of Ngn2 to suppress the expression of Sox3 (Fig. 6K,U) and blocked the establishment of Sox11 protein expression (Fig. 6L). However, a high proportion of the electroporated cells were post-mitotic, and many cells had also up-regulated the expression of p27Kip1 (Fig. 6M,N,U). In contrast, only few cells coelectroporated with Ngn2 and α-Sox4 and α-Sox11 siRNAs had induced the expression of Tuj1 and NF1 (Fig. 6O,P,U). Thus, in the absence of Sox4/11 expression, Ngn2 can force cells to exit the division cycle but is unable to direct cells to up-regulate the expression of neuronal markers.
Neuronal gene expression is subjected to negative regulation by REST (Chong et al. 1995; Schoenherr and Anderson 1995), a transcriptional repressor protein expressed in the ventricular zone. REST has been demonstrated to exert its function through a direct repressive interaction with neuronal gene enhancers. However, it has not been addressed whether REST also represses the expression of genes encoding activators of neuronal gene expression. To examine this possibility, chick embryos were transfected with full-length REST and analyzed for the expression of Sox4 and Sox11. Indeed, 48 h after REST overexpression the number of cells expressing Sox4 and Sox11 was significantly reduced (Fig. 6Q; data not shown). Moreover, studies in both chick and mouse embryos indicate that loss of REST function leads to derepression of neuronal markers within the ventricular zone (Chen et al. 1998). To examine whether the loss of REST activity also is followed by a derepression of Sox4/11, we next misexpressed a dominant-negative form of REST (dnREST) (Chen et al. 1998). Twenty hours after dnREST transfection, many electroporated cells had initiated the expression of Sox4/11 (Fig. 6R; data not shown), and this was followed by an up-regulation of neuronal markers (Fig. 6S; data not shown). Notably, the expression level of the proneural genes Ngn2 and Cash1 was not altered by misexpressed full-length REST or dnREST (Supplementary Fig. 6), and transfected progenitor cells responded similarly to dnREST as to Sox4/11 and up-regulated only a partial array of neuronal markers. Hence, dnREST-transfected cells up-regulated the expression of Tuj1 and MAP2 but not that of NF1 or NeuN (data not shown). In addition, the ability of electroporated dnREST to derepress neuronal marker expression was blocked when the accompanying up-regulation of Sox4/11 expression was prevented by coelectroporated α-Sox4 and α-Sox11 (Fig. 6T). Thus, the ability of REST to suppress neuronal gene expression appears, at least in part, to be dependent on its capacity to restrict the expression of Sox4/11 (Fig. 6V).
Discussion
Proneural bHLH transcription factors are essential for the progression of neurogenesis and can induce cell cycle exit and commit progenitors to a neurogenic program (Farah et al. 2000; Bertrand et al. 2002; Kintner 2002; Lo et al. 2002), but how these proteins promote differentiated progeny to obtain a neuronal phenotype has remained elusive. We have shown here that Sox4 and Sox11 function downstream from proneural bHLH protein as critical activators of both generic and subtype-specific neuronal properties. Elimination of Sox4 and Sox11 activity did not disrupt the ability of proneural bHLH proteins to promote cell cycle exit, but blocked their capacity to establish the expression of neuronal properties. Together, these data reveal a central regulatory role of group-C Sox proteins during neuronal maturation and suggest that the induction of Sox4 and Sox11 expression reflects a critical step in the acquisition of a neuronal phenotype.
The bHLH proteins Math3 and NeuroD represent examples of transcription factors that are expressed in post-mitotic neurons, downstream from proneural proteins. Mice deficient for Math3 or NeuroD display only minor neurogenic defects (Lee et al. 1995; Naya et al. 1997; Tomita et al. 2000). However, misexpression of these proteins in Xenopus embryos can convert epiblast and neural precursor cells into differentiated neurons (Lee et al. 1995; Perron et al. 1999). Hence, even if Math3 and NeuroD are expressed exclusively in post-mitotic neural cells, these factors appear to have the capacity to induce progenitor cells to complete a neurogenic program. In contrast, Sox4/11 could induce self-renewing precursor cells to up-regulate the expression of neuronal markers, but misexpression of these proteins did not promote cells to exit the cell cycle or suppress progenitor specific gene expression. Thus, the activity of Math3 and NeuroD differs from that of Sox4/11 and more resembles the function of proneural bHLH proteins. These observations also establish that the ability of Sox4/11 to induce the expression of neuronal proteins can be functionally separated from mechanisms whereby proneural bHLH proteins promote the initial steps of neurogenesis. Apart from Math3 and NeuroD, additional basic and non-basic HLH proteins are expressed in differentiating neurons, downstream from proneural proteins (Bertrand et al. 2002). While the activity of Sox4/11 is distinct from that of Math3 and NeuroD, our results do not argue against the possibility that other members of the HLH transcription factor family are required, together with Sox4/11, for the induction of neuronal traits in post-mitotic neurons (Fig. 6V).
Although Sox4/11 appear to be essential for neuronal protein expression, they could only induce a partial array of neuronal markers in self-renewing progenitors. The inductive capacity of Sox4/11 was not, however, dependent on the proliferative status of the transfected progenitor cells. For instance, Sox11 induced the same set of neuronal markers in cells that were in a self-renewing state, as in cells that had been forced to exit the division cycle by the CDK inhibitor p27Kip1 (Supplementary Fig. 7). Another possibility, which may explain why Sox4/11 could induce only a subset of neuronal markers at ectopic locations, is that neuronal genes may be subjected to active repression in progenitor cells. Thus, a relief of such repressive program may be a prerequisite for the establishment of a full neuronal phenotype. Finally, Sox proteins have been reported to act in concert with heterodimerizing partner factors (Kamachi et al. 2000). Hence, it is possible, and even likely, that Sox4/11 normally act in synergy with other factors during neuronal maturation and that the absence of such partner factor(s) in undifferentiated progenitor cells limits the inductive capacity of Sox4/11 (Fig. 6V).
We found that the expression of Sox4/11 is directed to post-mitotic neural cells by a combinatorial function of proneural proteins and REST/NRSF. The expression of Sox4/11 was induced by proneural proteins and suppressed by Id2, an inhibitor of proneural protein function (Yokota 2001). Moreover, full-length REST/NRSF repressed the expression of Sox4/11, whereas its dominant-negative form (dnREST) induced high levels of ectopic Sox4/11 expression, followed by an up-regulation of neuronal markers (Chen et al. 1998). Interestingly, in these experiments, the capacity of dnREST to activate the expression of neuronal proteins was dependent on Sox4/11 expression and was limited to neuronal markers that could be ectopically induced by Sox4/11 misexpression (e.g., Tuj1 and MAP2 but not NF1 or NeuN). Hence, in addition to its ability to directly repress neuronal gene enhancers (Lunyak et al. 2002), REST/NRSF also appears to prevent precocious expression of neuronal proteins in undifferentiated neural cells through its capacity to suppress the expression of Sox4/11 (Fig. 6V).
The expression of group-C Sox genes is not restricted to the developing CNS but can be detected in cells from several distinct origins (Uwanogho et al. 1995; de Martino et al. 2000; Maschhoff et al. 2003; Sock et al. 2004). For instance, Sox11 has been demonstrated to activate the muscle differentiation gene myogenin in C2C12 myoblasts (Schmidt et al. 2003). Furthermore, in zebrafish, one of the two Sox4 orthologs (Sox4b) is necessary for pancreatic endocrine cell differentiation and the expression of the hormone glucagon (Mavropoulos et al. 2005). Thus, as in the developing CNS, group-C Sox proteins can drive the expression of differentiation proteins both in mesodermal and endodermal cells. These findings suggest that the selection of downstream genes controlled by Sox4/11 ultimately depends on the linage from which the progenitor cells are originating.
Sox proteins have several regulatory functions during neurogenesis. The group-B Sox proteins (Sox1–3) maintain neural cells in an undifferentiated state by promoting progenitor-specific gene expression (Bylund et al. 2003; Graham et al. 2003). Another group-B member, Sox21, has the opposite activity and represses Sox1–3-activated gene expression (Sandberg et al. 2005). Interestingly, Sox21 operates downstream from proneural bHLH proteins and is required for their capacity to commit precursor cells to a neuronal differentiation program. The findings in this study show that also group-C Sox proteins are regulated by proneural bHLH proteins. Thus, by regulating the expression of different groups of Sox genes, proneural proteins can orchestrate both initial and later steps of neurogenesis. Notably, Sox and bHLH proteins have been demonstrated to play key roles in the regulation of progenitor cell differentiation also outside of the developing CNS (Kim et al. 2003; Schmidt et al. 2003). Thus, the functional interaction described in this study may establish a general paradigm for the mechanisms whereby progenitor cells are converted into more mature cell types.
Materials and methods
Expression constructs and in ovo electroporation
cDNAs encoding full-length chick Sox11, mouse Sox4, mouse REST, and the Zn-finger domain of REST (amino acids 203–440; dnREST), mouse p27kip1 were subcloned into the CMV-based expression vector pCAGGS (Niwa et al. 1991). Sox4 and Sox11 were tagged at their C termini with the myc epitope (EQKLISEEDL). Other Sox11 constructs included cDNA encoding amino acids 1–332 of chick Sox11 either fused to the VP16 activation domain (amino acids 400–488) or to a myc-tagged En repressor (amino acids 2–298 of the Drosophila Engrailed protein). Other expression constructs, Sox3-myc and Ngn2-myc, have been described previously (Bylund et al. 2003). RNA interference was performed by electroporation of 21-nucleotide (nt) RNA duplexes (siRNA; http://www.invitrogen.com) at a concentration of 2 μg/μL together with pCAGGS nls-GFP (kindly provided by Johan Ericson, Karolinska Institute, Stockholm, Sweden). The targeted sequences were Sox4 mRNA, 5′-GGC CCAGGAAGAAGGUGAA-3′; and Sox11 mRNA, 5′-GCUUU CAUGGUGUGGUCUA-3′. Electroporation was performed in chick embryos (Bylund et al. 2003) of HH stage 9–11 using expression vector concentration at 0.8–1.2 μg/μL. Electroporated embryos were incubated for 10–48 h before fixation in 4% paraformaldehyde and processing for in situ hybridization and immunohistochemistry. Statistical analysis was performed using two-tailed Student’s t-test based on six or more embryos from at least three individual experiments.
Immunohistochemistry and in situ hybridization
Antibody staining was performed as described previously (Tsuchida et al. 1994). Labeling of apoptotic cells, using an Apoptosis detection kit (Chemicon), was performed according to the manufacturer’s recommendation. Guinea pig anti-sera were generated against a chick Sox11 peptide (C terminus: GGRLYYSFKNITKQ). The mouse MAP2 antibodies were Abcam (ID no. Ab11267). Additional antibodies and BrdU labeling methods are described elsewhere (Briscoe et al. 2000; Bylund et al. 2003; Sandberg et al. 2005). In situ hybridization was performed as described previously (Tsuchida et al. 1994) using chick probes for Sox11, Sox4, SCG10 Cash1, and dnREST. cDNAs encoding SCG10 and chick Sox4 were obtained from MRC Geneservice (clone ID: chEST117f15 and chEST386n7).
Cell transfections and β-galactosidase assay
Mouse genomic fragments upstream of the Tubb3 transcriptional start base pairs +54 to −566 (Tubb3622) and −50 to −215 (Tubb3166) were amplified by PCR and subcloned into the pBGZA vector containing the minimal β-globin promoter and the reporter gene LacZ. Three mutated versions of Tubb3166 (S1M, S23M, and S123M) were generated by PCR. The distinct reporter constructs were transfected (200 ng/200,000 cells), with or without Sox3, Sox4, or Sox11 expression vectors (70–280 ng/200,000 cells) into COS1 cells. Twenty-four hours after cell transfection, the relative Luciferase and β-galactosidase activity was measured. Cell transfections and activity assay methods have been described elsewhere (Wang et al. 2003).
Gel shift assays
The oligos used for the gel shift assay were ordered from Invitrogen: S1: sense, 5′-GCCTGGGTTCTATTGTCCCCACCAGA GCGCTAG-3′; S1M: sense, 5′-GCCTGGGTTCTCCCGTCCC CAGAGCGCTAG-3′; S23: sense, 5′-AGCCTGCCGCATTGT GCGGCGCTCCACTAG-3′; S23M: sense, 5′-AGCCTGCCGC GGGGTGCGGCGCTCCACTAG-3′. Sox3, Sox4, and Sox11 proteins were produced in bacteria Escherichia coli strain BL21(DE3) Codon Plus (Stratagene) and purified according to Wingate et al. (2005). The gel shift assay procedure has been described previously (Aarnisalo et al. 2002).
Acknowledgments
We thank H. Kondoh for providing Sox11 cDNA, H. Cleavers for providing Sox4 cDNA, D. Anderson for providing REST cDNA, and B. Joseph for providing p27kip1 cDNA. We are grateful to J. Ericson and members of the Muhr laboratory for discussion and comments on the manuscript. J.M. is supported by The Swedish Natural Research Council, The Swedish Foundation for Strategic Research, and the Ake Wibergs, Magnus Bergvalls, and Jeanssons research foundations. J.M. and T.P. are supported by the Ludwig Institute for Cancer Research. M.W. is supported by the Swedish Brain Foundation.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.403406
References
- Aarnisalo, P., Kim, C.H., Lee, J.W., Perlmann, T. Defining requirements for heterodimerization between the retinoid X receptor and the orphan nuclear receptor Nurr1. J. Biol. Chem. 2002;277:35118–35123. doi: 10.1074/jbc.M201707200. [DOI] [PubMed] [Google Scholar]
- Berk, A.J., Boyer, T.G., Kapanidis, A.N., Ebright, R.H., Kobayashi, N.N., Horn, P.J., Sullivan, S.M., Koop, R., Surby, M.A., Triezenberg, S.J. Mechanisms of viral activators. Cold Spring Harb. Symp. Quant. Biol. 1998;63:243–252. doi: 10.1101/sqb.1998.63.243. [DOI] [PubMed] [Google Scholar]
- Bertrand, N., Castro, D.S., Guillemot, F. Proneural genes and the specification of neural cell types. Nat. Rev. Neurosci. 2002;3:517–530. doi: 10.1038/nrn874. [DOI] [PubMed] [Google Scholar]
- Briscoe, J., Pierani, A., Jessell, T.M., Ericson, J. A homeodomain code specifies progenitor cell identity and neuronal fate in the ventral neural tube. Cell. 2000;101:435–445. doi: 10.1016/s0092-8674(00)80853-3. [DOI] [PubMed] [Google Scholar]
- Bylund, M., Andersson, E., Novitch, B.G., Muhr, J. Vertebrate neurogenesis is counteracted by Sox1-3 activity. Nat. Neurosci. 2003;6:1162–1168. doi: 10.1038/nn1131. [DOI] [PubMed] [Google Scholar]
- Chen, Z.F., Paquette, A.J., Anderson, D.J. NRSF/REST is required in vivo for repression of multiple neuronal target genes during embryogenesis. Nat. Genet. 1998;20:136–142. doi: 10.1038/2431. [DOI] [PubMed] [Google Scholar]
- Cheung, M., Abu-Elmagd, M., Clevers, H., Scotting, P.J. Roles of Sox4 in central nervous system development. Brain Res. Mol. Brain Res. 2000;79:180–191. doi: 10.1016/s0169-328x(00)00109-1. [DOI] [PubMed] [Google Scholar]
- Chong, J.A., Tapia-Ramirez, J., Kim, S., Toledo-Aral, J.J., Zheng, Y., Boutros, M.C., Altshuller, Y.M., Frohman, M.A., Kraner, S.D., Mandel, G. REST: A mammalian silencer protein that restricts sodium channel gene expression to neurons. Cell. 1995;80:949–957. doi: 10.1016/0092-8674(95)90298-8. [DOI] [PubMed] [Google Scholar]
- Curmi, P.A., Andersen, S.S., Lachkar, S., Gavet, O., Karsenti, E., Knossow, M., Sobel, A. The stathmin/tubulin interaction in vitro. J. Biol. Chem. 1997;272:25029–25036. doi: 10.1074/jbc.272.40.25029. [DOI] [PubMed] [Google Scholar]
- de Martino, S., Yan, Y.L., Jowett, T., Postlethwait, J.H., Varga, Z.M., Ashworth, A., Austin, C.A. Expression of sox11 gene duplicates in zebrafish suggests the reciprocal loss of ancestral gene expression patterns in development. Dev. Dyn. 2000;217:279–292. doi: 10.1002/(SICI)1097-0177(200003)217:3<279::AID-DVDY6>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- Dennis, K., Uittenbogaard, M., Chiaramello, A., Moody, S.A. Cloning and characterization of the 5′-flanking region of the rat neuron-specific Class III β-tubulin gene. Gene. 2002;294:269–277. doi: 10.1016/s0378-1119(02)00801-6. [DOI] [PubMed] [Google Scholar]
- El Wakil, A., Francius, C., Wolff, A., Pleau-Varet, J., Nardelli, J. The GATA2 transcription factor negatively regulates the proliferation of neuronal progenitors. Development. 2006;133:2155–2165. doi: 10.1242/dev.02377. [DOI] [PubMed] [Google Scholar]
- Farah, M.H., Olson, J.M., Sucic, H.B., Hume, R.I., Tapscott, S.J., Turner, D.L. Generation of neurons by transient expression of neural bHLH proteins in mammalian cells. Development. 2000;127:693–702. doi: 10.1242/dev.127.4.693. [DOI] [PubMed] [Google Scholar]
- Fode, C., Gradwohl, G., Morin, X., Dierich, A., LeMeur, M., Goridis, C., Guillemot, F. The bHLH protein NEUROGENIN 2 is a determination factor for epibranchial placode-derived sensory neurons. Neuron. 1998;20:483–494. doi: 10.1016/s0896-6273(00)80989-7. [DOI] [PubMed] [Google Scholar]
- Fode, C., Ma, Q., Casarosa, S., Ang, S.L., Anderson, D.J., Guillemot, F. A role for neural determination genes in specifying the dorsoventral identity of telencephalic neurons. Genes & Dev. 2000;14:67–80. [PMC free article] [PubMed] [Google Scholar]
- Garcia-Dominguez, M., Poquet, C., Garel, S., Charnay, P. Ebf gene function is required for coupling neuronal differentiation and cell cycle exit. Development. 2003;130:6013–6025. doi: 10.1242/dev.00840. [DOI] [PubMed] [Google Scholar]
- Garel, S., Marin, F., Grosschedl, R., Charnay, P. Ebf1 controls early cell differentiation in the embryonic striatum. Development. 1999;126:5285–5294. doi: 10.1242/dev.126.23.5285. [DOI] [PubMed] [Google Scholar]
- Gradwohl, G., Fode, C., Guillemot, F. Restricted expression of a novel murine atonal-related bHLH protein in undifferentiated neural precursors. Dev. Biol. 1996;180:227–241. doi: 10.1006/dbio.1996.0297. [DOI] [PubMed] [Google Scholar]
- Graham, V., Khudyakov, J., Ellis, P., Pevny, L. SOX2 functions to maintain neural progenitor identity. Neuron. 2003;39:749–765. doi: 10.1016/s0896-6273(03)00497-5. [DOI] [PubMed] [Google Scholar]
- Grishok, A., Mello, C.C. RNAi (Nematodes: Caenorhabditis elegans) Adv. Genet. 2002;46:339–360. doi: 10.1016/s0065-2660(02)46012-9. [DOI] [PubMed] [Google Scholar]
- Guillemot, F., Joyner, A.L. Dynamic expression of the murine Achaete-Scute homologue Mash-1 in the developing nervous system. Mech. Dev. 1993;42:171–185. doi: 10.1016/0925-4773(93)90006-j. [DOI] [PubMed] [Google Scholar]
- Hargrave, M., Wright, E., Kun, J., Emery, J., Cooper, L., Koopman, P. Expression of the Sox11 gene in mouse embryos suggests roles in neuronal maturation and epithelio-mesenchymal induction. Dev. Dyn. 1997;210:79–86. doi: 10.1002/(SICI)1097-0177(199710)210:2<79::AID-AJA1>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Kamachi, Y., Uchikawa, M., Kondoh, H. Pairing SOX off: With partners in the regulation of embryonic development. Trends Genet. 2000;16:182–187. doi: 10.1016/s0168-9525(99)01955-1. [DOI] [PubMed] [Google Scholar]
- Karlsson, J.E., Rosengren, L.E., Haglid, K.G. A rapid HPLC method to separate the triplet proteins of neurofilament. J. Neurochem. 1987;49:1375–1378. doi: 10.1111/j.1471-4159.1987.tb01002.x. [DOI] [PubMed] [Google Scholar]
- Kim, J., Lo, L., Dormand, E., Anderson, D.J. SOX10 maintains multipotency and inhibits neuronal differentiation of neural crest stem cells. Neuron. 2003;38:17–31. doi: 10.1016/s0896-6273(03)00163-6. [DOI] [PubMed] [Google Scholar]
- Kintner, C. Neurogenesis in embryos and in adult neural stem cells. J. Neurosci. 2002;22:639–643. doi: 10.1523/JNEUROSCI.22-03-00639.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlbrodt, K., Herbarth, B., Sock, E., Enderich, J., Hermans-Borgmeyer, I., Wegner, M. Cooperative function of POU proteins and SOX proteins in glial cells. J. Biol. Chem. 1998;273:16050–16057. doi: 10.1074/jbc.273.26.16050. [DOI] [PubMed] [Google Scholar]
- Lee, J.E., Hollenberg, S.M., Snider, L., Turner, D.L., Lipnick, N., Weintraub, H. Conversion of Xenopus ectoderm into neurons by NeuroD, a basic helix–loop–helix protein. Science. 1995;268:836–844. doi: 10.1126/science.7754368. [DOI] [PubMed] [Google Scholar]
- Lo, L., Dormand, E., Greenwood, A., Anderson, D.J. Comparison of the generic neuronal differentiation and neuron subtype specification functions of mammalian achaete-scute and atonal homologs in cultured neural progenitor cells. Development. 2002;129:1553–1567. doi: 10.1242/dev.129.7.1553. [DOI] [PubMed] [Google Scholar]
- Lunyak, V.V., Burgess, R., Prefontaine, G.G., Nelson, C., Sze, S.H., Chenoweth, J., Schwartz, P., Pevzner, P.A., Glass, C., Mandel, G., et al. Corepressor-dependent silencing of chromosomal regions encoding neuronal genes. Science. 2002;298:1747–1752. doi: 10.1126/science.1076469. [DOI] [PubMed] [Google Scholar]
- Maschhoff, K.L., Anziano, P.Q., Ward, P., Baldwin, H.S. Conservation of Sox4 gene structure and expression during chicken embryogenesis. Gene. 2003;320:23–30. doi: 10.1016/j.gene.2003.07.002. [DOI] [PubMed] [Google Scholar]
- Mavropoulos, A., Devos, N., Biemar, F., Zecchin, E., Argenton, F., Edlund, H., Motte, P., Martial, J.A., Peers, B. sox4b is a key player of pancreatic α cell differentiation in zebrafish. Dev. Biol. 2005;285:211–223. doi: 10.1016/j.ydbio.2005.06.024. [DOI] [PubMed] [Google Scholar]
- Moody, S.A., Stein, D.B. The development of acetylcholinesterase activity in the embryonic nervous system of the frog, Xenopus laevis. Brain Res. 1988;467:225–232. doi: 10.1016/0165-3806(88)90026-0. [DOI] [PubMed] [Google Scholar]
- Naya, F.J., Huang, H.P., Qiu, Y., Mutoh, H., DeMayo, F.J., Leiter, A.B., Tsai, M.J. Diabetes, defective pancreatic morphogenesis, and abnormal enteroendocrine differentiation in BETA2/neuroD-deficient mice. Genes & Dev. 1997;11:2323–2334. doi: 10.1101/gad.11.18.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa, H., Yamamura, K., Miyazaki, J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- Novitch, B.G., Chen, A.I., Jessell, T.M. Coordinate regulation of motor neuron subtype identity and pan-neuronal properties by the bHLH repressor Olig2. Neuron. 2001;31:773–789. doi: 10.1016/s0896-6273(01)00407-x. [DOI] [PubMed] [Google Scholar]
- Perron, M., Opdecamp, K., Butler, K., Harris, W.A., Bellefroid, E.J. X-ngnr-1 and Xath3 promote ectopic expression of sensory neuron markers in the neurula ectoderm and have distinct inducing properties in the retina. Proc. Natl. Acad. Sci. 1999;96:14996–15001. doi: 10.1073/pnas.96.26.14996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pevny, L., Placzek, M. SOX genes and neural progenitor identity. Curr. Opin. Neurobiol. 2005;15:7–13. doi: 10.1016/j.conb.2005.01.016. [DOI] [PubMed] [Google Scholar]
- Pevny, L.H., Sockanathan, S., Placzek, M., Lovell-Badge, R. A role for SOX1 in neural determination. Development. 1998;125:1967–1978. doi: 10.1242/dev.125.10.1967. [DOI] [PubMed] [Google Scholar]
- Rao, M., Baraban, J.H., Rajaii, F., Sockanathan, S. In vivo comparative study of RNAi methodologies by in ovo electroporation in the chick embryo. Dev. Dyn. 2004;231:592–600. doi: 10.1002/dvdy.20161. [DOI] [PubMed] [Google Scholar]
- Riederer, B., Matus, A. Differential expression of distinct microtubule-associated proteins during brain development. Proc. Natl. Acad. Sci. 1985;82:6006–6009. doi: 10.1073/pnas.82.17.6006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roztocil, T., Matter-Sadzinski, L., Alliod, C., Ballivet, M., Matter, J.M. NeuroM, a neural helix–loop–helix transcription factor, defines a new transition stage in neurogenesis. Development. 1997;124:3263–3272. doi: 10.1242/dev.124.17.3263. [DOI] [PubMed] [Google Scholar]
- Sandberg, M., Kallstrom, M., Muhr, J. Sox21 promotes the progression of vertebrate neurogenesis. Nat. Neurosci. 2005;8:995–1001. doi: 10.1038/nn1493. [DOI] [PubMed] [Google Scholar]
- Schmidt, K., Glaser, G., Wernig, A., Wegner, M., Rosorius, O. Sox8 is a specific marker for muscle satellite cells and inhibits myogenesis. J. Biol. Chem. 2003;278:29769–29775. doi: 10.1074/jbc.M301539200. [DOI] [PubMed] [Google Scholar]
- Schoenherr, C.J., Anderson, D.J. The neuron-restrictive silencer factor (NRSF): A coordinate repressor of multiple neuron-specific genes. Science. 1995;267:1360–1363. doi: 10.1126/science.7871435. [DOI] [PubMed] [Google Scholar]
- Smith, S.T., Jaynes, J.B. A conserved region of engrailed, shared among all en-, gsc-, Nk1-, Nk2- and msh-class homeoproteins, mediates active transcriptional repression in vivo. Development. 1996;122:3141–3150. doi: 10.1242/dev.122.10.3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sock, E., Rettig, S.D., Enderich, J., Bosl, M.R., Tamm, E.R., Wegner, M. Gene targeting reveals a widespread role for the high-mobility-group transcription factor Sox11 in tissue remodeling. Mol. Cell. Biol. 2004;24:6635–6644. doi: 10.1128/MCB.24.15.6635-6644.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Y., Nadal-Vicens, M., Misono, S., Lin, M.Z., Zubiaga, A., Hua, X., Fan, G., Greenberg, M.E. Neurogenin promotes neurogenesis and inhibits glial differentiation by independent mechanisms. Cell. 2001;104:365–376. doi: 10.1016/s0092-8674(01)00224-0. [DOI] [PubMed] [Google Scholar]
- Tomita, K., Moriyoshi, K., Nakanishi, S., Guillemot, F., Kageyama, R. Mammalian achaete-scute and atonal homologs regulate neuronal versus glial fate determination in the central nervous system. EMBO J. 2000;19:5460–5472. doi: 10.1093/emboj/19.20.5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchida, T., Ensini, M., Morton, S.B., Baldassare, M., Edlund, T., Jessell, T.M., Pfaff, S.L. Topographic organization of embryonic motor neurons defined by expression of LIM homeobox genes. Cell. 1994;79:957–970. doi: 10.1016/0092-8674(94)90027-2. [DOI] [PubMed] [Google Scholar]
- Uwanogho, D., Rex, M., Cartwright, E.J., Pearl, G., Healy, C., Scotting, P.J., Sharpe, P.T. Embryonic expression of the chicken Sox2, Sox3 and Sox11 genes suggests an interactive role in neuronal development. Mech. Dev. 1995;49:23–36. doi: 10.1016/0925-4773(94)00299-3. [DOI] [PubMed] [Google Scholar]
- van Beest, M., Dooijes, D., De van Wetering, M., Kjaerulff, S., Bonvin, A., Nielsen, O., Clevers, H. Sequence-specific high mobility group box factors recognize 10–12-base pair minor groove motifs. J. Biol. Chem. 2000;275:27266–27273. doi: 10.1074/jbc.M004102200. [DOI] [PubMed] [Google Scholar]
- de van Wetering, M., Oosterwegel, M., van Norren, K., Clevers, H. Sox-4, an Sry-like HMG box protein, is a transcriptional activator in lymphocytes. EMBO J. 1993;12:3847–3854. doi: 10.1002/j.1460-2075.1993.tb06063.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z., Benoit, G., Liu, J., Prasad, S., Aarnisalo, P., Liu, X., Xu, H., Walker, N.P., Perlmann, T. Structure and function of Nurr1 identifies a class of ligand-independent nuclear receptors. Nature. 2003;423:555–560. doi: 10.1038/nature01645. [DOI] [PubMed] [Google Scholar]
- Wegner, M. From head to toes: The multiple facets of Sox proteins. Nucleic Acids Res. 1999;27:1409–1420. doi: 10.1093/nar/27.6.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiebe, M.S., Nowling, T.K., Rizzino, A. Identification of novel domains within Sox-2 and Sox-11 involved in autoinhibition of DNA binding and partnership specificity. J. Biol. Chem. 2003;278:17901–17911. doi: 10.1074/jbc.M212211200. [DOI] [PubMed] [Google Scholar]
- Wingate, A.D., Campbell, D.G., Peggie, M., Arthur, J.S. Nur77 is phosphorylated in cells by RSK in response to mitogenic stimulation. Biochem J. 2005;393:715–724. doi: 10.1042/BJ20050967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota, Y. Id and development. Oncogene. 2001;20:8290–8298. doi: 10.1038/sj.onc.1205090. [DOI] [PubMed] [Google Scholar]