Abstract
The regulation of iron is critical for maintaining homeostasis in the tsetse fly (Diptera:Glossinidae), in which both adult sexes are strict blood feeders. We have characterized the cDNAs for two putative iron-binding proteins (IBP) involved in transport and storage; transferrin (GmmTsf1) and ferritin from Glossina morsitans morsitans. GmmTsf1 transcripts are detected in the female fat body and in adult reproductive tissues, and only in the adult developmental stage in a bloodmeal independent manner. In contrast, the ferritin heavy chain (GmmFer1HCH) and light chain (GmmFer2LCH) transcripts are expressed ubiquitously, suggesting a more general role for these proteins in iron transport and storage. Protein domain predictions for each IBP suggest both the conservation and loss of several motifs present in their vertebrate homologues. In concert with many other described insect transferrins, putative secreted GmmTsf1 maintains 3 of the 5 residues necessary for iron-binding in the N-terminal lobe, but exhibits a loss of this iron-binding ability in the C-terminal role as well as a loss of large sequence blocks. Both putative GmmFer1HCH and GmmFer2LCH proteins have signal peptides, similar to other insect ferritins. GmmFer2LCH has lost the 5’UTR iron-responsive element (IRE) and, thus, translation is no longer regulated by cellular iron levels. On the other hand, GmmFer1HCH maintains both the conserved ferroxidase center and the 5’UTR IRE; however, transcript variants suggest a more extensive regulatory mechanism for this subunit.
Keywords: Iron, ferritin, transferrin, Glossina morsitans morsitans
1. Introduction
Iron-binding proteins (IBPs) are ubiquitous in nature because iron transport and delivery to cells for utilization or storage are essential functions in all organisms. IBPs are also vital in sequestering iron where an overabundance can quickly lead to oxidative stress, a process potentially destructive towards membranes, proteins, and nucleic acids. Hematophagous (blood-feeding) insects, such as the tsetse (Diptera: Glossinidae), are particularly vulnerable to iron toxicity. Tsetse are the vectors of the protozoan African trypanosomes, the agents of human sleeping sickness, and both sexes are strict blood feeders. Tsetse has a viviparous reproductive biology where the mother gives rise to a fully mature larva that pupates soon after deposition. Hence, all developmental stages of tsetse rely on the vertebrate blood meal diet for their nutritional needs.
Transferrins (Tfs) and ferritins make up the two major groups of IBPs (Dunkov and Georgieva, 2006, Harizanova et al., 2005, Winzerling and Pham, 2006). Vertebrate transferrins include diverse proteins such as serum Tfs, lactoferrin and ovotransferrin and regulate extracellular iron levels by their capacity to sequester two iron atoms. Acting as a shuttle, transferrin distributes iron to various tissues in the body. The iron-transferrin complex is internalized by cell surface receptor-mediated endocytosis at various tissue sites. The iron is released from transferrin upon acidification of the endosome, and the apo-transferrin is then recycled to the cell surface to resume its role in iron-shuttling (Thorstensen and Romslo, 1990). Other transferrin family members, such as lactoferrin, found in milk, tears, and extracellular fluids, and ovotransferrin, found in egg-whites, can also act as antimicrobial proteins by withholding tightly sequestered ferric ions from pathogens (Ibrahim et al., 1998, Farnaud and Evans, 2003). Similar to vertebrates, insect transferrins have been postulated to have roles in iron transport (Bartfield and Law, 1990), immunity (Yoshiga et al., 1997b, Yoshiga et al., 1999, Yun et al., 1999), and in addition in vitellogenesis (Kurama et al., 1995, Hirai et al., 2000a). The majority of known insect transferrins maintain iron-binding capabilities in the N-terminal lobe of transferrin only. Furthermore, insect genomes have yet to reveal a homolog of the vertebrate transferrin uptake receptor (Law, 2002).
The ferritin molecule is composed of a total of 24 subunits arranged as a spherical structure that can store up to 4,500 iron atoms as an Fe(III) inorganic complex (Chasteen and Harrison, 1999). The ferritin subunits come in the form of either a heavy chain (HC) or a light chain (LC), and the ratio of these subunits will vary according to the requirements of the cell. The heavy chain subunit is characterized by a ferroxidase center that facilitates the rapid oxidation and uptake of iron. The light chain subunit is the site of iron core nucleation and works in concert with the heavy chain subunit to complex iron. Insect ferritins are produced by most tissues and, in contrast to vertebrate ferritins that are cytosolic, are found in the secretory pathways and circulate in the hemolymph (Nichol et al., 2002). The secretion of insect ferritins suggests that they may share the role of extracellular iron-transport proteins with Tfs. Two types of secreted ferritin subunits have been described in insects. The first subunit is a homologue of the vertebrate heavy chain (HCH) and contains a ferroxidase center. The second subunit is homologous to the vertebrate ferritin light chain (LCH), with the characteristic absence of the ferroxidase center.
Here we report on the iron-binding protein homologues, transferrin (GmmTsf1) and both the heavy (GmmFer1HCH) and light (GmmFer2LCH) chain subunits of ferritin, identified from Glossina morsitans morsitans. We describe the molecular characteristics of the cDNAs, patterns of expression for GmmTsf1, GmmFer1HCH and GmmFer2LCH in relation to sex, developmental stage, and tissue specificity. We have investigated the transcriptional response of these IBPs during the blood meal acquisition and digestion process. We discuss our findings in light of tsetse’s restricted diet (vertebrate blood), viviparous reproductive biology and its multiple microbial partners, which are symbiotic and parasitic in nature.
2. Materials and methods
2.1 Tsetse colony
The G. m. morsitans colony maintained at Yale University originated from puparia collected in Zimbabwe, and for colonization puparia were obtained from the International Atomic Energy Agency Laboratories in Seibersdorf, Austria. Flies are maintained at 24 ± 1°C with 50-55% relative humidity and receive defibrinated bovine blood every other day using an artificial membrane feeding system (Moloo, 1971).
2.2 Sequence analysis of Glossina IBPs
GmmTsf1, GmmFer1HCH, and GmmFer2LCH homologues were identified from a G. m. morsitans fat body-specific EST library through a search of Genbank non-redundant database (National Center for Biotechnology Information, Bethesda, Maryland) using BlastX (Altschul et al., 1990, Attardo et al., 2006). Alignment of EST sequences identified in the fat body EST library and G. m. morsitans midgut EST libraries provided the full-length cDNAs for GmmTsf1 and GmmFer1HCH. The fat body EST library provided the full-length cDNA for GmmFer2LCH. Comparative sequence analyses of the full length cDNAs were conducted using the DNASTAR software programs (Lasergene, Madison, WI), SignalPv1.1, NetNGlyc 1.0, and NetOGlyc 3.1 Server (Center for Biological Sequence Analysis, Technical University of Denmark, http://www.cbs.dtu.dk/services/) (Julenius et al., 2005). Molecular weight and isolectric point estimations were performed using the Sequence Manipulation Suite (Stothard, 2000).
Multiple sequence alignments of the deduced amino acid sequences of GmmTsf1, GmmFer1HCH, and GmmFer2LCH with homologous protein sequences were performed with ClustalW of the European Bioinformatics Institute, (http://www.ebi.ac.uk/clustalw). The translated sequences of transferrins from the following taxa were selected for the GmmTsf1 multiple sequence alignment based on the BLASTX alignment output: Drosophila melanogaster (Genbank Accession CG6186), Aedes aegypti (AAB87414), Anopheles gambiae (EAA06303), Manduca sexta (AAA29338), and Blaberus discoidalis (AAA27820). For the GmmFer1HCH multiple sequence alignment, the amino acid sequences from the following taxa were used: D. melanogaster (AAB70121), Ae. aegypti (AAF82348), Manduca sexta (AAK39636), and Caldope ethlius (AAD50239). Lastly, for the GmmFer2LCH multiple sequence alignment, the following amino acid sequences were used: D. melanogaster (AAF07879), Ae. aegypti (AAO41698), M. sexta (AAF44717), and C. ethlius (AAD50241). The molecular phylogeny of insect transferrins was examined by pairwise alignment for GmmTsf1. Pairwise alignment and tree generation was performed using the CLUSTALW program (http://clustalw.genome.jp/). Accession numbers and species names for sequences used in the analysis can be found in Figure 1.
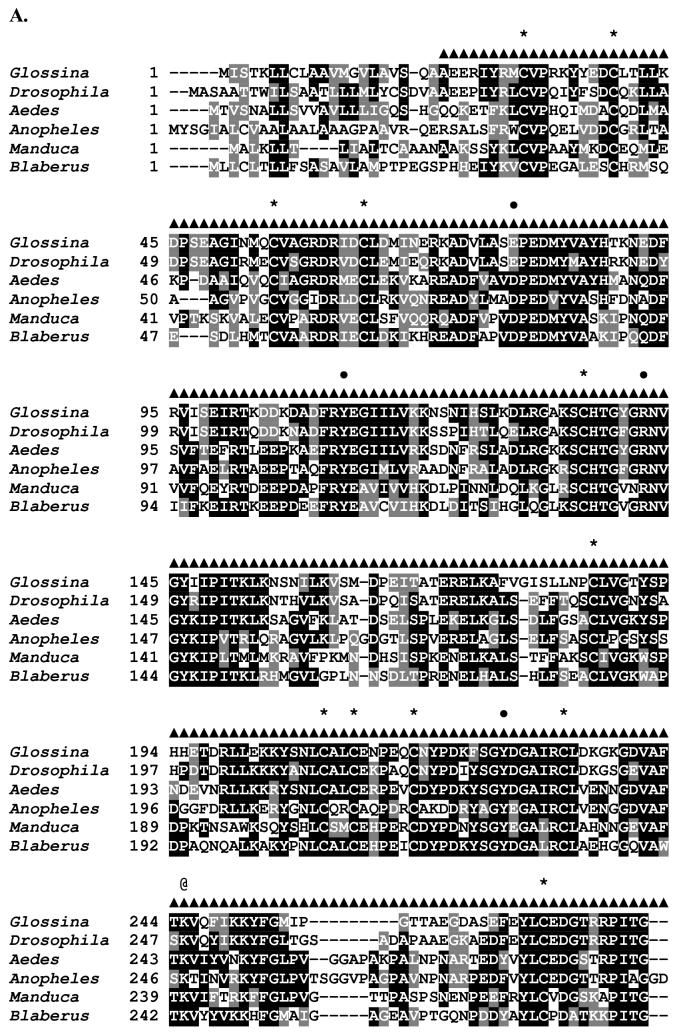
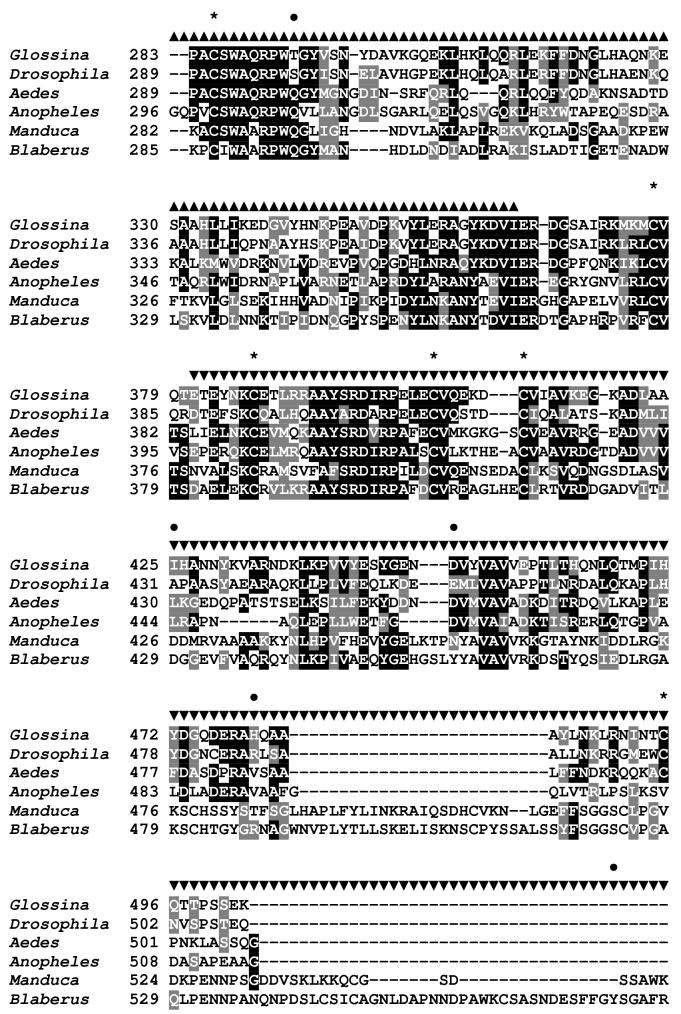
Figure 1.
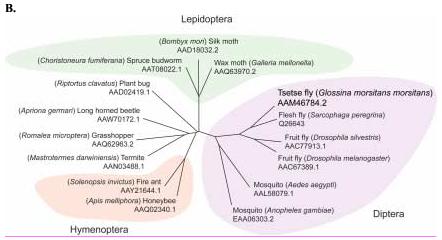
Sequence analysis of the G. m. morsitans transferrin homologue (GmmTsf1).
(A) Multiple alignment of transferrin N-terminal lobe amino acid sequences from the following: G. m. morsitans; D. melanogaster (Genbank Accession: CG6186); Ae. aegypti (AAB87414), An. gambiae (EAA06303), M. sexta (AAA29338), and B. discoidalis (AAA27820). The output for multiple sequence alignments is based on a ClustalW consensus sequence and indicates ≥50% identity (black shading) or ≥50% similarity (gray shading). Amino acid residues necessary for iron binding in vertebrate transferrins are indicated by a ‘•’, a conserved lysine with@, and conserved cysteine residues by ‘*”. (B) Molecular phylogeny of dipteran transferrins. Pairwise alignment and tree generation for GmmTsf1 was performed using the CLUSTALW program (http://clustalw.genome.jp/). Accession numbers for sequences used in the analysis have been included.
2.3 Isolation of RNA and Northern blot analysis.
Total RNA was isolated from all samples using the TRIzol®Reagent (Invitrogen, Carlsbad, CA) according to manufacturer’s instructions. Total RNA was isolated from pooled fat body dissected from 5 individual flies for 3-week old males or 3-week old females. In addition, total RNA was isolated from early larvae (1st and 2nd instars) and late larvae (3rd instars) that were removed surgically from the uterus of pregnant females; from early-pupae (one day after larvaposition) and late-pupae (25-30 day after larvaposition). For post-bloodmeal analysis, total RNA was prepared from individual flies at designated times.
For Northern analysis, 10 μg total RNA was analyzed on a denaturing 2% agarose formaldehyde gel prepared in 1x 3-(N-morpholino) propane-sulfonic acid (MOPS) buffer and transferred to a nylon membrane (Hybond-N+, Amersham Biosciences, Piscataway, NJ) by capillary blotting. A 242 bp GmmTsf1 amplicon, a 1048 bp GmmFer1HCH amplicon and a 407 bp amplicon for glyceraldehyde 3-phosphate dehydrogenase I (GmmGAP), a housekeeping gene, were PCR amplified from fat body cDNA and were used as hybridization probes. PCR amplification reactions contained 50mM KCl, 10mM Tris-HCl (pH 9 at 25°C), 1.5 mM MgCl2, 0.1% TrixonX-100, 0.4 μmol of each primer, 100mM of each dNTP, and 2.5 units of Taq polymerase (Promega, Madison, WI). Reactions were conducted at 94°C 4 min for 1 cycle; 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min for 35 cycles; 72°C for 5 min for 1 cycle. The gene specific primers used for amplification were as follows: for GmmTsf1: TF655F 5’-CCTACTCTCCCCATCACGAA-3’ and TF897R 5’-ATTCAAATTCGGAAGCATCG-3’; for GmmFer1HCH: FHC28F 5’-GCGGGCTGTAATAAACGAAA-3’ and FHC1075R 5’-TGGATAGACCCCATCGCATA-3’; for GmmGAP: GAP23F 5’-TAGATTGGACTGTGCGCTTG-3’ and GAP429R 5’-ATGGGTGGATGGTGAGAGTC-3’ and for GmmTub: TUBβF 5’-ATGAGAGAAATCGTTCATATTCAAGC-3’ and TUBβR 5’-TAGTTCTCTTCAACTTCAGCCTCTT-3’.
The gel-purified amplification products were radiolabeled using Redivue™ [α-32P] dCTP (10mCi/ml) (Amersham Biosciences, Piscataway, NJ) and the Random Primed Labeling Kit (Roche Applied Science, Indianapolis, IN) according to manufacturer’s instructions. Pre-hybridized membranes were incubated at 60°C with each denatured probe in 7% SDS/1% BSA/0.5mM EDTA in 0.5M phosphate buffered solution overnight. The membranes were washed twice for 30 minutes first with a low stringency wash buffer (5% SDS, 40mM phosphate buffer, 1mM EDTA), next with a high stringency wash buffer (1% SDS, 40mM phosphate buffer, 1mM EDTA), and exposed to a phosphorimager screen (Molecular Dynamics, Sunnyvale, CA, USA) overnight. All blots were hybridized to either the GmmGAP or GmmTub probe as internal loading controls, and hybridization signals for GmmTsf1 and GmmFer1HCH were normalized to the control gene signal using Kodak 1D 3.6.1 Imaging Software.
2.4 Assay for transcript abundance in adult tissues using RT-PCR.
For the semi-quantitative RT-PCR assay of transcript abundance, midgut, fat body/milk gland tubules, reproductive tract (ovary, uterus and milk gland) tissues of females and midgut, fat body, and reproductive tissues (including testes and accessory glands) of males were dissected. Total RNA was purified as previously described, and treated with RNase-free DNase I. No DNA could be detected by PCR amplification after treatment (data not shown). Synthesis of cDNAs from RNA of each of the three tissues was performed with the Advantage RT-for-PCR kit (Clontech) using 1 μg of total RNA and oligo (dT)18 according to the manufacturer’s instructions. Initially, PCR-products corresponding to GmmGAP were analyzed from each template cDNA pool serially diluted (1:1, 1:10, 1:100) following amplification for 25, 28, 30, and 35 cycles, respectively. PCR reaction contents were described in the previous section, and the cycling parameters were as follows: 1 cycle at 94°C for 5 min, 92°C for 1 min, 55°C for 1 min, and 72°C for 5 min for 20, 25, or 30 cycles, and a final cycle at 72°C for 5 min. Quantitation of transcript abundance of GmmGAP was performed with the Kodak 1D 3.6.1 Imaging Software. The cDNA dilution and number of cycles to be used for final analysis was chosen empirically where comparable band intensities for GmmGAP amplification were observed in each experiment while avoiding saturation. Amplification was performed for 25 cycles for GmmTsf1 using described oligonucleotides, for GmmFer1HCH with the primers FHC942F:5’-TTGCATGGTCAACGTGATTT-3’ and FHC1418R:5’-TGCTTGCATACAGACCAAGG-3’, which amplify a 477 bp amplicon, and for GmmFer2LCH with the primers FLC147:5’-TTTTTGTTGCATTGCTTGCT-3’ and FLC845:5’-CCTGAAAAGCCATCCTAGTCC-3’, which amplify a 699 bp amplicon. Products were visualized on a 2% agarose gel by ethidium bromide staining.
2.5 Cloning and sequencing of GmmFer1HCH PCR-amplified products
Amplification of fat body GmmFer1HCH was performed using the PCR reaction and cycling parameters described in the previous section and using a primer set designed to amplify a 656 bp product in the 5’-region of the cDNA: FHC28F 5’-GCGGGCTGTAATAAACGAAA-3’and FHC683R 5’-ACTTTTTCAAGGCAGCGAAA -3’. Three products were generated with this primer set at approximately 700 bp, 600 bp, and 450 bp. The three amplicons were excised from a 2.5% agarose gel and purified using the QIAquick Gel Extraction Kit (Qiagen, Valencia, CA) according to manufacturer’s instructions. Purified products were cloned into the pCR 2.1 TOPO vector (Invitrogen, Carlsbad, CA) according to manufacturer’s instructions. Upon confirmation of correct insert size, sequences from two representative clones were obtained using both the forward and reverse M13 vector primers, then aligned with the full-length GmmFer1HCH transcript sequence.
3. Results
3.1 Characterization of G. m. morsitans transferrin (GmmTsf1) cDNA
The 2108 bp full-length cDNA sequence for GmmTsf1 encodes a deduced 634-aa peptide with a molecular mass of approximately 72 kDa and an estimated pI value of 6.90 (Genbank Accession: AF368908). A 21 residue signal sequence is predicted at the N-terminal end of the GmmTsf1 cDNA. A multiple sequence alignment of the deduced amino acid sequence of GmmTsf1 with Tf amino acid sequences of three other dipterans, D. melanogaster, Ae. aegypti, and An. gambiae, as well as the lepidopteran M. sexta and the blattarian B. discoidalis is shown (Figure 1A). In the human Tf amino acid sequence, there are five iron-binding residues (Asp82, Tyr115, Arg142, Tyr207, His268), and two lysines involved in iron-release (Lys225 and Lys315). GmmTsf1 maintains 3 of the 5 conserved N-terminal lobe iron-binding residues (Tyr112, Arg142, Tyr227), but none of the pertinent residues in the C-terminal lobe. The loss of the C-terminal iron-binding residues is a trait shared among the majority of described insect Tfs, except that of the cockroach B. discoidalis (Jamroz et al., 1993) (represented in our alignment) and that of the termite, Mastotermes darwiniensis (Thompson et al., 2003b). The loss of iron-binding in the C-terminal lobe of insect Tfs has been proposed as a means to subvert the theft of iron by microbial pathogens since many of these bacterial transferrin binding receptors interact principally with the C-terminal and not the N-terminal lobe (Yoshiga et al., 1997a). Also typical of insect Tfs, in GmmTsf1 only the first of the two lysine residues (Lys245) is conserved; the second has been replaced with an isoleucine (Ile336). Similarly to D. melanogaster (Yoshiga et al., 1999), GmmTsf1 shows conservation of 13 Cys residues in the N-terminus and 7 in the C-terminus.
A number of iron binding residues in the N-terminal lobe of insect Tfs have been replaced in comparison to their vertebrate homologs. The N-terminal His residue (His268) of vertebrate transferrins has been replaced with Gln in insects, including Ae. aegypti and An. gambiae. Gln is similarly neutral but a poor electron donor, thus, its substitution for His results in an easier release of iron. Despite the weaker iron binding properties of Gln, it is presumed that the lack of the pH-sensitive dilysine interaction in insect Tfs counterbalances this weaker binding. S. peregrina and G. m. morsitans both have a Thr in place of the His (Thr293 in GmmTsf1), while D. melanogaster and D. silvestris have a Ser in this position. The binding capacity of the Thr and Ser residues in place of His has not been determined quantitatively. His contributes to the binding of Fe by donating lone pairs of electrons to the coordinate bond, which Ser and Thr cannot do. Thus, iron would likely still bind to this site, but with a much lowered affinity. The second type of replacement is that of the conserved Asp of vertebrate Tfs with Glu in G. m. morsitans (Glu79) and in two other higher order dipterans, D. melanogaster (Glu83) (9) and S. peregrina (Glu78) (7). Both Asp and Glu are negatively charged, polar amino acids that easily substitute for one another and, together with the two Tyr residues, would balance the charge of the Fe (III) ion (52).
The molecular phylogeny of insect Tfs was examined using amino acid sequences of Tfs from a number of insect orders including, Diptera, Lepidoptera, Hymenoptera, Orthoptera, Coleoptera, Hemiptera and Isoptera (Figure 1B). Within the dipteran transferrins, G. m. morsitans Tf shows the highest amino acid identity to S. peregrina (76.6%) and D. silvestris (62.5%), followed by Ae. aegypti (45.3%) and An. gambiae (37.5%). The phylogeny of the Tfs closely follows the dipteran phylogeny, clustering the three dipterans D. melanogaster, G. m. morsitans, and S. peregrina of the suborder Brachycera, and clustering Ae. aegypti and An. gambiae of the suborder Nematocera. Within the Brachyceran dipteran cluster, the two members of the Calyptrata section of the S. peregrina and G. m. morsitans are grouped separately from D. melanogaster, which is a member of the Acalyptrata section. This division refers to the development of the calypteres (small lobes(s) at the base of the wing), which are well developed in the Calyptrate section (Borror et al., 1992).
3.2 Characterization of G. m. morsitans GmmFer2LCH and GmmFer1HCH homologues.
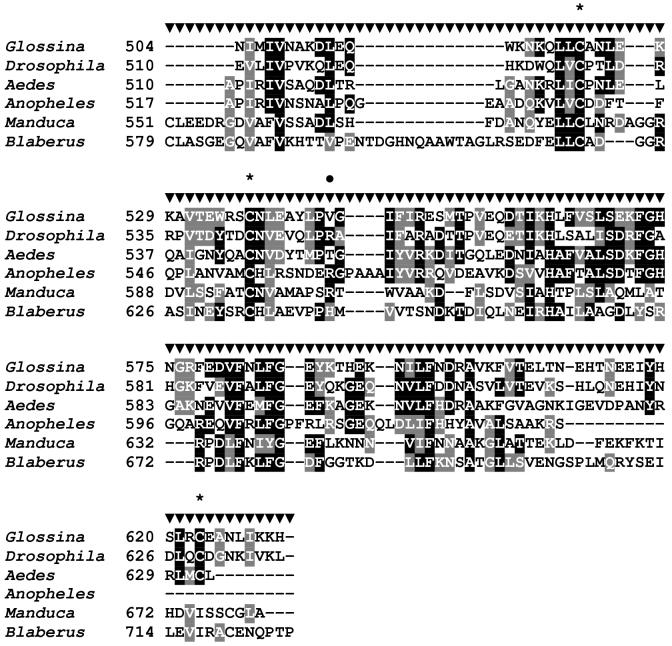
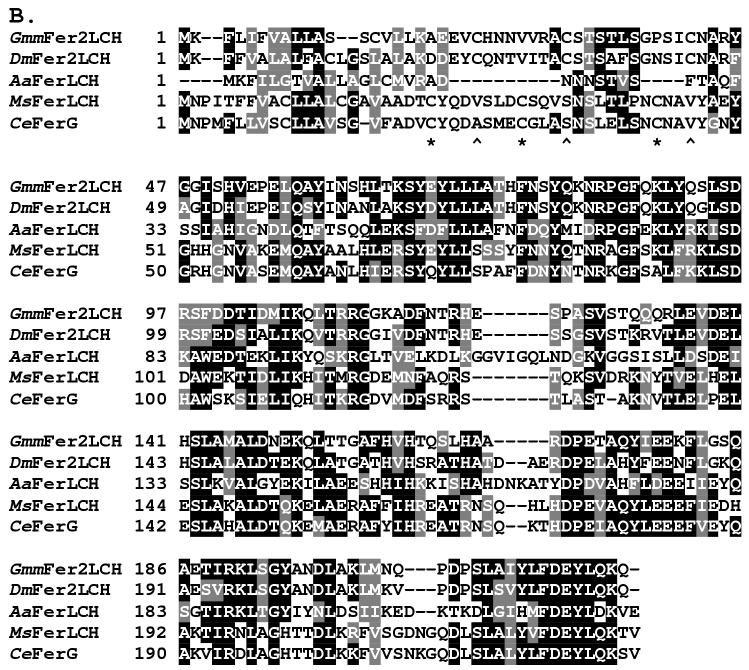
The 1,167 bp cDNA sequence of the GmmFer2LCH homologue encodes a 222 aa protein, with a 19 aa signal peptide predicted at the N-terminus, an estimated molecular weight of 25.04 kDa and an estimated pI value of 6.79 (Genbank Accession:AF368916) (Figure 2A). Insect LCHs typically lack the ferroxidase center that defines the vertebrate and insect heavy chain subunits (Dunkov and Georgieva, 1999). Multiple sequence alignment of GmmFer2LCH with other insect LCH homologues shows the highest identity with D. melanogaster (DmFer2LCH, 72%). All other identities fall within a similar range, 34.2 % Ae. aegypti (AaFerLCH), 40.5 % M. sexta (MsFerLCH), and 39.6% C. ethlius (CeFerG). In M. sexta (Asp135) (Pham et al., 1996) and C. ethlius (Asp133) (Nichol and Locke, 1999) LCH homologues have a functional glycosylation site (Figure 2B). With the exception of the unusual placement of an N-glycosylation site at the beginning of the Ae. aegypti mature LCH peptide (Asp22/23) (Geiser et al., 2003), known dipteran LCH homologues lack glycosylation, including G. m. morsitans (Lind et al., 1998a). Previously published alignments of insect LCHs, including that of D. melanogaster, maintain conservation of three Cys residues (Kim et al., 2004b, Kim et al., 2004a). In our alignment, GmmFer2LCH maintains the three conserved Cys residues in the N-terminus in alignment with D. melanogaster; however, in our alignment, the D. melanogaster sequence does not align with the conserved Cys residues of other insect sequences as was described previously (Kim et al., 2004b).
Figure 2.
Sequence analysis of the G. m. morsitans ferritin light chain homologue (GmmFer2LCH). (A) Full length cDNA and deduced amino acid sequence for GmmFLC. The signal peptide sequence is underlined, the two polyadenylation signals are indicated by a string of asterisks ‘*’, and the oligonucleotide sequences used in the tissue-specific semi-quantitative RT-PCR analysis are in bold and labeled as FLC149F (forward primer) and FLC847R (reverse primer). (B) Multiple sequence alignment of amino acid sequences for ferritin light chain homologues from G. m. morsitans (GmmFLC), D. melanogaster (DmFer2LCH) (Genbank Accession: AAF07879), Ae. aegypti (AaFerLCH) (AAO41698), M. sexta (MsFerLCH) (AAF44717), and C. ethlius (CeFerG) (AAD50241). The output for multiple sequence alignments is based on a ClustalW consensus sequence and indicates ≥50% identity (black shading) or ≥50% similarity (gray shading). Conserved cysteine residues are marked with asterisks ‘*” and the corresponding cysteine residues in D. melanogaster and G. m. morsitans with karats ‘∧’.
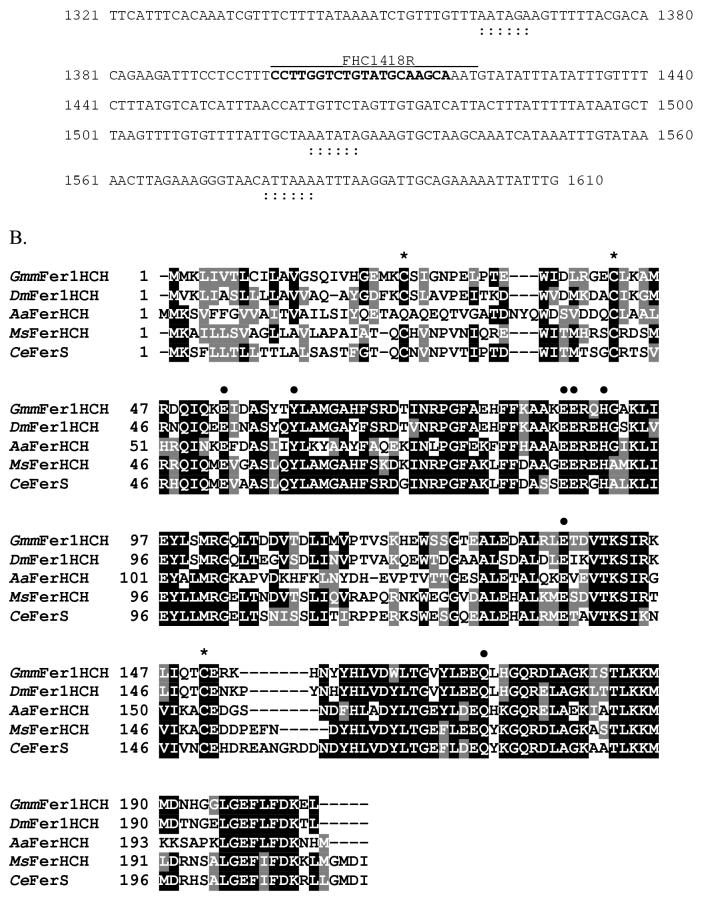
The 1610 bp full-length cDNA sequence for GmmFer1HCH encodes a 205 aa peptide, with a 20 aa signal peptide at the N-terminus, an estimated molecular weight of 23.42 kDa, and an estimated pI value of 6.23 (Genbank Accession: DQ294233) (Figure 3A). The defining feature of HCH subunits is the conservation of seven amino acid residues, including four Glu, one Tyr, one His, and one Gln, that form the ferroxidase center in the assembled molecule (Lawson et al., 1989). All of these residues are conserved in GmmFer1HCH (Figure 3B, Glu53, Tyr60, Glu87, Glu88, His91, Glu137, and Gln172). In addition, there are typically 3 conserved Cys residues in the heavy chain, 2 in the N-terminus and one in the C-terminus, all of which are present in GmmFer1HCH (Cys24, 42, and 151). Multiple sequence alignment of GmmFer1HCH with other insect heavy chain homologues shows an identity of 71% with D. melanogaster DmFer1HCH, 55.1% with M. sexta MsFerHCH, 54.6% with C. ethlius CeFerS and 42.9% with Ae. aegypti AaFerHCH.
Figure 3.
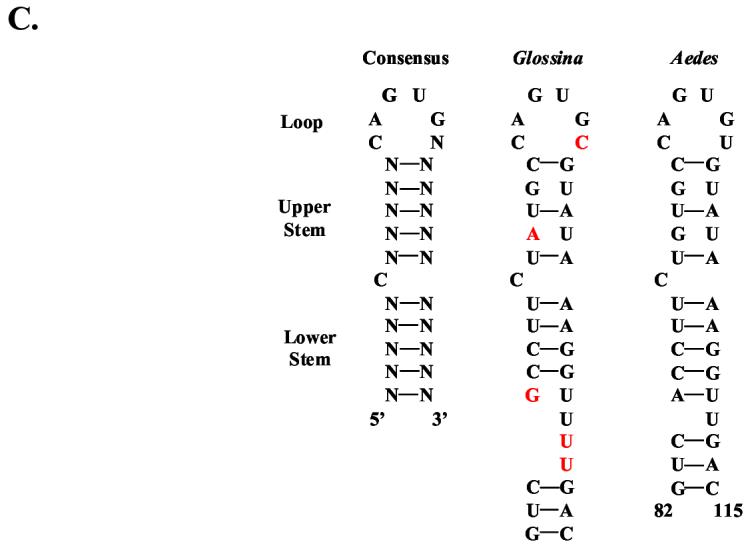
Sequence analysis of the G. m. morsitans ferritin heavy chain homologue (GmmFer1HCH). (A) Full-length cDNA and deduced amino acid sequence for GmmFer1HCH. The IRE is indicated by a string of karats ‘∧’ and the consensus loop is in bold and double-underlined. The signal peptide sequence is underlined, the polyadenylation signal is indicated by a string of asterisks ‘*’, and the putative non-canonical polyadenylation signals (Georgieva et al., 2002, Geiser et al., 2003) are indicated by a string of colons ‘:’. The oligonucleotide sequences used in the tissue-specific semi-quantitative RT-PCR analysis are in bold and labeled as FHC942F (forward primer) and FHC1418R (reverse primer). The oligonucleotide sequences used to amplify the 656 bp region that resulted in multiple transcripts are in bold and labeled as FHC28F (forward primer) and FHC683R (reverse primer). (B) Multiple alignment of amino acid sequences for ferritin heavy chain homologues from G. m. morsitans (GmmFer1HCH), D. melanogaster (DmFer1HCH) (Genbank Accession: AAB70121), A. aegypti (AaFerHCH) (AAF82348), M. sexta (MsFerHCH) (AAK39636), and C. ethlius (CeFerS) (AAD50239). The output for multiple sequence alignments is based on a ClustalW consensus sequence and indicates ≥50% identity (black shading) or ≥50% similarity (gray shading). The seven conserved amino acid residues that form the ferroxidase center are marked with dark circles ‘•’, and conserved cysteine residues are marked with asterisks ‘*’. (C) A comparison of the iron-responsive elements (IREs) of G. m. morsitans and Ae. aegypti with the consensus IRE motif. The motif consists of a hexanucleotide loop (5’-CAGUGN-3’) and a stem, interrupted by a bulge with an unpaired C residue.
In many organisms, ferritin mRNAs are post-transcriptionally regulated by the interaction of a stem loop structure in the 5’UTR that acts as an iron-responsive element (IRE) (Pantopoulos, 2004). When cellular iron is low, the an iron-regulatory protein (IRP) binds to the IREs and inhibits translation of the mRNAs. The IRE motif consists of two elements that are both required for the interaction with the IRP, a hexanucleotide loop (5’-CAGUGN-3’) and a stem interrupted by a bulge with an unpaired C nucleotide (Hentze and Kuhn, 1996, Huang et al., 1996, Pham, 2002, Theil, 1994). GmmFer1HCH contains an IRE that is highly similar in sequence to the stem loop structure in the Ae. aegypti HCH homologue (Figure 3C). In contrast, the GmmFer2LCH cDNA sequence does not contain an IRE sequence (Figure 2A). The lack of an IRE in the GmmFer2LCH cDNA sequence is in agreement with LCH homologues characterized from two other dipteran species, D. melanogaster and Ae. aegypti (Georgieva et al., 2002, Geiser et al., 2003).
Modified putative polyadenylation signals have been identified in cDNAs encoding HC and/or LC ferritin subunits from M. sexta (Pham et al., 1996, Zhang et al., 2001), C. ethlius (Nichol and Locke, 1999), Ae. aegypti (Dunkov et al., 2002), Galleria mellonella (Kim et al., 2002) and the moth Hyphantria cunea (Kim et al., 2004b). The cDNA sequence of the Ae. aegypti HCH subunit shows four overlapping, non-canonical, putative polyadenylation signals proximal to the coding region (Dunkov et al., 2002), and D. melanogaster uses two polyadenylation signals in both HC and LC subunits to produce messages of different lengths (Georgieva et al., 1999, Dunkov et al., 2002). Similar to D. melanogaster, the GmmFer2LCH cDNA sequence indicates two possible polyadenylation signals (Figure 2A). Conversely, only one canonical polyadenylation signal was identified in GmmFer1HCH, though several non-canonical sites were identified (Figure 3A), one of which is similar to a non-canonical site in Ae. aegypti and the rest of which are similar to non-canonical sites identified in H. cunea (Kim et al., 2004a). The use of these dual polyadenylation signals in GmmFer2LCH and of the non-canonical sites in GmmFer1HCH for the production of messages of different lengths has not yet been confirmed.
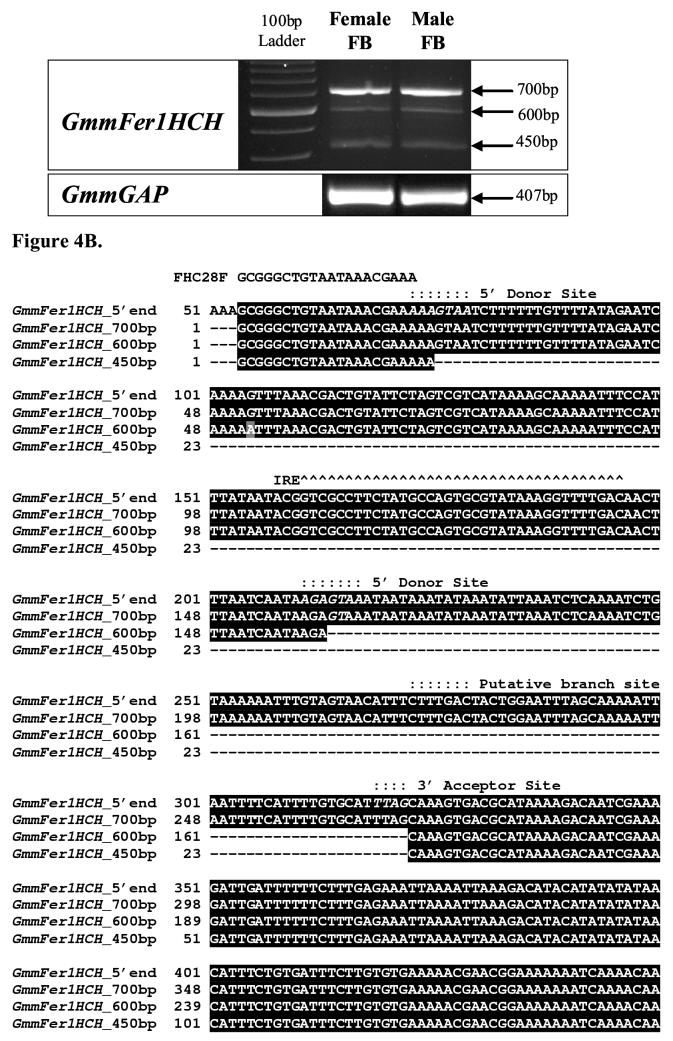
3.3 Presence of multiple transcripts of GmmFer1HCH.
Three GmmFer1HCH transcripts at amplicon sizes of approximately 450 bp, 600 bp, and 700 bp were amplified from male and female fat body using a primer set designed to amplify a 656 nt region spanning the 5’-untranslated region of GmmFer1HCH cDNA (Figure 4A). Multiple-sized transcripts for the HCH subunit have been identified in both D. melanogaster and Ae. aegypti and are a result of a complex mechanism of transcriptional regulation involving multiple transcriptional start sites, alternative splicing and/or multiple polyadenylation signals (Lind et al., 1998a, Pham et al., 1999, Dunkov et al., 2002, Geiser et al., 2003b). Thus, the presence of multiple GmmFer1HCH transcripts was suggestive of a potentially similar transcriptional regulation mechanism.
Figure 4.
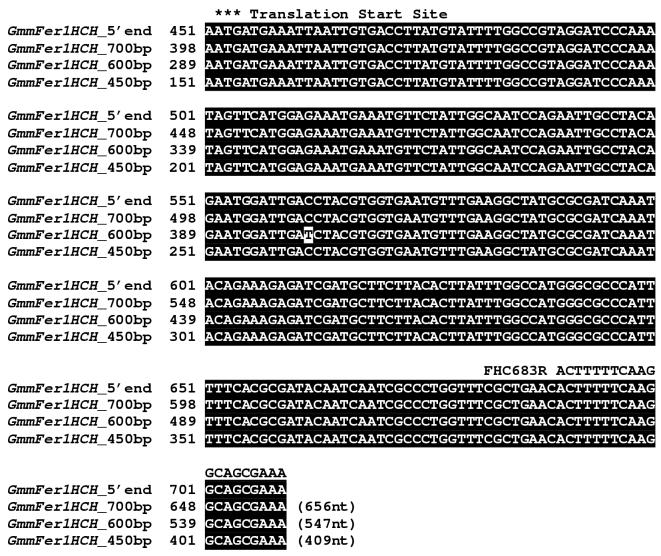
Multiple transcripts of G. m. morsitans ferritin heavy chain homologue. (A) Semi-quantitative RT-PCR of G. m. morsitans fat body (FB) mRNA using a primer set designed to amplify 656 bp region of the GmmFer1HCH cDNA sequence produced three different cDNA products. GmmGAP specific primers were used as a control. (B) Alignment of the complete GmmFer1HCH nucleotide sequence with sequences of the multiple RT-PCR amplification products at approximately 700, 600, and 450bp. The oligonucleotide sequences used in RT-PCR of the multiple GmmFer1HCH transcripts are underlined and labeled as FHC28F (forward primer) and FHC683R (reverse primer). The iron-responsive element is marked with karats, ∧, the translation start site with asterisks, *, the putative branch site with colons,:, and the putative donor and acceptor sites are in italics.
Sequencing results indicated that the largest of the amplified fragments corresponds to the 656 bp full-length GmmFer1HCH transcript containing the IRE sequence (Figure 4B). The approximately 450bp amplified fragment corresponds to a 409 nt region of the GmmFer1HCH transcript, which, as predicted, no longer contains the IRE stem-loop sequence. Unexpectedly, the ∼600bp band represents a 547nt region of the GmmFer1HCH transcript that maintains the IRE stem-loop sequence, but lacks a 109nt sequence downstream of the IRE stem-loop (Figure 4B). Both the 547nt and the 409nt sequences resume at nucleotide position 297 of the 656 nt region matching the full-length transcript. Similar to the alternative splicing described for D. melanogaster, it appears that the IRE is maintained within an intron, and both the 656nt and the 547nt cDNAs retain this intron.
All three splice sites follow the GT-AG rule (M, 1992, Mount, 1982). The two competing 5’ donor splice sites both begin with GT at the 5’end of the intron, and the acceptor splice site ends with AG at the 3’ end of the intron (in italics in Figure 4B). The GmmFer1HCH 5’ and 3’ splice sites are not wholly consistent with the consensus sequences typically surrounding the conserved GT-AG (Mount, 1982). The reported consensus 5’ donor splice site is (A/C)AG-GT(A,G)AGT, while the first GmmFer1HCH 5’ splice site is AAA-GTAATC and the second site is AGA-GTAAAT. The consensus sequence for the 3’ acceptor site is N(C/T)AG and is preceded by a pyrimidine rich region, (C/T)n. GmmFer1HCH’s 3’ acceptor splice site is TTAG, which is preceded by a pyrimidine rich region between the branch site and the 3’ acceptor site. The branch site, which contains a conserved A that is the site of lariat formation with the 5’G of the intron, is approximately 20-50bp upstream of the acceptor site. In the case of GmmFer1HCH a putative branch site, CTTTGAC, is located 50bp upstream and agrees with the consensus sequence reported for branch sites, (C/T)N(C/T)T(A/G)A(C/T) (Mount, 1982, Helfman and Ricci, 1989).
3.4 Transcript abundance of GmmTsf1 and GmmFer1HCH during the life cycle of tsetse.
The expression of GmmTsf1 and GmmFer1HCH was assessed in adult male and female G. m. morsitans and in developmental stages. Detection of GmmFer2LCH transcripts was attempted on Northern blots; however, the transcript abundance was presumably too low for detection. Significant differences in expression were detected for GmmTsf1 and GmmFer1HCH (Figure 5A). In particular, GmmTsf1 transcripts were detected only in the adult stage of the tsetse, not in any of the developmental stages. In contrast, GmmFer1HCH transcripts were found in all stages of development, from early larva to adult.
Figure 5.
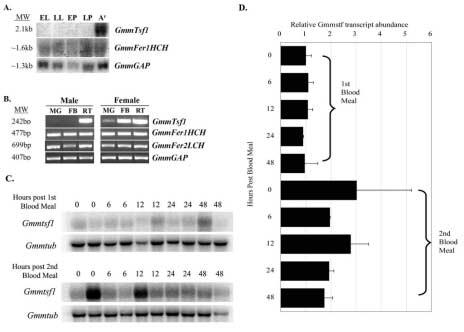
Transcript abundance of transferrin and ferritin during life stages and in isolated tissues of G. m. morsitans. (A). Northern blot analysis of G. m. morsitans RNA from early larva (EL), late larva (LL), early pupa (EP), late pupa (LP), and adult female fat body (AF). (B). Tissue-specific expression of GmmTsf1, GmmFer1HCH, and GmmFer2LCH in adult male and female tsetse identified by semi-quantitative RT-PCR analysis. Amplification products were generated using cDNAs from midgut (MG), fat body (FB) and reproductive tissues (RT) as templates. (C). Northern blot analysis of GmmTsf1 transcript abundance in whole flies analyzed at 0, 6, 12, 24 and 48 hours after blood feeding. Flies were collected from points after their first blood meal post eclosion and acquisition of their second blood meal. The tsetse tubulin gene GmmTub was used in hybridizations as a loading control. (D). Quantification of GmmTsf1 expression levels. Transcript abundance for the Northern data in (C) was quantified and then normalized against the GmmTub loading control.
A semi-quantitative RT-PCR analysis using different tissues indicated that GmmTsf1 transcripts were abundant in adult female fat body and reproductive tract (Figure 5B). Unlike GmmTsf1, GmmFer1HCH and GmmFer2LCH transcripts could be detected in both male and female midgut, fat body and reproductive tissues. To evaluate whether GmmTsf1 expression was regulated by bloodmeal both in newly emerged (teneral) flies and during the process of digestion, GmmTsf1 expression was analyzed in a time course study. Results indicate that GmmTsf1 expression can be detected before blood meal acquisition in teneral adults but its abundance increases upon multiple blood feeding (Figure 5C and D). It remains to be seen whether translational control for the expression of transferrin protein exists in teneral flies and during bloodmeal acquisition and digestion.
4. Discussion
We report here on the molecular characterization of cDNAs encoding two major iron-binding proteins, ferritin and transferrin, as well as the associated features of their putative translated products, their expression patterns and their inferred biological roles in G. m. morsitans. The organisms residing within an adult tsetse can include three bacterial symbionts (Wigglesworthia, Sodalis, and Wolbachia), viviparous progeny, as well as invading trypanosomes. Iron, which is an essential nutrient for nearly all living organisms, is required by the fly itself and by all of these organisms that develop within it. Presumably, iron is abundant in both male and female tsetse as a result of their blood meal. Nevertheless, the fly must be able to balance its own iron requirements with those of its dependents. Iron-binding proteins are fundamentally iron transporters and storage proteins. Beyond these basic functions, these proteins may play a larger role in the protection against cytotoxicity associated with tsetse’s hematophagous lifestyle and in the defense against microbial theft of valuable iron resources.
Given that both adult sexes of tsetse are strictly hematophagous, the female-specific nature of transferrin expression in fat body tissue was unexpected and suggested additional roles for this protein in tsetse biology. GmmTsf1 exhibits a bias towards tissues associated with reproduction, specifically the ovaries and fat body in the female. Despite careful dissections, it is possible that the fat body material analyzed may have been contaminated by milk gland tubules. This maternal transferrin perhaps plays a role in reproductive success by supplying iron to the intrauterine larva during development. Insect embryos exhibit a varying level of maternal dependency where, in addition to the substances supplied by nurse cells and/or follicle cells during oogenesis, many proteins (vitellogenin, lipophorin, insecticyanin, heme-binding protein) are taken up into the eggs from the maternal hemolymph (Kang et al., 1995, van Antwerpen et al., 1998, Machado et al., 1998, Van Antwerpen et al., 1993). Transferrin was identified as one such hemolymph protein transferred to eggs during vitellogenesis in the fleshfly, S. peregrina (Kurama et al., 1995). Once transported to the oocyte, the transferrin probably releases the iron, and a larger protein, putatively ferritin, binds and stores the iron in the egg. In concert with this idea of a maternal supply of transferrin are the patterns of expression in D. melanogaster (Harizanova et al., 2005) and in Ae. aegypti (Yoshiga et al., 1999). Transferrin mRNA is detected in neither the embryonic stage of either insect nor in the early mosquito larval stage. Furthermore, the association of transferrin expression with certain insects’ reproductive cycles, which are regulated by the hormones ecdysone and juvenile hormone (two hormones critical in development), suggests that transferrin could be regulated by these hormones (Hirai et al., 2000b). Given GmmTsf1’s association with reproductive tissues, the hormonal regulation of GmmTsf1 is a compelling avenue for future research. Given the viviparous reproductive biology of tsetse, it also remains to be seen if GmmTsf protein is maternally transmitted to the intrauterine larva since no transcripts were detected in the immature developmental stages. Initially, transferrin was thought inconsequential to adult male tsetse because the expression of GmmTsf1 in male fat body was quite low. However, subsequent RT-PCR analysis of specific tissues isolated from adult males showed a significant amount of transcript in the reproductive tissues. Transferrin in male tsetse may possibly act as an iron shuttle in reproductive tissues similarly to transferrin in mammalian testes (Sylvester and Griswold, 1994). Mammalian transferrin synthesized by Sertoli cells, or nurse cells, has been hypothesized to act as an iron shuttle system designed to transport ferric ions to germ cells by crossing the blood-testis barrier through tight cellular junctions (Sylvester and Griswold, 1994), which is further supported by the mutant mouse model that is unable to synthesize transferrin and, as a result, is defective in spermatogenesis (Bernstein, 1987).
Several lines of evidence suggest that transferrin acts as an antimicrobial agent by sequestering iron from invading microbes (Ibrahim et al., 1998, Farnaud and Evans, 2003, Aguilera et al., 2003). Transferrin, in particular, has been identified as an immune-responsive molecule following microbial challenge (Yoshiga et al., 1997b, Yoshiga et al., 1999, Yun et al., 1999, Thompson et al., 2003a, Beerntsen et al., 1994, Ampasala et al., 2004). The identification of motifs associated with the binding of the transcriptional control element, NF-κB, identified in the Ae. aegypti transferrin gene, further implicates transferrin as a component of immune defense. It remains to be seen if parasite challenge or established infections in tsetse influence GmmTsf transcription. The concept of transferrin’s role in immune defense is appealing not only for its potential role in host-microbe interactions, but also as prophylaxis during copulation and reproduction, particularly given the association of GmmTsf1 expression with the reproductive tissues of tsetse. Without an active defense mechanism, microbes that are introduced into the female reproductive tract during mating (Mueller et al., 2004) could result in infections that negatively impact reproductive success. Male Drosophila are known to transfer at least four antimicrobial peptides or proteins to females during copulation (Samakovlis et al., 1991, Lung et al., 2001). Similarly, if transferrin is supplied by male tsetse to female tsetse during copulation, it could act as a prophylactic agent within the female genital tract.
In contrast to transferrin, ferritin mRNA is expressed in both male and female tsetse more ubiquitously and during all developmental stages. Similarly ferritin is detected commonly throughout the life history of insects such as H. cunea (Kim et al., 2004b), D. melanogaster (Georgieva et al., 2002, Lind et al., 1998a, Georgieva et al., 1999), and Ae. aegypti (Geiser et al., 2003b, Pham et al., 2003). Furthermore, the signal peptide sequences identified for both GmmFer1HCH and GmmFer2LCH signify that tsetse ferritin serves to not only store, but also to transport iron; a role reserved for transferrin in vertebrate organisms.
GmmFer1HCH and GmmFer2LCH cDNAs show significant similarities to both D. melanogaster and Ae. aegypti. Multiple transcripts detected for GmmFer1HCH reveal several transcript variants, including one without an IRE. The production of transcript variants in tsetse may closely mimic what has been observed in Drosophila. Three different 3’ splice acceptor sites in D. melanogaster’s ferritin heavy chain transcript allow for four mRNA transcripts, only one of which retains the IRE sequence (Lind et al., 1998a). Moreover, iron enrichment shifts the population of mRNAs towards those lacking the IRE (Georgieva et al., 1999). An unexpected find was the GmmFer1HCH transcript variant that maintained the IRE, but was missing a portion of the 5’UTR downstream of the IRE. It has been shown that the proximity of the IRE to the mRNA cap site influences the translational repression of ferritin by the IRE/IRP interaction (Muckenthaler et al., 1998, Paraskeva et al., 1999). When the IRE is less than 60 nt away from the mRNA cap site, IRP binding of the IRE prevents recruitment of the eIF4F complex, inhibiting translation completely (Muckenthaler et al., 1998). When the IRE is farther than 60 nt from the mRNA cap site, ribosomes can bind to the mRNA and scan for a translational start codon, thus translation is repressed only partially (Paraskeva et al., 1999). What is not known, however, is whether the proximity of the IRE to the translational start codon can also affect the efficiency of translational repression by the IRP/IRE interaction. It remains to be seen in tsetse whether there is equal binding efficiency of the IRE by the IRP in the truncated GmmFer1HCH transcript maintaining the IRE sequence. Once available, an analysis of the GmmFer1HCH genomic DNA would aid in the determination of intron placement and confirm the location of these potential splicing factors.
The upregulatory response of ferritin in Ae. aegypti females during a bloodmeal, but not during a sugar meal, supports the hypothesis that ferritin reduces the threat of iron toxicity (Geiser et al., 2003a). Furthermore, the subunits of the ferritin shell have different functions. The heavy chain’s ferroxidase center catalyzes the oxidation of ferrous iron, thus playing a role in iron detoxification, while the light chain serves in iron core nucleation, mineralization, and long-term iron storage (Lawson et al., 1991). Thus, an effective way to thwart iron toxicity would be to produce more of the ferritin heavy chain that rapidly oxidizes free iron. D. melanogaster’s diet is rarely rich in iron and the expression of the ferritin light chain subunit tends to exceed that of the heavy chain subunit, in order to store rapidly the iron that is made available (Georgieva et al., 2002). In contrast, transcript levels of the ferritin heavy chain subunit are greater than that of the light chain in Ae. aegypti. Similarly normalization of GmmFer2LCH transcripts in semi-quantitative RT-PCR analysis against GmmFer1HCH indicates lower GmmFer2LCH transcript levels (data not shown). Furthermore, repeated analysis of GmmFer2LCH expression on the identical Northern blots used for GmmFer1HCH expression analysis failed to detect GmmFer2LCH, indicating a possible bias towards higher transcript levels for the heavy chain subunit in tsetse.
The acquisition of iron is essential for most pathogens. Trypanosomes have adapted elegantly to the microenvironments of both the human and tsetse. Bloodstream form trypanosomes use a transferrin uptake receptor to steal iron from the vertebrate host’s transferrin (Borst and Fairlamb, 1998). The lack of detection of this transferrin receptor expression in insect stage trypanosomes may not be surprising given the evolutionary loss of the C-terminal lobe in insect transferrins. However, since all organisms require iron, trypanosomes within tsetse may utilize an alternative and uncharacterized iron-acquiring mechanism. Although the modes of iron acquisition used by tsetse’s symbionts, Wigglesworthia glossinidia and Sodalis glossinidius, remain to be characterized, both genomes have been sequenced and provide clues for how iron metabolism occurs in these symbionts. Despite the drastic reduction in genome size in the obligate intracellular Wigglesworthia, it retains the potential to synthesize protoheme (Akman et al., 2002). If functional, this protein would require a source of iron, which we speculate might come from iron-storage proteins such as ferritin. The Sodalis genome sequence revealed two possible mechanisms for iron acquisition, including a heme transport system and a siderophore-mediated iron-binding system (Toh et al., 2006). The heme transport system possibly could be used for scavenging heme during bloodmeal digestion or to scavenge iron from the heme-producing Wigglesworthia during periods of low iron availability. The siderophore-mediated iron acquisition system might be used during blood digestion when free iron is readily available in the gut. It remains to be seen whether these two iron recruitment systems are functional.
Acknowledgements
This research was supported by grants from the NIH/NIAID-AI51584 and Robert Leet and Clara Gutherie Patterson Trust awards to S.A. The authors would like to thank Dr. Rhoel R. Dinglasan and Brian Weiss for their critical reading of the manuscript.
References
- Aguilera O, Quiros LM, Fierro JF. Transferrins selectively cause ion efflux through bacterial and artificial membranes. FEBS Letters. 2003;548:5–10. doi: 10.1016/s0014-5793(03)00719-1. [DOI] [PubMed] [Google Scholar]
- Akman L, Yamashita A, Watanabe H, Oshima K, Shiba T, Hattori M, Aksoy S. Genome sequence of the endocellular obligate symbiont of tsetse, Wigglesworthia glossinidia. Nat. Genet. 2002;32:402–407. doi: 10.1038/ng986. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of Molecular Biology. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Ampasala DR, Zheng SC, Retnakaran A, Krell PJ, Arif BM, Feng QL. Cloning and expression of a putative transferrin cDNA of the spruce budworm, Choristoneura fumiferana. Insect Biochemistry and Molecular Biology. 2004;34:493–500. doi: 10.1016/j.ibmb.2004.03.002. [DOI] [PubMed] [Google Scholar]
- Attardo G, Strickler-Dinglasan P, Perkin S, Caler E, Bonaldo M, Soares M, El-Sayeed N, Aksoy S. Analysis of fat body transcriptome from the adult tsetse fly, Glossina morsitans morsitans. insect Mol Biology. 2006;15:411–424. doi: 10.1111/j.1365-2583.2006.00649.x. [DOI] [PubMed] [Google Scholar]
- Bartfield NS, Law JH. Isolation and molecular cloning of transferrin from the tobacco hornworm, Manduca sexta. Journal of Biological Chemistry. 1990;265:21684–21691. [PubMed] [Google Scholar]
- Beerntsen BT, Severson DW, Christensen BM. Aedes aegypti: Characterization of a hemolymph polypeptide expressed during melanotic encapsulation of filarial worms. Experimental Parasitology. 1994;79:312–321. doi: 10.1006/expr.1994.1094. [DOI] [PubMed] [Google Scholar]
- Bernstein SE. Hereditary hypotransferrinemia with hemosiderosis, a murine disorder resembling human atransferrinemia. Journal of Laboratory and Clinical Medicine. 1987;110:690–705. [PubMed] [Google Scholar]
- Borror DJ, Triplehorn CA, Johnson NF. In An Introduction to the Study of Insects. Harcourt Brace College Publishers; Fort Worth: 1992. pp. 499–575. [Google Scholar]
- Borst P, Fairlamb AH. Surface receptors and transporters of Trypanosoma brucei. Annual Review in Microbiology. 1998;52:745–78. doi: 10.1146/annurev.micro.52.1.745. [DOI] [PubMed] [Google Scholar]
- Chasteen ND, Harrison PM. Mineralization in ferritin: An efficient means of iron storage. Journal of Structural Biology. 1999;126:182–194. doi: 10.1006/jsbi.1999.4118. [DOI] [PubMed] [Google Scholar]
- Dunkov B, Georgieva T. Insect iron binding proteins: Insights from the genomes. Insect Biochem Mol Biol. 2006;36:300–9. doi: 10.1016/j.ibmb.2006.01.007. [DOI] [PubMed] [Google Scholar]
- Dunkov BC, Georgieva T. Organization of the ferritin genes in Drosophila melanogaster. DNA Cell Biol. 1999;18:937–44. doi: 10.1089/104454999314791. [DOI] [PubMed] [Google Scholar]
- Dunkov BC, Georgieva T, Yoshiga T, Hall M, Law JH. Aedes aegypti ferritin heavy chain homologue: feeding of iron or blood influences message levels, lengths, and subunit abundance. Journal of Insect Science. 2002;2:1–9. doi: 10.1093/jis/2.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnaud S, Evans RW. Lactoferrin--a multifunctional protein with antimicrobial properties. Molecular Immunology. 2003;40:395–405. doi: 10.1016/s0161-5890(03)00152-4. [DOI] [PubMed] [Google Scholar]
- Geiser DL, Chavez CA, Flores-Munguia R, Winzerling JJ, Pham DQD. Aedes aegypti ferritin. A cytotoxic protector against iron and oxidative challenge? European Journal of Biochemistry. 2003;270:3667–3674. doi: 10.1046/j.1432-1033.2003.03709.x. [DOI] [PubMed] [Google Scholar]
- Georgieva T, Dunkov BC, Dimov S, Ralchev K, Law JH. Drosophila melanogaster ferritin: cDNA encoding a light chain homologue, temporal and tissue specific expression of both subunit types. Insect Biochemistry and Molecular Biology. 2002;32:295–302. doi: 10.1016/s0965-1748(01)00090-x. [DOI] [PubMed] [Google Scholar]
- Georgieva T, Dunkov BC, Harazanova N, Ralchev K, Law JH. Iron availability dramatically alters the distribution of ferritin subunit messages in Drosophila melanogaster. Proceedings of the National Academy of Sciences USA. 1999;96:2716–2721. doi: 10.1073/pnas.96.6.2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harizanova N, Georgieva T, Dunkov BC, Yoshiga T, Law JH. Aedes aegypti transferrin. Gene structure, expression pattern, and regulation. Insect Mol Biol. 2005;14:79–88. doi: 10.1111/j.1365-2583.2004.00533.x. [DOI] [PubMed] [Google Scholar]
- Helfman DM, Ricci WM. Branch point selection in alternative splicing of tropomyosin pre-mRNAs. Nucleic Acids Res. 1989;17:5633–50. doi: 10.1093/nar/17.14.5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentze MW, Kuhn LC. Molecular control of vertebrate iron metabolism: mRNA-based regulatory circuits operated by iron, nitric oxide and oxidative stress. Proceedings of the National Academy of Sciences USA. 1996;93:8175–8182. doi: 10.1073/pnas.93.16.8175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai M, Watanabe D, Chinzei Y. A juvenile hormone-repressible transferrin-like protein from the Bean Bug, Riptortus clavatus: cDNA sequence analysis and protein identification during diapause and vitellogenesis. Archives of Insect Biochemistry and Physiology. 2000a;44:17–26. doi: 10.1002/(SICI)1520-6327(200005)44:1<17::AID-ARCH3>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Hirai M, Watanabe D, Chinzei Y. A juvenile hormone-repressible transferrin-like protein from the bean bug, Riptortus clavatus: cDNA sequence analysis and protein identification during diapause and vitellogenesis. Arch Insect Biochem Physiol. 2000b;44:17–26. doi: 10.1002/(SICI)1520-6327(200005)44:1<17::AID-ARCH3>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Huang TS, Law JH, Soderhall K. Purification and cDNA cloning of ferritin from hepatopancreas of the freshwater crayfish Pacifastacus leniusculus. European Journal of Biochemistry. 1996;236:450–456. doi: 10.1111/j.1432-1033.1996.00450.x. [DOI] [PubMed] [Google Scholar]
- Ibrahim HR, Iwamori E, Sugimoto Y, Aoki T. Identification of a distinct antibacterial domain within the N-lobe of ovotransferrin. Biochimica et Biophysica Acta. 1998;1401:289–303. doi: 10.1016/s0167-4889(97)00132-8. [DOI] [PubMed] [Google Scholar]
- Jamroz RC, Gasdaska JR, Bradfield JY, Law JH. Transferrin in a cockroach: molecular cloning, characterization, and suppression by juvenile hormone. Proc Natl Acad Sci U S A. 1993;90:1320–4. doi: 10.1073/pnas.90.4.1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julenius K, Molgaard A, Gupta R, Brunak S. Prediction, conservation analysis, and structural characterization of mammalian mucin-type O-glycosylation sites. Glycobiology. 2005;15:153–64. doi: 10.1093/glycob/cwh151. [DOI] [PubMed] [Google Scholar]
- Kang Y, Kulakosky PC, van Antwerpen R, Law JH. Sequestration of insecticyanin, a blue hemolymph protein, into the egg of the hawkmoth Manduca sexta. Evidence for receptor-mediated endocytosis. Insect Biochem Mol Biol. 1995;25:503–10. doi: 10.1016/0965-1748(94)00090-l. [DOI] [PubMed] [Google Scholar]
- Kim BS, Lee CS, Seol JY, Yun CY, Kim HR. Cloning and expression of 32 kDa ferritin from Galleria mellonella. Archives of Insect Biochemistry and Physiology. 2002;47:8–17. doi: 10.1002/arch.10050. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Yun CY, Cheon HM, Chae B, Lee IH, Park SJ, Kang YJ, Seo SJ. Hyphantria cunea Ferritin Heavy Chain Homologue: cDNA sequence and mRNA expression. Archives of Insect Biochemistry and Physiology. 2004a;56:21–33. doi: 10.1002/arch.10141. [DOI] [PubMed] [Google Scholar]
- Kim SR, Lee KS, Yoon HJ, Park NS, Lee SM, Kim I, Seo SJ, Sohn JD, Jin BR. Molecular cloning, expression and characterization of cDNAs encoding the ferritin subunits from the beetle, Apriona germari. Comparative Biochemistry and Physiology, Part B. 2004b;138:423–433. doi: 10.1016/j.cbpc.2004.05.008. [DOI] [PubMed] [Google Scholar]
- Kurama T, Kurata S, Natori S. Molecular characterization of an insect transferrin and its selective incorporation into eggs during oogenesis. European Journal of Biochemistry. 1995;228:229–235. [PubMed] [Google Scholar]
- Law JH. Insects, oxygen, and iron. Biochem Biophys Res Commun. 2002;292:1191–5. doi: 10.1006/bbrc.2001.2015. [DOI] [PubMed] [Google Scholar]
- Lawson DM, Artymiuk PJ, Yewdall SJ, Smith JM, Livingstone JC, Treffry A, Luzzago A, Levi S, Arosio P, Cesareni G, Thomas CD, Shaw WV, Harrison PM. Solving the structure of human H ferritin by genetically engineering intermolecular crystal contacts. Nature. 1991;349:541–544. doi: 10.1038/349541a0. [DOI] [PubMed] [Google Scholar]
- Lawson DM, Treffry A, Artymiuk PJ, Harrison PM, Yewdall SJ, Luzzago A, Cesareni G, Levi S, Arosio P. Identification of the ferroxidase centre in ferritin. FEBS Letters. 1989;254:207–210. doi: 10.1016/0014-5793(89)81040-3. [DOI] [PubMed] [Google Scholar]
- Lind MI, Ekengren S, Melefors O, Soderhall K. Drosophila ferritin mRNA: alternative RNA splicing regulates the presence of the iron-responsive element. FEBS Letters. 1998a;436:476–482. doi: 10.1016/s0014-5793(98)01186-7. [DOI] [PubMed] [Google Scholar]
- Lind MI, Ekengren S, Melefors O, Soderhall K. Drosophila ferritin mRNA: alternative RNA splicing regulates the presence of the iron-responsive element. FEBS Lett. 1998b;436:476–82. doi: 10.1016/s0014-5793(98)01186-7. [DOI] [PubMed] [Google Scholar]
- Lung O, Kuo L, Wolfner MF. Drosophila males transfer antibacterial proteins from their accessory gland and ejaculatory duct to their mates. Journal of Insect Physiology. 2001;47:617–622. doi: 10.1016/s0022-1910(00)00151-7. [DOI] [PubMed] [Google Scholar]
- M M. Alternative mRNA Splicing. Annual Review of Cell Biology. 1992;8:133–155. doi: 10.1146/annurev.cb.08.110192.001025. [DOI] [PubMed] [Google Scholar]
- Machado EA, Oliveira PL, Moreira MF, de Souza W, Masuda H. Uptake of Rhodnius heme-binding protein (RHBP) by the ovary of Rhodnius prolixus. Archives of Insect Biochemistry and Physiology. 1998;39:133–143. doi: 10.1002/(SICI)1520-6327(1998)39:4<133::AID-ARCH1>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Moloo SK. An artificial feeding technique for Glossina. Parasitology. 1971:63. doi: 10.1017/s0031182000080021. [DOI] [PubMed] [Google Scholar]
- Mount SM. A catalogue of splice junction sequences. Nucleic Acids Res. 1982;10:459–72. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muckenthaler M, Gray NK, Hentze MW. IRP-1 binding to ferritin mRNA prevents the recruitment of the small ribosomal subunit by the cap-binding complex eIF4F. Molecular Cell. 1998;2:383–8. doi: 10.1016/s1097-2765(00)80282-8. [DOI] [PubMed] [Google Scholar]
- Mueller JL, Ripoll DR, Aquadro CF, Wolfner MF. Comparative structural modeling and influence of conserved protein classes in Drosophila seminal fluid. Proceedings of the National Academy of Sciences USA. 2004;101:13542–13547. doi: 10.1073/pnas.0405579101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichol H, Law JH, Winzerling JJ. Iron Metabolism in Insects. Annual Review of Entomology. 2002;47:535–559. doi: 10.1146/annurev.ento.47.091201.145237. [DOI] [PubMed] [Google Scholar]
- Nichol H, Locke M. Secreted ferritin subunits are of two kinds in insects: Molecular cloning of cDNAs encoding two major subunits of secreted ferritin from Calpodes ethlius. Insect Biochemistry and Molecular Biology. 1999;29:999–1013. doi: 10.1016/s0965-1748(99)00076-4. [DOI] [PubMed] [Google Scholar]
- Pantopoulos K. Iron Metabolism and the IRE/IRP Regulatory System. Annals of the New York Academy of Sciences. 2004;1012:1–13. doi: 10.1196/annals.1306.001. [DOI] [PubMed] [Google Scholar]
- Paraskeva E, Gray NK, Schlager B, Wehr K, Hentze MW. Ribosomal pausing and scanning arrest as mechanisms of translational regulation from cap-distal iron-responsive elements. Molecular and Cellular Biology. 1999;19:807–16. doi: 10.1128/mcb.19.1.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham DQD. Molecular modeling of insect ferritins. In Silico Biology. 2002;2:S31–S44. [PubMed] [Google Scholar]
- Pham DQD, Shaffer JJ, Chavez CA, Douglass PL. Identification and mapping of the promoter for the gene encoding the ferritin heavy-chain homologue of the yellow fever mosquito Aedes aegypti. Insect Biochemistry and Molecular Biology. 2003;33:51–62. doi: 10.1016/s0965-1748(02)00167-4. [DOI] [PubMed] [Google Scholar]
- Pham DQD, Winzerling JJ, Dodson MS, Law JH. Transcriptional control is relevant in the modulation of mosquito ferritin synthesis by iron. European Journal of Biochemistry. 1999;266:236–240. doi: 10.1046/j.1432-1327.1999.00849.x. [DOI] [PubMed] [Google Scholar]
- Pham DQD, Zhang D, Hufnagel DH, Winzerling JJ. Manduca sexta hemolymph ferritin: cDNA sequence and mRNA expression. Gene. 1996;172:255–259. doi: 10.1016/0378-1119(96)00012-1. [DOI] [PubMed] [Google Scholar]
- Samakovlis C, Kylsten P, Kimbrell DA, Engstrom A, Hultmark D. The andropin gene and its product, a male-specific antibacterial peptide in Drosophila melanogaster. The EMBO Journal. 1991;10:163–9. doi: 10.1002/j.1460-2075.1991.tb07932.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stothard P. The sequence manipulation suite: JavaScript programs for analyzing and formatting protein and DNA sequences. Biotechniques. 2000;28:1102–1104. doi: 10.2144/00286ir01. [DOI] [PubMed] [Google Scholar]
- Sylvester SR, Griswold MD. The testicular iron shuttle: a “nurse” function of the Sertoli cells. Journal of Andrology. 1994;15:381–5. [PubMed] [Google Scholar]
- Theil EC. Iron Regulatory elements (IREs): a family of mRNA non-coding sequences. Biochemical Journal. 1994;304:1–11. doi: 10.1042/bj3040001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson GJ, Ching Crozier Y, Crozier RH. Isolation and characterization of a termite transferrin gene up-regulated on infection. Insect Molecular Biology. 2003a;12:1–7. doi: 10.1046/j.1365-2583.2003.00381.x. [DOI] [PubMed] [Google Scholar]
- Thompson GJ, Crozier YC, Crozier RH. Isolation and characterization of a termite transferrin gene up-regulated on infection. Insect Mol Biol. 2003b;12:1–7. doi: 10.1046/j.1365-2583.2003.00381.x. [DOI] [PubMed] [Google Scholar]
- Thorstensen K, Romslo I. The role of transferrin in the mechanism of cellular iron uptake. Biochem J. 1990;271:1–9. doi: 10.1042/bj2710001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh H, Weiss BL, Perkin SA, Yamashita A, Oshima K, Hattori M, Aksoy S. Massive genome erosion and functional adaptations provide insights into the symbiotic lifestyle of Sodalis glossinidius in the tsetse host. Genome Res. 2006;16:149–56. doi: 10.1101/gr.4106106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Antwerpen R, Conway R, Law JH. Protein and lipoprotein uptake by developing oocytes of the hawkmoth Manduca sexta. An ultrastructural and immunocytochemical study. Tissue Cell. 1993;25:205–18. doi: 10.1016/0040-8166(93)90020-l. [DOI] [PubMed] [Google Scholar]
- van Antwerpen R, Salvador K, Tolman K, Gentry C. Uptake of lipids by developing oocytes of the hawkmoth Manduca sexta. The possible role of lipoprotein lipase. Insect Biochem Mol Biol. 1998;28:399–408. doi: 10.1016/s0965-1748(98)00012-5. [DOI] [PubMed] [Google Scholar]
- Winzerling JJ, Pham DQ. Iron metabolism in insect disease vectors: Mining the Anopheles gambiae translated protein database. Insect Biochem Mol Biol. 2006;36:310–21. doi: 10.1016/j.ibmb.2006.01.006. [DOI] [PubMed] [Google Scholar]
- Yoshiga T, Georgieva T, Dunkov BC, Harizanova N, Ralchev KL,JH. Drosophila melanogaster transferrin. Cloning, deduced protein sequence, expression during the life cycle, gene localization and up-regulation on bacterial infection. European Journal of Biochemistry. 1999;260:414–420. doi: 10.1046/j.1432-1327.1999.00173.x. [DOI] [PubMed] [Google Scholar]
- Yoshiga T, Hernandez VP, Fallon AM, Law JH. Mosquito transferrin, an acute-phase protein that is up-regulated upon infection. Proc Natl Acad Sci U S A. 1997a;94:12337–42. doi: 10.1073/pnas.94.23.12337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshiga T, Hernandez VP, Fallon AM, Law JH. Mosquito transferrin, an acute-phase protein that is up-regulated upon infection. Proceedings of the National Academy of Sciences USA. 1997b;94:12337–12342. doi: 10.1073/pnas.94.23.12337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun EY, Kang SW, Hwang JS, Goo TW, Kim SH, Jin BR, Kwon O-Y, Kim KY. Molecular cloning and characterization of a cDNA encoding a transferrin homolog from Bombyx mori. Biological Chemistry. 1999;380:1455–1459. doi: 10.1515/BC.1999.188. [DOI] [PubMed] [Google Scholar]
- Zhang D, Albert DW, Kohlhepp P, DQ DP, Winzerling JJ. Repression of Manduca sexta ferritin synthesis by IRP1/IRE interaction. Insect Molecular Biology. 2001;10:531–9. doi: 10.1046/j.0962-1075.2001.00293.x. [DOI] [PubMed] [Google Scholar]