Abstract
TRPM2 channels, activated by adenosine diphosphoribose (ADPR) and related molecules, are assembled as oligomers and most likely tetramers. However, the molecular determinants driving the subunit interaction and assembly of the TRPM2 channels are not well defined. Here we examined, using site-directed mutagenesis in conjunction with co-immunoprecipitation and patch clamp recording, the role of a coiled-coil domain in the intracellular C-terminus of TRPM2 subunit in the subunit interaction and the channel assembly. Deletion of the coiled-coil domain resulted in severe disruption of the subunit interaction and substantial loss of the ADPR-evoked channel currents. Individual or combined mutations to glutamine of the hydrophobic residues at positions a and d of the abcdef heptad repeat, key residues for protein-protein interaction, significantly reduced the subunit interaction and the channel currents; the mutational effects on the subunit interaction and the channel currents were clearly correlated. Furthermore, deletion of the coiled-coil domain in a pore mutant subunit abolished its dominant negative phenotypic functional suppression. These results provide strong evidence that the coiled-coil domain is critically engaged in the TRPM2 subunit interaction and such interaction is required for assembly of functional TRPM2 channel. The coiled-coil domain, which is highly conserved within the TRPM subfamily, may serve as a general structural element governing the assembly of TRPM channels.
Mammalian transient receptor potential (TRP) proteins, homologues of the Drosophila TRP proteins (1), form a large group of cation channels that are activated via diverse mechanisms and serve numerous physiological functions. They are subdivided into TRPC, TRPV, TRPM, TRPP, TRPML and TRPA subfamilies based on sequence relatedness (2–5). TRPM2 channel, the second member of TRPM subfamily, is activated by ADPR and related molecules, and also by exposure to oxidative stress and to warm temperature (6–11). High expression of the TRPM2 channels is well documented in excitable cells and in particular in the brain (12–14), although the physiological roles are largely elusive. TRPM2 shows widespread expression in non-excitable cells, where the TRPM2 channels constitute a calcium entry system (7, 8, 11, 15–17).
All the TRP family proteins including TRPM2 have a membrane topology similar to the voltage-gated potassium channels, calcium-activated channels and cyclic nucleotide-gated channels; each subunit comprises six transmembrane segments (S1–S6), a pore loop between the S5 and S6, and intracellular N- and C-termini (2–4,18). TRPM2 subunit contains an N-terminal calmodulin binding site, which mediates functional regulation of the TRPM2 channel by calmodulin (19), and a TRP motif in the proximal C-terminal part. Unique to the TRPM2 subunit, the distal C-terminal part shares substantial homology to the NUDT9 proteins (thus termed NUDT9-H domain). Between the TRP motif and NUDT9-H domain is a region showing a strong propensity of coiled-coil formation (Fig.1A).
Fig. 1. TRPM2 coiled-coil domain is not required for protein expression.
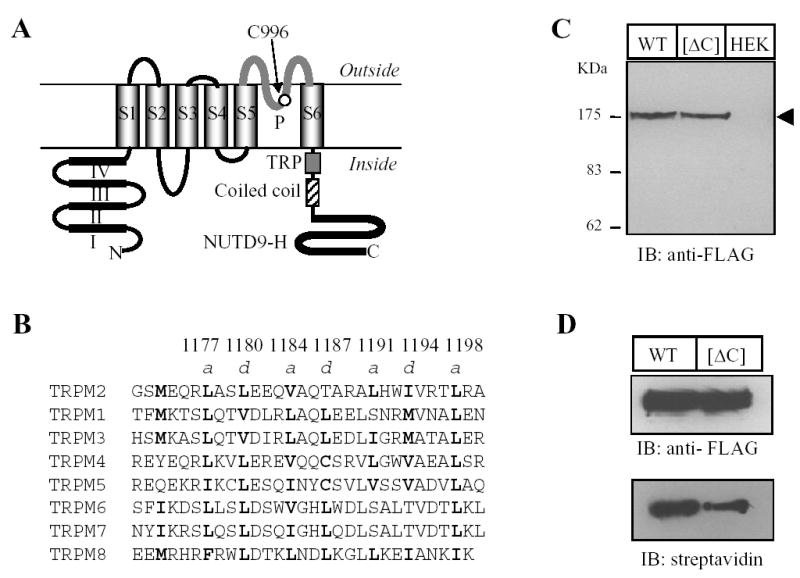
A. Schematic representation of the membrane topology of the human TRPM2 subunit, showing the cytosolic N- and C-termini, six membrane-spanning segments (S1 to S6) and the pore region (P). The N-terminus contains four domains (I – IV) that are highly conserved with the TRPM subfamily, and the C-terminus has a TRP motif in the proximal C-terminal region, a coiled-coil domain (Gly1171 to Ala1200, see panel B) and a NUTD9-H domain in the distal C-terminal region. Cys996 in the pore region is indicated. B. Sequence alignment of the human TRPM subunit C-terminal coiled-coil domains. Highlighted in bold are the residues at the position a and d of the abcdef heptad repeats. The numbers above denotes the residues in the TRPM2 subunit. C–D. Western blot of whole cell lysates (C) or total and biotin-labelled TRPM2 (D) from cells expressing the FLAG-tagged wild type (WT) or coiled-coil domain deletion mutant ([ΔC]) or untransfected cells (HEK). The arrow head indicates the TRPM2 protein. Similar results were observed in two dependent experiments. The arrow heads indicate the TRPM2 proteins (also applied in other figures). E–F. Confocal images showing the immunoreactivities of TRPM2 and calreticulin (E), or TRPM2 and TGN46 proteins (F). The scales are 10 μM. Note that there was clear co-localization of TRPM2 proteins with calreticulin but not with TGN46.
TRPM2 channels presumably form tetramers, like the TRPV channels (20–22). Despite the overall structural similarity, rather surprisingly studies on the TRPV subfamily have revealed distinct molecular domains within different locations that mediate the TRPV subunit interaction and the channel assembly. Thus, for example, a C-terminal coiled-coil domain, located immediately after S6, directs the subunit interaction and tetramerization of the TRPV1 channel (23), but a later study points the importance towards both the transmembrane and the C-terminal domains in the TRPV1 channel assembly (24). For the TRPV4 channel, the N-terminal ankyrin domains play a key role in subunit multimerization (25), whereas both N- and C-terminal domains contribute to the TRPV5 channel assembly (26). Compared to the TRPV channels, the molecular domains mediating the subunit interaction and the assembly of TRPM channels (and other TRP subfamilies) are less clear. A previous study implied a role of the C-terminus in the subunit interaction and assembly of the functional TRPM4 channel (27). Coiled-coil, a common protein-protein interaction domain (28, 29), has been shown to direct the subunit interaction and assembly of several ion channels, including the ether-a-go-go potassium channel (30), the KCNQ potassium channel (31), the TRPV1 channel (23), and very recently the TRPM8 channel (32). Here, by using molecular biology in conjunction with biochemistry and electrophysiology approaches, we provide evidence to indicate that the C-terminal coiled-coil domain is engaged in the subunit interaction and the assembly of the TRPM2 channel.
EXPERIMENTAL PROCEDURES
Cell and molecular biology
Human embryonic kidney (HEK) 293 cells were maintained in Dulbecco’s modified Eagle medium supplemented with 2 mM glutamine and 10% heat-inactivated foetal bovine serum (Invitrogen) at 37ºC under 5% CO2 humidified conditions. Transient transfection was performed using lipofectamin2000 reagents according to the manufacturer’s instructions (Invitrogen). The cDNA encoding the hTRPM2 protein with a C-terminal Glu-Glu (or EE) epitope tag (EYMPME) was subcloned in pcDNA3 vector in a previous study (33). The plasmid expressing a C-terminal FLAG-tagged hTRPM2 protein was made as follows. The sequence from the initiation codon to the SfiI site (fragment A) of the TRPM2 subunit was amplified using a forward primer (5’-ATG GAG CCC TCA GCC CTG AGG AAA GC) and a reverse primer (5’-GCG CTC GTT GTG GAT GAG G), and the sequence from the SfiI site to the stop codon (fragment B) was amplified using a forward primer (5’-GTG TGG GTG GTG TCC TTC) and a reverse primer (5’-AAAC TCA CTT GTC ATC GTC GTC CTT GTA GTC TCT AGA GTA GTG AGC CCC GG, the underlined sequence for the FLAG tag). The PCR products were separately inserted into pCR2.1 vector (Invitrogen). The fragment A was excised using HindIII and SfiI, and the fragment B using SfiI and EcoRV, and then ligated with pcDNA 3.1/Myc-His/A vector (Invitrogen), which was prior restricted using HindIII and PmeI. The C-terminal cMyc-tagged TRPM2 construct was made using similar protocols. Deletion of the coiled-coil domain (residues from 1167 to 1201) was performed by overlapping extension PCR (34), and site-directed mutagenesis using the QuikChange II procotols (Stratagene). All constructs were confirmed by sequencing.
Electrophysiological recording
As previously described, whole cell recordings were carried out using an Axopatch 200B amplifier at room temperature (32). Patch pipettes were fabricated from borosilicate glass capillaries with a resistance of 4–6 MΩ. Extracellular solutions contained 147 mM NaCl, 2 mM KCl, 1 mM MgCl2, 2 mM CaCl2, 10 mM HEPES and 13 mM glucose. Intracellular solution contained 147 mM NaCl, 50 μM EGTA, 10 mM HEPES, 1 mM ATP and 1 mM ADPR. Cells were held at −40 mV, and voltage ramps from −120 mV to 80 mV with 1 s duration were applied every 5–10 s. Recordings started before changing the cell attached configuration into whole cell configuration, seen as an increase in the transient capacitance currents at the beginning and the end of the voltage ramp, and thus no capacitance and resistance compensation was made. Flufenamic acid (FFA) (0.5 mM) was used via a rapid solution changer RSC-160 system (Biologic Science Instruments, France).
Co-immunoprecipitation, biotin-labeling and Western blotting
These experiments were performed as detailed previously (33). In brief, HEK293 cells were prepared in T25 flask (~ 3 x 106 cells), transfected with 3 μg plasmid, and were used 24 hr later. For co-immunoprecipitation, cells were collected and lysed in 300 μl lysis buffer (50 mM Tris-HCl pH8.0, 150 mM NaCl, 2 mM EGTA, 1% Triton X-100 and 5% glycerol) supplemented with protease inhibitor cocktail (Roche) at 4°C for 30 min. After cleared by centrifugation, the cell lysates were mixed with 3 μl anti-FLAG M2 antibody (Sigma) and then incubated at 4°C for 2 hr. Upon addition of 20 μl Protein A/G-agarose bead suspension (Santa Cruz), the mix continued to incubate overnight. Following extensive washing with the lysis buffer, the agarose beads were re-suspended in 40 μl electrophoresis buffer (50 mM Tris-HCl pH 7.4, 100 mM DTT, 2% SDS, 10% glycerol and 0.05% bromophenol blue). Protein in the cell lysates or immunoprecipitated samples were separated on 10% SDS-PAGE gels, transferred onto nitrocellular membranes, and detected using mouse anti-FLAG M2 (1:1000, Sigma), rabbit anti-EE (1:2000, Bethyl), or mouse anti-cMyc antibody (1:500, Santa Cruz) primary antibodies, and horseradish peroxidase (HRP) conjugated anti-mouse or anti-rabbit IgG secondary antibodies (1:2000, Santa Cruz). Proteins were visualized using a supersignal west pico chemiluminescence system (Pierce), and the protein intensities were quantified using a gel doc XR quantity system (Biorad). The subunit interaction was determined by the intensity of the EE-tagged wild type subunit co-immunoprecipitated with the FLAG-tagged mutant subunits, normalized to the intensity of the EE-tagged wild type subunit co-immunoprecipitated with the FLAG-tagged wild type subunit. The variation in the protein expression level was corrected by normalizing the intensity of the mutant subunits to that of the parental wild type subunit in each parallel experiment.
For biotin-labelling, cells were washed extensively with phosphate buffer solution (PBS) (136 mM NaCl, 1.4 mM KCl, 10 mM Na2HPO4, 1.7 mM KH2PO4, pH 8.0) and incubated with in PBS solution containing 1 mg/ml sulfo-NHS-biotin (Pierce) for 30 min at 4°C. Then the cells were washed with PBS containing 50 mM glycine and lysed in 300 μl lysis buffer (see above) supplemented with protease inhibitor cocktail (Roche) at 4°C for 30 min. The cell lysates were immunoprecipitated with anti-FLAG M2 antibody (Sigma) and the immunoprecipitated proteins were analysed by Western blotting as described above using HRP conjugated streptavidin (1:1000, Pierce) and mouse anti-FLAG M2 antibody (1:1000, Sigma) to detect the proteins at the cell surface and the total amount of proteins respectively.
Immunostaining
Cells were fixed in Zamboni’s fixative at room temperature for 30 min. For double staining of TRPM2 and calreticulin (a ER protein marker), cells expressing the FLAG-tagged wild type or [ΔC] subunit, after permeabilized with 2% Triton X-100 and blocked with 5% goat serum, were incubated with mouse anti-calreticulin antibody (1:500, Stressgen) at 4°C overnight, and then with a FITC-conjugated goat anti-mouse IgG antibody (1:500, Sigma) for 2 hr. After extensively rinsed, the cells were incubated with a rabbit anti-FLAG antibody (1:500, Sigma) for 3–4 hr at room temperature, and then with a TRITC-conjugated goat anti-rabbit IgG antibody (1:500, Sigma) for 2 hr. For double staining of FLAG-tagged TRPM2 and TGN46 (a trans-Golgi network protein), cells expressing the FLAG-tagged wild type or [ΔC] subunits, after permeabilized with Triton X-100 and blocked with 5% donkey serum (Sigma), were incubated with a sheep anti-TGN46 antibody (1:400) at 4°C overnight, and then with FITC-conjugated a donkey anti-sheep IgG antibody (1:500, Jackson ImmunoResearch Laboratories) for 2 hr. After extensively rinsed, the cells were incubated with a mouse anti-FLAG antibody (1:500, Sigma) for 3–4 hr at room temperature, and then with a TRITC-conjugated goat anti-mouse IgG antibody (1:500, Sigma) for 2 hr. For double staining of cMyc-tagged wild type and FLAG-tagged [ΔC] mutant TRPM2 proteins, cells were permeabilized with Triton X-100 and blocked with 5% goat serum (Sigma). Cells, co-expressing cMyc-tagged wild type and FLAG-tagged [ΔC] mutant subunits, were incubated with a mouse anti-cMyc antibody (1:80, Santa Cruz.) at 4°C overnight, and then with FITC-conjugated a goat anti-mouse IgG antibody (1:500, Sigma) for 2 hr. After extensively rinsed, the cells were incubated with a rabbit anti-FLAG antibody (1:500, Sigma) for 3–4 hr at room temperature, and then with a TRITC-conjugated goat anti-rabbit IgG antibody (1:500, Sigma). The images were captured using a Zeiss AxioVert 200M confocal microscope and LSM510 META software.
All data, where appropriate, are presented as mean ± SEM, and statistical analysis was carried out using Student’s t test.
RESULTS
TRPM2 coiled-coil domain mediates subunit interaction
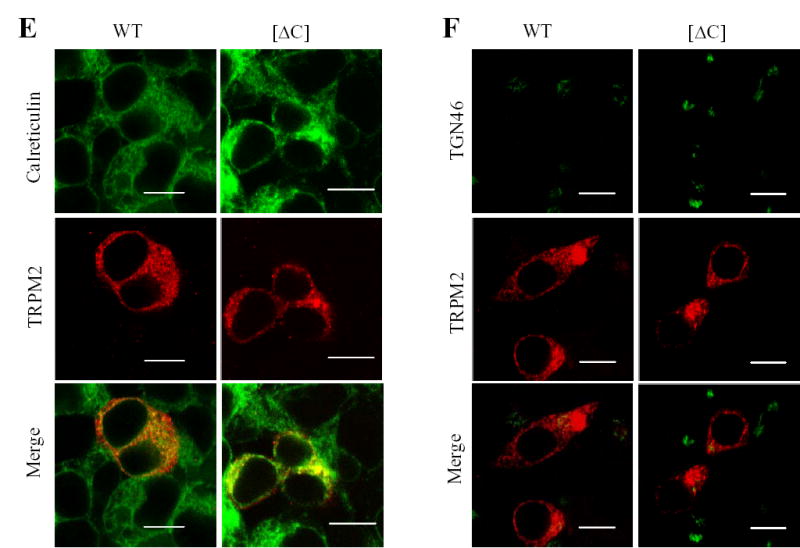
Analysis of the TRPM2 subunit sequence using the coiled-coil domain prediction program (www.ch.embnet.org) indicates that a stretch of residues in the C-terminus between the TRP motif and the NUDT9-H domain (Gly1171 to Ala1200 in the human TRPM2 subunit; Fig.1A) exhibit a strong probability of coiled-coil formation. To determine the potential role of this coiled-coil domain in the TRPM2 channel, we firstly generated a mutant subunit in which the coiled-coil domain was deleted ([ΔC] mutant subunit). The total protein expression level of the [ΔC] mutant subunit was virtually the same as the parental wild type subunit determined by Western blotting (Fig.1C), but the surface expression level was about half of the wild type subunit shown by biotin-labelling assay (Fig.1D). As shown by double labelling with antibodies against calreticulin (an endoplasmic reticulum or ER protein) or TGN46 (a trans-Golgi network protein), the TRPM2 proteins were strongly co-localized with calreticulin (Fig.1E), but lacked noticeable co-localization with TGN46 (Fig.1F). These results suggest that deletion of the coiled-coil domain seems to not significantly alter the targeting of the TRPM2 protein from the ER to the plasma membrane. For multi-subunit channels, reduction in the surface expression could result alternatively from defects in assembly of multi-subunit protein complex. Subunit interaction is the first step in the assembly of multi-subunit channel. To address the possible role of the coiled-coil domain in the TRPM2 subunit interaction, C-terminal FLAG-tagged [ΔC] mutant were co-expressed with C-terminal EE-tagged wild type subunit or with C-terminal cMyc-tagged [ΔC] mutant, and the subunit interaction was examined by co-immunoprecipitation. The interaction between wild type and [ΔC] mutant subunits were severely disrupted (Fig.2A); the average residual subunit interaction was less than 20% of the interaction between two wild type subunits (Fig.2B). The interaction between two mutant subunits was also dramatically attenuated (Fig.2C), and the reduction was slightly greater than that seen between wild type and mutant subunits (data not shown). In cells co-expressing wild type and mutant subunits, double staining shows strong co-localization rather than segregate distribution of two subunits (Fig.2D), suggesting that loss of co-immunoprecipitation was due to reduced interaction between wild type and mutant subunits rather than potential preferential homo-subunit interaction. All these results indicate that the coiled-coil domain is important in the TRPM2 subunit interaction.
Fig. 2. TRPM2 coiled-coil domain supports subunit interaction.
A. Co-immunoprecipitation analysis of interaction between EE-tagged wild type and FLAG-tagged wild type or deletion mutant TRPM2 subunits. Cells were co-transfected with plasmids in a 1:1 ratio encoding EE-tagged wild-type TRPM2 subunit and FLAG-tagged wild type or coiled-coil deletion mutant ([ΔC]). Western blots of the EE-tagged wild type TRPM2 protein in whole cell lysate (left), the FLAG-tagged wild type or mutant TRPM2 protein (middle), and the EE-tagged wild type TRPM2 protein immunopreciptated using an anti-FLAG antibody (right). The arrow heads, * and ** (middle) indicate the TRPM2 proteins, non-specific band and the antibody heavy chain respectively. B. Mean data from 3 independent co-immunoprecipitation experiments are shown in A. *** p < 0.001. C. Co-immunoprecipitation analysis of TRPM2 subunit interaction between FLAG-tagged and cMyc-tagged deletion mutant TRPM2 subunits, and between the parental wild type subunits. Western blots using an anti-cMyc antibody of the cMyc-tagged TRPM2 protein co-immunoprecipitated using an anti-FLAG antibody. Similar results were observed in two independent experiments. D. Confocal images showing double immunostaining of cMyc-tagged wild type and FLAG-tagged deletion mutant subunits. The scale is 10 μM. Note that wild type and mutant subunits were substantially co-localized.
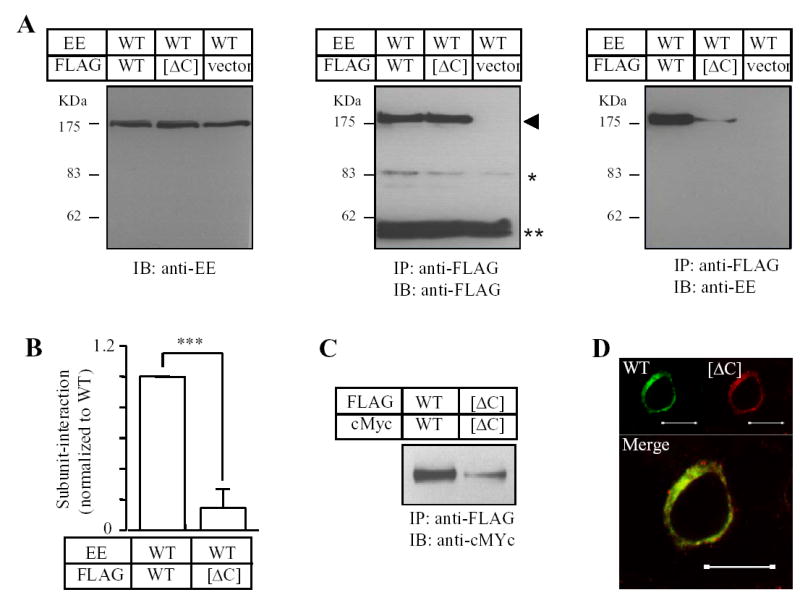
Canonical coiled-coil domain comprises a heptad repeats denoted by abcdefg where the positions a and d are preferably occupied by hydrophobic residues and are important to the protein-protein interaction (28, 29). Therefore, to testify further the importance of the coiled-coil domain in the TRPM2 subunit interaction, we introduced substitutions of these hydrophobic residues (Fig.1B) in the FLAG-tagged TRPM2 subunit with the hydrophilic residue glutamine, co-expressed the resultant mutant subunit with the EE-tagged wild type subunit, and performed co-immunoprecipitation to assess the mutational effect on the subunit interaction. Fig.3 shows the results. Similar to the co-expression of deletion mutant and wild type subunits, co-expression of any of the FLAG-tagged point mutants with EE-tagged wild type subunit resulted in no significant alteration in the protein expression level for both proteins. However, as expected, mutation of each of these hydrophobic residues (Leu1177, Leu1180, Val1184, Leu1191, Ile1194 and Leu1198) indeed strongly reduced the subunit interaction, although the reduction was variable (the order of the potency: Ilu1194 > Leu1180 > Leu1198 > Leu1177 ≥ Leu 1191 ≥ Val1184). In contrast, substitution of the hydrophilic residue Thr1187, which was predicted not to alter the coiled-coil formation, had no effect on the subunit interaction (Fig.3), arguing against the possibility that the effects of mutating the hydrophobic residues on the subunit interaction were non-specific. We then introduced combined mutations in the four most outstanding positions (Ilu1194, Leu1180, Leu1198 and Leu1177) to see to which extent mutation of these residues could mimic the effect of the [ΔC] mutant. The double mutants have substitutions in two close ([L1177/L1180] and [I1194/L1198]) or distant positions ([L1177/L1194] and [L1177/L1198]), the triple mutants contain additional substitution in distant positions ([L1177/L1180/I1194], [L1177/L1180/L1198], [L1177/I1194/L1198] and [L1180/I1194/L1198]), and the quadruple mutant includes mutations of all the four residues. Introduction of two substitutions enhanced the reduction in the subunit interaction. Of all the triple and quadruple mutants, the subunit interaction was not different from that of the [ΔC] mutant subunit (Fig.3C). These results clearly strengthen the argument that the C-terminal coiled-coil domain plays an important role in mediating the TRPM2 subunit interaction.
Fig. 3. Effects of mutating the hydrophobic residues in the coiled-coil domain on the subunit interaction.
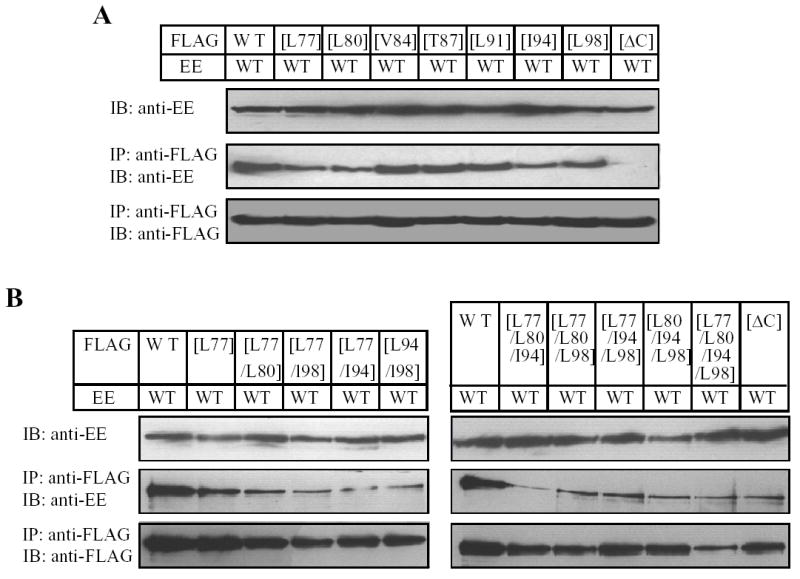
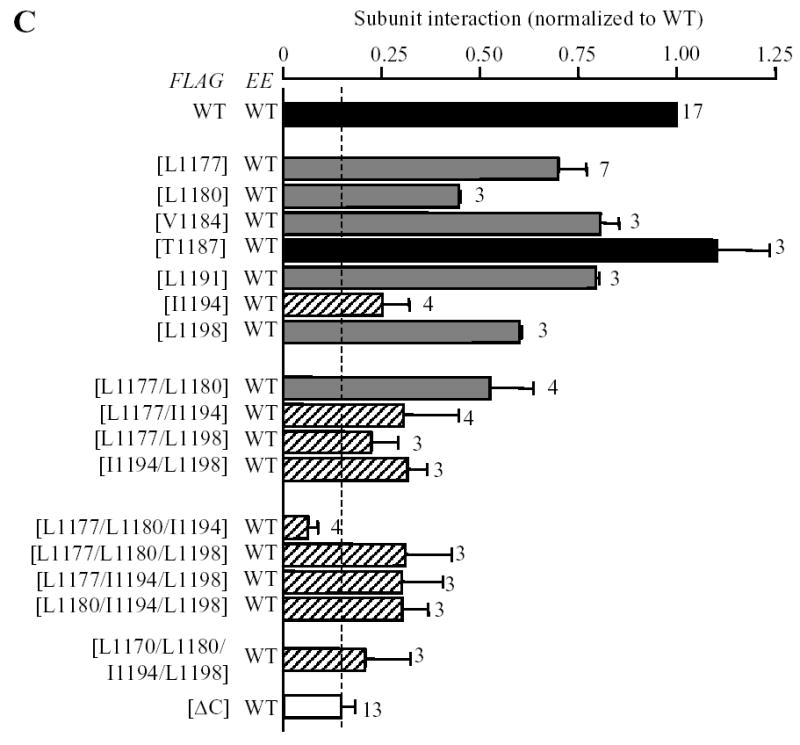
A–B. Western blots of EE-tagged wild type TRPM2 protein in cell lysates (top blot in each panel) and co-immunopreciptated (middle blot in each panel) with FLAG-tagged wild type or mutant TRPM2 (bottom blot in each panel). Experiments examined single (A), double, triple and quadruple mutants (B). C. Mean data from experiments as shown in A–B. The number of independent experiments in each case is indicated next to the bar. Statistical analysis results are denoted by different color or pattern of the bars: black, similar to the wild type (p > 0.1); gray, significantly smaller than the wild type, but higher than the deletion mutant [ΔC] (p < 0.05); hatched, significantly smaller than the wild type (p < 0.01), and not significantly different to the [ΔC] mutant (p > 0.1). The dotted line denotes the reduction in the coiled-coil domain deletion mutant. Note the progressive tendency of reducing the subunit interaction by increasing the number of mutations.
Coiled-coil domain is important for functional TRPM2 channel assembly
Subunit interaction is the prerequisite for functional channel assembly. Thus, we next investigated whether the subunit interaction mediated by the coiled-coil domain is essential for the assembly of functional TRPM2 channel. We used whole cell patch clamp recording to measure ADPR-evoked TRPM2 channel currents, a functional assay widely used in similar studies of other ion channels (e.g., 23, 27, 30–32, 36, 37). Robust TRPM2 channel currents were rapidly evoked by a supermaximal concentration of ADPR (1 mM) (10, 17), and blocked by flufenamic acid as expected for the TRPM2 channels (33, 35). We firstly compared the ADPR-evoked currents in cells expressing the wild type channel or the [ΔC] mutant channel. The ADPR-evoked currents were readily detected in cells expressing the [ΔC] mutant channel. However, the average current amplitude was substantially smaller than that of the wild type channel recorded in parallel experiments (Fig.4). The decrease in the current amplitude was about 75%, approximating to the decrease seen in the subunit interaction (Fig.2). We performed another set of experiments to compare the currents evoked by 1 mM and 3 mM ADPR in cells expressing the wild type or [ΔC] mutant channel. The currents evoked by 1 mM ADPR (WT: 4382 ± 238 pA, n = 3; [ΔC]: 709 ± 322 pA, n = 8) reached the maximal values at both the wild type and mutant channels, as there was no significant increase in currents evoked by 3 mM ADPR (WT: 3805 ± 505 pA, n = 3; [ΔC]: 650 ± 197, n = 5; in both cases p > 0.3). In addition, the mutant channel showed a typical I/V relationship like the wild type channel (Fig.4A). These results indicate that the agonist sensitivity and the I/V relationship are not responsible for the reduced currents.
Fig. 4. The coiled-coil domain is important for formation of functional TRPM2 channel.
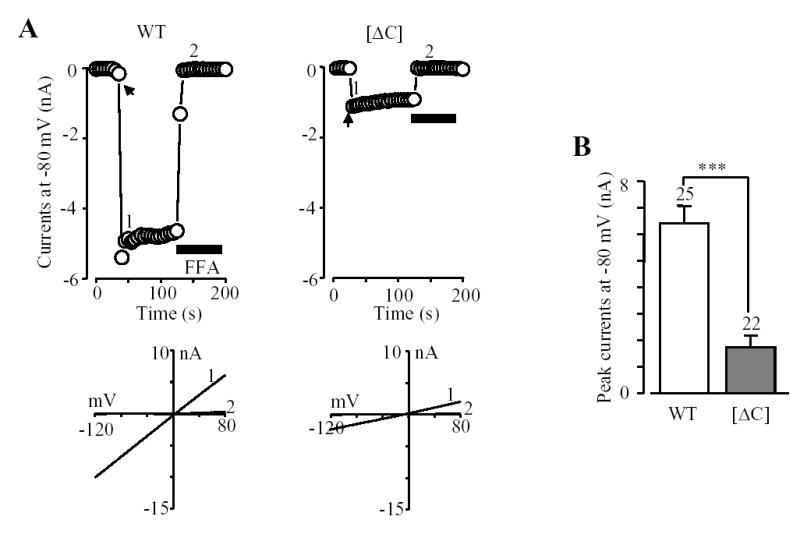
A. Representative ADPR-evoked currents at a membrane potential of −80 mV (top) and the I/V relationship (bottom) before and after FFA application (at time points indicated by 1 and 2) in a cell expressing the wild type or coiled-coil deletion mutant ([ΔC]) TRPM2 channel. The arrows indicate the first recordings in the whole cell configuration. Flufenamic acid (FFA) (0.5 mM) was used to block the TRPM2 channel currents. B. Mean amplitude of the ADPR-evoked peak currents from experiments shown in A. The number of cells examined in each case is indicated above the bar. *** p < 0.001.
Fig.5 summarizes the ADPR-evoked currents recorded in cells expressing the point mutant channels. The current amplitude for single mutant channel [L1177Q], [1180Q], [I1194Q] or [L1198Q] was significantly reduced compared to that of the wild type channel. The current amplitude for the [L1191Q] mutant channel showed a tendency of reducing but the reduction was not statistically significant, whereas there was no difference between the [V1184Q] mutant and the wild type channels. Thus the order of potency of reducing the expression of the functional TRPM2 channels is: Leu1180 ~ Leu1198 > Leu1177 ≥ Ile1194 > Leu1191 ~ Val1184. Of notice, both biochemical and functional approaches unequivocally identified Leu1177, Leu1180, Ile1194 and Leu1198 as the most important residues (Fig.3C and Fig.5B), although the orders were slightly different. The mean ADPR-evoked current of the [T1187Q] mutant channel was not significantly different from that of the wild type channel, again in line with the lack of effect on the subunit interaction. Combined mutations of the most outstanding four hydrophobic residues caused further decrease in the ADPR-evoked currents; there were no significant differences between the triple and quadruple mutant channels and the wild type channel (Fig.5B). Furthermore, there was clear correlation between the mutational effects on the subunit interaction and the ADPR-evoked channel currents (Fig.5C). Thus the biochemical and functional results are consistent with the notion that the coiled-coil domain is critical in mediating the TRPM2 subunit interaction and in the assembly of the functional TRPM2 channel.
Fig. 5. Effects of mutating the hydrophobic residues in the coiled-coil domain on formation of functional TRPM2 channel.
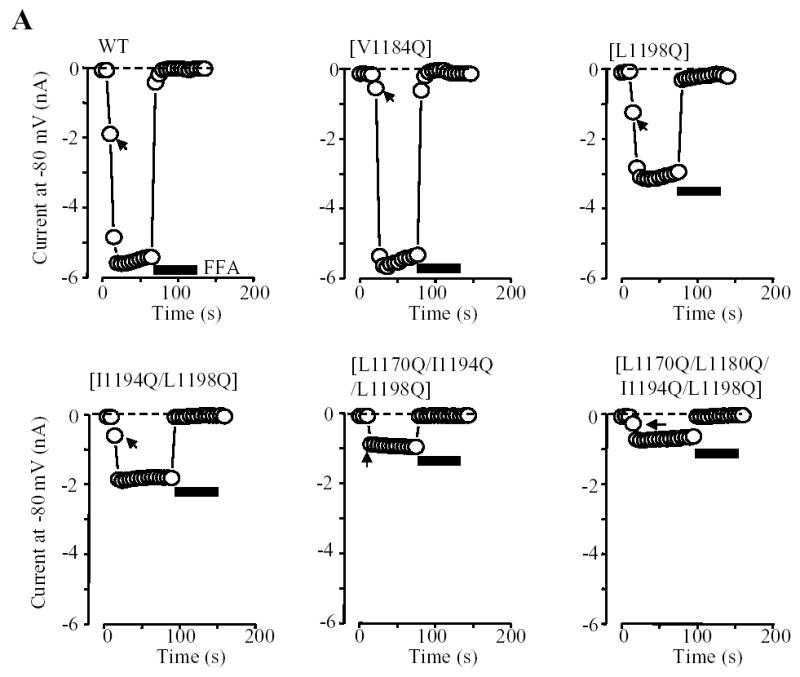
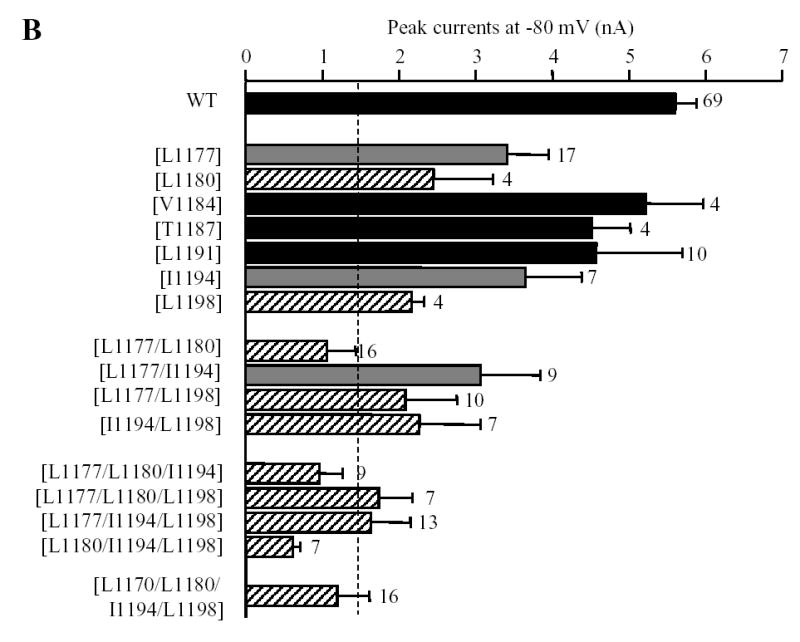
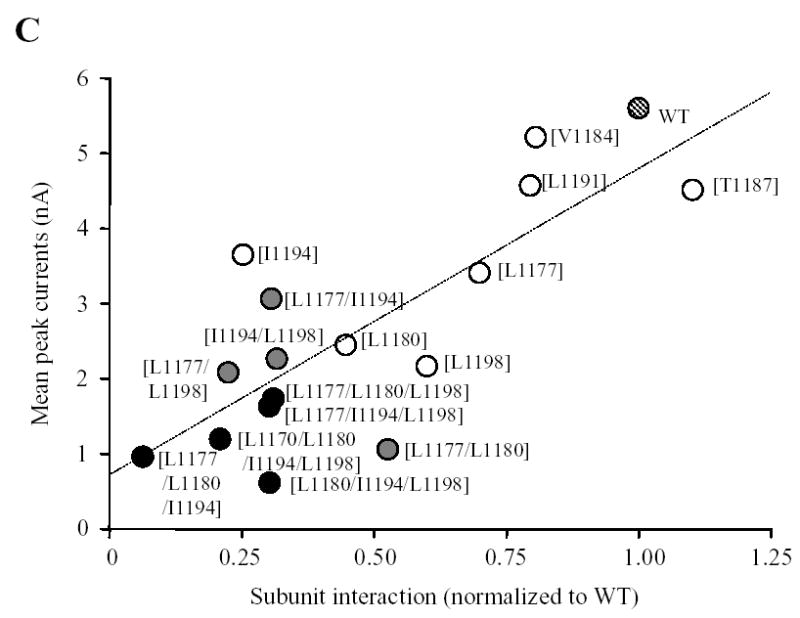
A. Examples of ADPR-evoked whole cell currents in cells expressing the wild type or indicated point mutant TRPM2 channels. The membrane potential was −80 mV. The arrows indicate the first recordings in the whole cell configuration. B. Mean peak currents for the wild type and mutant TRPM2 channels as shown in A. The number of cells examined in each case is indicated next to the bar. Statistical analysis results are denoted by different color or pattern of the bars: black, similar to the wild type (p > 0.1); gray, significantly smaller than the wild type, but higher than the deletion mutant [ΔC] (p < 0.05); hatched, significantly smaller than the wild type (p < 0.01), and not significantly different to the [ΔC] mutant (p > 0.1). The dotted line indicates the currents for the coiled-coil domain deletion mutant (scaled from Fig.4B). Note the progressive tendency of reducing the currents by increased the number of mutations. C. The amplitude of ADPR-evoked peak currents for the wild type and all the mutant TRPM2 channels is plotted against the subunit interaction (from Fig.3C). The hatched, open, grey and black circles indicate the wild type, single, double, or triple and quadruple mutants respectively. Note that there is clear correlation (r = 0.8) between the subunit interaction and functional channel currents.
Deletion of coiled-coil domain removes functional suppression of TRPM2 subunit carrying a phenotypic pore mutation
Mutation of the completely conserved residue Cys996 in the pore region of the TRPM2 subunit (Fig.1A) confers dominant negative phenotypic functional suppression (33). To seek further evidence for the role of the coiled-coil domain in the TRPM2 subunit interaction and the channel assembly, we asked whether deletion of the coiled-coil domain can abolish the functional suppression by C996S mutation. Similar approach was successfully applied in previous studies to attest to the role of the coiled-coil domains in the assembly of the large conductance calcium-activated potassium channel (38) and the TRPV1 channel (23). We introduced deletion of the coiled-coil domain into the [C996S] mutant, and co-expressed the [C996S] or [C996S/ΔC] mutant subunit with the wild type subunit. We examined the assembly of functional channels by recording ADPR-evoked currents and the subunit interaction by co-immunoprecipitation. Fig.6 shows the results. In striking contrast with [C996S] mutant subunit, the [C996S/ΔC] mutant subunit almost entirely lost its dominant negative phenotypic suppression of the ADPR-evoked currents (Fig.6A), and its ability of subunit interaction (Fig.6B). These results further support the critical role of the coiled-coil domain in both subunit interaction and assembly of the TRPM2 channel.
Fig. 6. Deletion of coiled-coil domain removes functional suppression of pore mutation C996S.
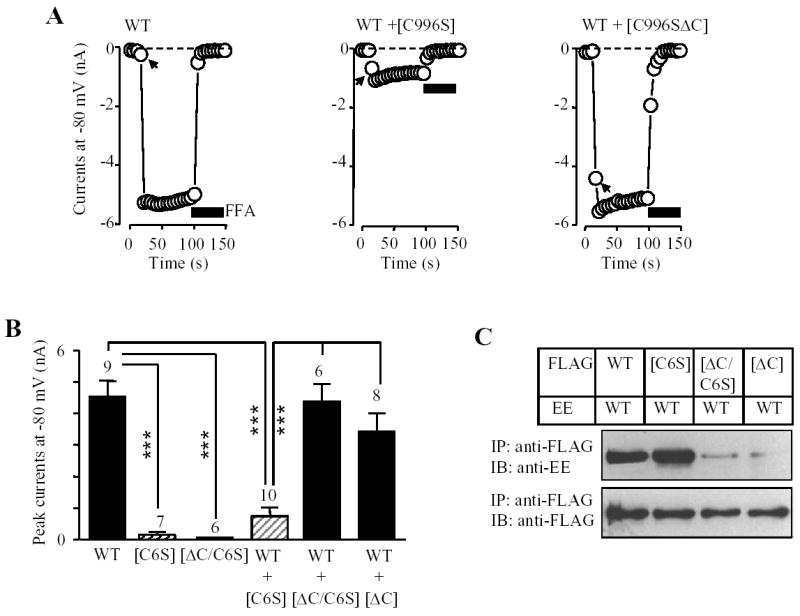
A. Examples (top) and mean ADPR-evoked whole cell currents (bottom left) in cells expressing the wild type (WT) or indicated mutant subunits alone, or co-expressing wild type and indicated mutant subunits. The arrows indicate the first recordings in the whole cell configuration. The number of cells examined in each case is indicated above the bar. *** p < 0.001. There were no significantly differences between WT subunit plus mutant subunit lack of the coiled-coil domain ([ΔC]) and WT subunit plus mutant subunit containing both C996S and coiled-coil domain deletion ([ΔC/C6S]). Note that the currents were significantly reduced in cells co-expressing wild type with mutant containing C996S ([C6S]) and deletion of the coiled-coil domain results in loss of the functional suppression by C996S mutation. B. Co-immunoprecipitation analysis of the subunit interaction between the FLAG-tagged wild type, [C6S], [ΔC] or [ΔC/C6S] and the EE-tagged wild type subunits. Western blots of the EE-tagged wild type TRPM2 protein (top) co-immunoprecipitated with the FLAG-tagged wild type or mutant TRPM2 protein (bottom). Similar results were observed in two independent experiments.
DISCUSSION
The key finding of this study is that the C-terminal coiled-coil domain is important in the subunit interaction and the assembly of the TRPM2 channel, supported by the following major results obtained by using molecular, biochemical and functional approaches. Firstly, we showed that deletion of this coiled-coil domain resulted in severe disruption of the subunit interaction and significant loss of the channel currents. Secondly, progressive introduction of glutamine mutation of the hydrophobic residues at the positions a and d of the abcdef heptad repeats in the TRPM2 coiled-coil domain gave rise to gradual loss of the subunit interaction and channel currents. The mutational effects on the subunit interaction and the channel currents were clearly correlated. Thirdly, the dominant negative functional suppression by a pore mutation C996S was abolished as a result of removing the coiled-coil domain. These results are in good agreement with the demonstration by Tsuruda et al (32) that the TRPM2 coiled-coil domain is able to form tetramers, and together offer convincing evidence indicating that the coiled-coil domain represents the key molecular determinant for efficient assembly of the TRPM2 channel.
Subunit interaction is the first step in the assembly of multi-subunit functional channels. The best understood example of the channel assembly is the voltage-gated potassium channels. However, even though these channels bear strong structural relatedness, studies have revealed channel subunit specific assembly domains with distinct locations and/or features. Thus, the hydrophilic domain governing tetramerization of the Shaker voltage-gated potassium channel is localized to the beginning of the N-terminus (39, 40), whereas the hydrophilic tetramerization domain is present in the C-terminus of the large-conductance calcium-activated channel (38). A C-terminal coiled-coil domain is engaged in the assembly of the KCNQ channels (31, 35, 41) and the ether-a-go-go potassium channels (30, 34), and the cyclic nucleotide-gated channels (42, 43). Among the TRP superfamily, the assembly of the TRPV subfamily has been subjected to extensive investigation. Surprisingly these studies have also identified diverse regions and/or domains mediating the subunit interaction and the assembly of the TRPV channels (2–4) (see Introduction). Identification of naturally occurring truncated or mutant subunits with functional phenotypes suggests involvement of the N-terminus and/or transmembrane domains in the TRPM subunit interaction (44–46). The requirement of the C-terminus in TRPM subunit interaction and functional channel assembly was firstly demonstrated in the TRPM4 channel; deletion of the majority of its C-terminus including the coiled-coil domain results in loss of subunit interaction and functional channel activity, determined by co-immunoprecipitation and patch clamp current recording (27). Sequence analysis indicates that a coiled-coil domain is present in the C-termini of the TRPM channel subfamily (Fig.1B). The results presented here have demonstrated the importance of the coiled-coil domain in the subunit interaction and assembly of the TRPM2 channel. In a very recent study, Tsuruda et al have nicely demonstrated that the coiled-coil domain is necessary to direct the TRPM8 channel assembly (32). Moreover, they have also demonstrated that the coiled-coil domains from several TRPM channels including TRPM2 and TRPM8 channels can form tetramers or oligomers (32). Therefore, it is tempting to hypothesize that this coiled-coil domain may serve as a common tetramerization motif in the TRPM subfamily channels. The relative contributions for each of the residues in the abcdefg heptad repeats to the specificity of the subunit interaction and selective assembly of the homomeric and heteromeric TRPM channels remain to be determined. Along with previous reports on the ether-a-go-go potassium channel (30), KCNQ potassium channel (31), cyclic nucleotide-gated channel (42, 43), and TRPV1 channel (23), studies on the TRPM8 channel (32) and the TRPM2 channel (this study) demonstrate that the coiled-coil domain play a general role in directing the subunit interaction and the assembly of multi-subunit channels.
The present study on the TRPM2 channel and the study on the TRPM8 channel (32) are converging to support the importance of the C-terminal coiled-coil in the TRPM channel assembly. There are differences between the two channels in the role of the coiled-coil domains illustarted by the effects of manipulating the coiled-coil domain, although the exact reasons for the discrepancies are currently unclear. Removal and mutation of the coiled-coil domain in the TRPM8 subunit leads to complete loss of the protein expression and the channel activity, implying additional roles in channel maturation and/or membrane trafficking (32). Thus, the first major different finding from this study was that deletion of the TRPM2 coiled-coil domain caused no obvious change in the global protein expression (Fig.1C), and significant but incomplete loss in the surface expression (Fig.1D). TRPM2 channel is similar to the TRPM4 channel in a sense that the latter channel lack of most of its C-terminus including the coiled-coil domain can express a substantial level of protein that reaches the cell surface (27). We have performed a number of experiments and made the following observations. Firstly, double staining with antibodies recognizing the ER (calreticulin) and the trans-Golgi network (TGN46) proteins showed no detectable difference in the targeting of mutant TRPM2 protein from the ER to the plasma membrane conferred by deleting the coiled-coil domain (Fig.1E–F). Secondly, co-immunoprecipitation analysis shows that removal of the TRPM2 coiled-coil domain resulted in dramatic reduction in the subunit interaction (Fig.2). Introduction of point mutations in the key hydrophobic residues in the coiled-coil domain nicely recapitulated the effects on the subunit interaction of deleting the coiled-coil domain (Fig.5). Thirdly, removal of the coiled-coil domain led to loss of the phenotypic functional suppression by the pore mutation C996S (Fig.6). Fourthly, double immunostaining shows no substantial segregation but co-localization of wild type and deletion mutant proteins (Fig.2C). Finally, there was similar degree of loss by deleting the coiled-coil domain (Fig.2 and Fig.4) and clear correlation of the decrease by mutating the coiled-coil domain (Fig.3 and Fig.5) in both the subunit interaction and the channel functional activity. The most consistent and simplest interpretation of all these results is that the coiled-coil domain is important in the TRPM2 subunit interaction and, although minor defect in the membrane trafficking remains possible, the apparent reduction in the surface expression of the deletion mutant subunit is primarily attributable or secondary to the impaired subunit interaction and channel assembly.
The second finding that differs from the TRPM8 channel was that loss of the TRPM2 coiled-coil domain strongly reduced but did not completely abolish the formation of functional TRPM2 channels. It has been well established that the tetramerization domain is required for efficient assembly of the Shaker voltage-gated potassium channels and that the subunits lack of the N-terminal tetramerization domain can still form but with a low efficiency functional channels (e.g., 39, 40, 47, 48). Of relevance, replacement with a coiled-coil domain can restore efficient assembly of the voltage-gated potassium channel lacking the N-terminal tetramerization domain (48). Our results describe a similar role of the TRPM2 coiled-coil domain in the TRPM2 channel assembly. A short TRPM2 subunit isoform comprising the N-terminus and first two transmembrane segments identified in a previous study can interact with the full length subunit (45). In our hands, the expression of this short subunit was very poor. Nevertheless, its interaction with the full length wild type subunit was also detectable, which was largely abolished by deleting the transmembrane domain (data not shown), suggesting that the transmembrane domain is able to mediate the subunit interaction, as proposed for the TRPV1 subunit (24). To reconcile with these results, it is very likely that the C-terminal coiled-coil domain is the key molecular determinant that is essential for efficient assembly of the TRPM2 channel, whereas the subunit interactions in other parts may be required at the different stages of the TRPM2 channel assembly and could mediate with a low efficiency assembly of the TRPM2 channel lack of the C-terminal coiled-coil domain as in the case of the voltage-gated potassium channels (48–50).
TRPM2 channel is activated by binding of ADPR and related molecules to the NUTD9-H domain in the distal C-terminus (6–11), which is linked to the pore forming domain via the coiled-coil domain (Fig.1A). Thus, in the sense of structural domain arrangement, the TRPM2 channel is analogous to that of the cyclic nucleotide-gated channels. In the latter channels, structural rearrangements upon binding of agonist (e.g., cAMP) to the distal C-terminal domain change the energy of the coupling between the linker region and the pore forming domain and thereby lead to the channel opening (51). The location of the coiled-coil domain between the agonist binding NUDT9-H domain and the channel pore domain raises the possibility that a similar mechanism controls the TRPM2 channel gating. Thus, the coiled-coil domain may play a role in the channel gating as well as in the channel assembly. This could explain in part the differences in the cell surface and functional activity of the channels lacking the coiled-coil domain (Fig.2D and Fig.4B) and in potency of the effects of mutating the hydrophobic residues in the coiled-coil domain on the subunit interaction and the channel function (Fig.5C).
In conclusion, we have provided biochemical and functional evidence supporting that the coiled-coil domain in the C-terminus of the TRPM2 subunit mediates the subunit interaction and the assembly of the functional TRPM2 channel. This study gives insight into a general structural domain governing the TRPM channel assembly.
Acknowledgments
This work was supported by the Wellcome Trust (to L-H Jiang). R Xia is a recipient of the UK Overseas Research Scholarship. The authors are grateful to Dr AM Scharenberg (Washington University, Seattle, USA) for providing the human TRPM2 cDNA clone, and Dr A Sivaprasadarao (University of Leeds) for kind advice in biochemical studies and providing the anti-TGN46 antibody, and Dr TK Taneja and Dr G Howell for assistance in using confocal microscope.
References
- 1.Montell C, Rubin GM. Neuron. 1989;2:1313–1323. doi: 10.1016/0896-6273(89)90069-x. [DOI] [PubMed] [Google Scholar]
- 2.Clapham DE. Nat Rev Neurosci. 2001;2:387–396. doi: 10.1038/35077544. [DOI] [PubMed] [Google Scholar]
- 3.Clapham DE. Nature. 2003;426:517–524. doi: 10.1038/nature02196. [DOI] [PubMed] [Google Scholar]
- 4.Montell C, Birnbaumer L, Flockerzi V. Cell. 2002;108:595–598. doi: 10.1016/s0092-8674(02)00670-0. [DOI] [PubMed] [Google Scholar]
- 5.Moran MM, Xu H, Clapham DE. Curr Opin Neurobiol. 2004;14:362–369. doi: 10.1016/j.conb.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 6.Perraud A, Fleig A, Dunn CA, Bagley LA, Launay P, Schmitz C, Stokes AJ, Xhu Q, Bessman MJ, Penner R, Kinet JP, Scharenberg AM. Nature. 2001;411:595–599. doi: 10.1038/35079100. [DOI] [PubMed] [Google Scholar]
- 7.Sano Y, Inamura K, Miyake A, Mochizuki S, Yokoi H, Matsushime H, Furuichi K. Science. 2001;293:1270–1271. doi: 10.1126/science.1062473. [DOI] [PubMed] [Google Scholar]
- 8.Hara Y, Wakamori M, Ishii M, Maeno E, Nishida M, Yoshida T, Yamada H, Shimizu S, Mori E, Kudoh J, Shimizu N, Kurose H, Okada Y, Imoto K, Mori Y. Mol Cell. 2002;9:163–173. doi: 10.1016/s1097-2765(01)00438-5. [DOI] [PubMed] [Google Scholar]
- 9.Wehage E, Eisfeld J, Heiner I, Jungling E, Zitt C, Luckhoff A. J Biol Chem. 2002;277:23150–23156. doi: 10.1074/jbc.M112096200. [DOI] [PubMed] [Google Scholar]
- 10.Kolisek M, Beck A, Fleig A, Penner R. Mol Cell. 2005;18:61–69. doi: 10.1016/j.molcel.2005.02.033. [DOI] [PubMed] [Google Scholar]
- 11.Togashi K, Hara Y, Tominaga T, Higashi T, Konishi Y, Mori Y, Tominaga M. EMBO J. 2006;25:1804–1815. doi: 10.1038/sj.emboj.7601083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hill K, Tigue NJ, Kelsell RE, Benham CD, McNulty S, Schaefer M, Randall AD. Neuropharmacology. 2006;50:89–97. doi: 10.1016/j.neuropharm.2005.08.021. [DOI] [PubMed] [Google Scholar]
- 13.Fonfria E, Murdock PR, Cusdin FS, Benham CD, Kelsell RE, McNulty S. J Recept Signal Transduct Res. 2006;26:159–178. doi: 10.1080/10799890600637506. [DOI] [PubMed] [Google Scholar]
- 14.Fonfria E, Marshall IC, Boyfield I, Skaper SD, Hughes JP, Owen DE, Zhang W, Miller BA, Benham CD, McNulty S. J Neurochem. 2005;95:715–723. doi: 10.1111/j.1471-4159.2005.03396.x. [DOI] [PubMed] [Google Scholar]
- 15.Inamura K, Sano Y, Mochizuki S, Yokoi H, Miyake A, Nozawa K, Kitada C, Matsushime H, Furuichi K. J Membr Biol. 2003;191:201–207. doi: 10.1007/s00232-002-1057-x. [DOI] [PubMed] [Google Scholar]
- 16.Kraft R, Grimm C, Grosse K, Hoffmann A, Sauerbruch S, Kettenmann H, Schultz G, Harteneck C. Am J Physiol Cell Physiol. 2004;286:C129–C137. doi: 10.1152/ajpcell.00331.2003. [DOI] [PubMed] [Google Scholar]
- 17.Beck A, Kolisek M, Bagley LA, Fleig A, Penner R. FASEB J. 2006;20:E47–E54. doi: 10.1096/fj.05-5538fje. [DOI] [PubMed] [Google Scholar]
- 18.Hille B. Ion channels of excitable membranes. third edition. Sinauer; Sunderland, MA: 2001. [Google Scholar]
- 19.Tong Q, Zhang W, Conrad K, Mostoller K, Cheung JY, Peterson BZ, Miller BA. J Biol Chem. 2006;281:9076–9085. doi: 10.1074/jbc.M510422200. [DOI] [PubMed] [Google Scholar]
- 20.Kedei N, Szabo T, Lile JD, Treanor JJ, Loah Z, Iadarola MJ, Blumberg PM. J Biol Chem. 2001;276:28613–28619. doi: 10.1074/jbc.M103272200. [DOI] [PubMed] [Google Scholar]
- 21.Kuzhikandathil EV, Wang H, Szabo T, Morozova N, Blumberg PM, Oxford GS. J Neurosci. 2001;21:8697–8706. doi: 10.1523/JNEUROSCI.21-22-08697.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoendrop JGJ, Voets T, Hoefs S, Weidema F, Prenen J, Niulius B, Bindels RJM. EMBO J. 2003;22:776–785. doi: 10.1093/emboj/cdg080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garcia-Sanz N, Fernandez-Carvajal A, Morenilla-Palao C, Planells-Cases R, Fajardo-Sanchez E, Fernandez-Ballester G, Ferrer-Montiel A. J Neurosci. 2004;24:5307–5313. doi: 10.1523/JNEUROSCI.0202-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hellwig N, Albrecht N, Harteneck C, Schultz G, Schaefer M. J Cell Sci. 2005;118:917–928. doi: 10.1242/jcs.01675. [DOI] [PubMed] [Google Scholar]
- 25.Arniges M, Fernandez-Fernandez JM, Albrecht N, Schaefer M, Valverde MA. J Biol Chem. 2006;281:1580–1586. doi: 10.1074/jbc.M511456200. [DOI] [PubMed] [Google Scholar]
- 26.Chang Q, Gyftogianni E, Van de Graaf SFJ, Hoefs S, Weidema FA, Bindels RJM, Hoenderop JGJ. J Biol Chem. 2004;279:54304–54311. doi: 10.1074/jbc.M406222200. [DOI] [PubMed] [Google Scholar]
- 27.Launay P, Cheng H, Srivatsan S, Penner R, Fleig A, Kinet JP. Science. 2004;306:1374–1377. doi: 10.1126/science.1098845. [DOI] [PubMed] [Google Scholar]
- 28.Lupas A, Van Dyke M, Stock J. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 29.Lupas A. Trends Biochem Sci. 1996;21:375–382. [PubMed] [Google Scholar]
- 30.Jenke M, Sanchez A, Monje F, Stuhmer W, Weseloh RM, Pardo LA. EMBO J. 2003;22:395–403. doi: 10.1093/emboj/cdg035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwake M, Athanasiadu D, Beimgraben C, Blanz J, Beck C, Jentsch TJ, Saftig P, Friedrich T. J Neurosci. 2006;26:3757–3766. doi: 10.1523/JNEUROSCI.5017-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsuruda PR, Julius D, Minor DL. Neuron. 2006;51:201–212. doi: 10.1016/j.neuron.2006.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mei ZZ, Mao HJ, Jiang LH. Am J Physiol Cell Physiol. 2006;291:C1022–C1028. doi: 10.1152/ajpcell.00606.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiang LH, Gawler DJ, Hodson N, Milligan CJ, Pearson HA, Porter A, Wray D. J Biol Chem. 2000;275:6235–6143. doi: 10.1074/jbc.275.9.6135. [DOI] [PubMed] [Google Scholar]
- 35.Hill K, Benham CD, McNulty S, Randall AD. Neuropharmacology. 2004;47:450–460. doi: 10.1016/j.neuropharm.2004.04.014. [DOI] [PubMed] [Google Scholar]
- 36.Ludwig J, Owen D, Pongs O. EMBO J. 1997;16:6337–6345. doi: 10.1093/emboj/16.21.6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schwake M, Jentsch TJ, Friedrich T. EMBO Rep. 2003;4:76–81. doi: 10.1038/sj.embor.embor715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Quirk JC, Reinhart PH. Neuron. 2001;31:13–23. doi: 10.1016/s0896-6273(01)00444-5. [DOI] [PubMed] [Google Scholar]
- 39.Li M, Yan YN, Jan LY. Science. 1992;257:1225–1230. doi: 10.1126/science.1519059. [DOI] [PubMed] [Google Scholar]
- 40.Shen NV, Pfaffinger PJ. Neuron. 1995;14:625–633. doi: 10.1016/0896-6273(95)90319-4. [DOI] [PubMed] [Google Scholar]
- 41.Schmitt N, Schwarz M, Peretz A, Abitbol I, Attali B, Pongs O. EMBO J. 2000;19:332–340. doi: 10.1093/emboj/19.3.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhong H, Molday LL, Molday RS, Yau KW. Nature. 2002;420:193–198. doi: 10.1038/nature01201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhong H, Lai J, Yau KW. Proc Natl Acad Sci USA. 2003;100:5509–5513. doi: 10.1073/pnas.0931279100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu XZ, Moebius F, Gill DL, Montell C. Proc Natl Acad Sci USA. 2001;98:10692–10697. doi: 10.1073/pnas.191360198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang W, Chu X, Tong Q, Cheung JY, Conrad K, Masker K, Miller BA. J Biol Chem. 2003;278:16222–16229. doi: 10.1074/jbc.M300298200. [DOI] [PubMed] [Google Scholar]
- 46.Chubanov V, Waldegger S, Mederos Y, Schnitzler M, Vitzthum H, Sassen MC, Seyberth HW, Konrad M, Gudermann T. Proc Natl Acad Sci USA. 2004;101:2894–2899. doi: 10.1073/pnas.0305252101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kobertz WR, Miller C. Nat Struct Biol. 1999;6:1122–1125. doi: 10.1038/70061. [DOI] [PubMed] [Google Scholar]
- 48.Zerangue N, Jan YN, Jan LY. Proc Natl Acad Sci USA. 2000;97:3591–3595. doi: 10.1073/pnas.060016797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tu LW, Santarelli V, Sheng ZF, Skach W, Pain D, Deutsch C. J Biol Chem. 1996;271:18904–18911. doi: 10.1074/jbc.271.31.18904. [DOI] [PubMed] [Google Scholar]
- 50.Schuleis CT, Nagaya N, Papazian DM. J Biol Chem. 1998;273:26210–26217. doi: 10.1074/jbc.273.40.26210. [DOI] [PubMed] [Google Scholar]
- 51.Zagotta WN, Olivier NB, Black KD, Young EC, Olson R, Gouaux E. Nature. 2003;425:200–205. doi: 10.1038/nature01922. [DOI] [PubMed] [Google Scholar]


