Abstract
The c-Jun N-terminal kinases (JNKs) are members of a larger group of serine/threonine (Ser/Thr) protein kinases from the mitogen-activated protein kinase family. JNKs were originally identified as stress-activated protein kinases in the livers of cycloheximide-challenged rats. Their subsequent purification, cloning, and naming as JNKs have emphasized their ability to phosphorylate and activate the transcription factor c-Jun. Studies of c-Jun and related transcription factor substrates have provided clues about both the preferred substrate phosphorylation sequences and additional docking domains recognized by JNK. There are now more than 50 proteins shown to be substrates for JNK. These include a range of nuclear substrates, including transcription factors and nuclear hormone receptors, heterogeneous nuclear ribonucleoprotein K, and the Pol I-specific transcription factor TIF-IA, which regulates ribosome synthesis. Many nonnuclear substrates have also been characterized, and these are involved in protein degradation (e.g., the E3 ligase Itch), signal transduction (e.g., adaptor and scaffold proteins and protein kinases), apoptotic cell death (e.g., mitochondrial Bcl2 family members), and cell movement (e.g., paxillin, DCX, microtubule-associated proteins, the stathmin family member SCG10, and the intermediate filament protein keratin 8). The range of JNK actions in the cell is therefore likely to be complex. Further characterization of the substrates of JNK should provide clearer explanations of the intracellular actions of the JNKs and may allow new avenues for targeting the JNK pathways with therapeutic agents downstream of JNK itself.
INTRODUCTION
Protein kinases comprise a large enzyme family in eukaryotes and prokaryotes. Protein kinases catalyze the transfer of the terminal phosphoryl group of ATP to their specific protein substrates. It has been recognized for >50 years that protein phosphorylation regulates many aspects of cellular function, such as metabolism, division, movement, survival, and death. Thus, any process that disrupts normal phosphorylation can disrupt cell function and cause disease (61). Conserved sequence motifs have allowed the identification of 518 protein kinases within the human genome, with these being grouped into 20 families based on sequence similarities (205). Additional analyses are being increasingly undertaken with a range of eukaryotes and prokaryotes, revealing striking conservation of some protein kinases across a range of organisms as well as protein kinase family members specific to particular organisms (42, 49, 112, 314).
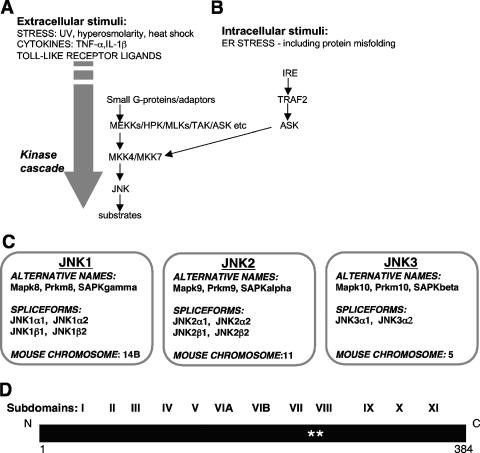
The c-Jun N-terminal kinases (JNKs) are members of a larger group of serine/threonine (Ser/Thr) protein kinases known as the mitogen-activated protein kinase (MAPK) family. The MAPK family is one subgroup of the CMGC class of protein kinases (where CMGC is the class name derived from the major kinase members of this class, namely, cyclin-dependent kinases [CDKs], MAPKs, glycogen synthase kinase 3 [GSK3], and casein kinase 2-related protein kinases). Within the classification of all protein kinases, the CMGC class represents one of the three major protein kinase classes, in addition to classical Ser/Thr kinases and tyrosine (Tyr) kinases (122). The JNKs act within a protein kinase cascade (Fig. 1A and B). They are themselves activated by dual phosphorylation, by the MAPK kinases MKK4 and MKK7, on a specific Thr and a specific Tyr in a typical Thr-X-Tyr motif within their “activation/phosphorylation loop” sequences (for a review, see reference 73).
FIG. 1.
Overview of the JNK pathway. (A) The classical JNK pathway was considered to be activated following the exposure of cells to extracellular stresses, such as UV irradiation, hyperosmolarity, and heat shock. Subsequently, JNK activation was also demonstrated following the exposure of cells to some proinflammatory cytokines, including TNF-α and interleukin-1β (IL-1β), as well as following the activation of Toll-like receptors. The pathway has been shown to involve the activation of various small G proteins and the engagement of adaptor proteins, followed by a protein kinase cascade. This cascade includes various members of the MAPK kinase kinase family (such as MEKKs, hematopoietic progenitor kinase [HPK], the mixed-lineage kinases [MLKs], transforming growth factor β-activated kinase [TAK], and apoptosis signal-regulating kinase [ASK]) and the MAPK kinases MKK4 and MKK7 and leads to JNK activation. (B) Any disruption of protein processing and folding within the ER leading to ER stress can also activate JNKs, and this is mediated by an ER stress transmembrane sensor protein kinase (IRE), the adaptor protein TNF receptor-associated factor 2 (TRAF2), and the upstream kinase ASK1. (C) In mammalian systems, there are three genes that encode the JNKs, namely, jnk1, jnk2, and jnk3. The alternative names for these JNK isoforms are provided, alongside information on their splice forms and positions on the mouse chromosomes. (D) Linear representation of the JNK1α1 protein highlighting conserved features of protein kinases such as the JNKs, with 11 regions of sequence similarity (I to XI), as originally identified by Hanks and colleagues (123). In addition, the positions of two residues in the “activation/phosphorylation loop” are indicated (**).
The diversity of mediators upstream of MKK4 and MKK7 (Fig. 1A) may allow JNK pathway activation by a range of external stimuli (for a review, see reference 73). As studies of JNK pathways have progressed, the range of initiating signals has been expanded to include a diversity of stimuli. Of particular interest is the activation of the JNK pathway following the exposure of cells to a range of proinflammatory cytokines, such as tumor necrosis factor-α (TNF-α) and interleukin-1 (for a review, see reference 81). Furthermore, the JNK pathway is activated in the innate immune response following the activation of various members of the Toll-like receptor family by invading pathogens (e.g., see references 13, 15, 178, 182, 216, 259, and 272) (Fig. 1A). The JNK pathway therefore appears to act as a critical intermediate in signaling in the immune system (81). As also shown in Fig. 1B, there are increasing links between the endoplasmic reticulum (ER) stress response and JNK activation (for a review, see reference 317). This provides at least one mechanism of activation of JNKs following the sensing of internal stress events, such as protein misfolding. There is also an increasing body of literature showing that JNK activation follows bacterial, fungal, prion, parasitic, or viral infections. Under these circumstances, JNK activation may influence important cellular consequences, such as alterations in gene expression (1, 53, 59, 162, 167, 176, 199, 294, 325, 326, 346), cell death (58, 89, 137, 139, 169, 193, 243, 293), viral replication, persistent infection or progeny release (215, 224, 251, 260), or altered cellular proliferation (178). The exact mechanism of JNK activation under each of these circumstances remains to be elucidated fully, although there may be involvement of Toll-like receptors, direct pathway modulation through interaction with upstream protein regulators, or the activation following an ER stress response (79, 87, 110, 124, 143, 191, 253, 261, 279, 294, 312).
Originally identified as stress-activated protein kinases (SAPKs) in the livers of cycloheximide-challenged rats (177), the subsequent purification, cloning, and naming of the JNKs have emphasized their ability to phosphorylate and activate the transcription factor c-Jun (77, 222, 257). The JNK-mediated phosphorylation of both Ser63 and Ser73 within the transactivation domain of c-Jun (Table 1) potentiates its transcriptional activity through the loss of repression mediated by an inhibitory complex associated with histone deacetylase 3 (316). Other events that follow c-Jun phosphorylation include its increased interaction with other binding partners, such as the transcription factor TCF4 or the E3 ubiquitin ligase Fbw7 (240, 241). The importance of c-Jun phosphorylation has been emphasized in studies of transgenic mice expressing the c-Jun mutant c-Jun Ser63→Ala Ser73→Ala, which lacks the two major sites for phosphorylation by JNK (22, 23, 72, 130, 150, 311). Increasing attention has been directed towards JNK as an activator of a wide range of c-Jun-dependent events, including apoptotic cell death and oncogenic transformation (22, 23, 188).
TABLE 1.
Summary of nuclear substrates of JNKs and their phosphorylated sequencesa
| Nuclear protein (function [GenBank accession no.]) | Phosphorylation site sequence(s) | Reference(s) |
|---|---|---|
| c-Jun (transcription factor [AAA37419]) | D-L-L-T-S63-P-D-V-G | 77, 230 |
| L-K-L-A-S73-P-E-L-E | ||
| H-I-T-T-T91-P-T-P-T | ||
| JunB (transcription factor [NP_032442]) | I-T-T-T-T102-P-T-P*-P | 190 |
| T-T-T*-P-T104-P-P-G-Q | ||
| JunD (transcription factor [CAA40010]) | G-L-L-A-S90-P-D-L-G | 339 |
| L-K-L-A-S100-P-E-L-E | ||
| L-V-T-T-T117-P-T-S-T | ||
| ATF2 (transcription factor [AAH26175]) | V-A-D-Q-T69-P-T*-P-Tb | 118, 197 |
| D-Q-T*-P-T71-P-T-R-Fb | ||
| JDP2 (transcription factor [NP_446346]) | D-S-V-R-T148-P-S-E-S | 155, 156 |
| Elk-1 (transcription factor [AAH56150]) | W-S-T-L-S383-P-I-A-P | 321, 337 |
| I-A-P-R-S389-P-A-K-L | ||
| Net (transcription factor [CAA83676]) | V-S-S-V-S239-P-S-S-S | 86 |
| S-S-S-R-S245-P-S-L-S*b | ||
| S*-P-S-L-S249-P-D-S*-Pb | ||
| L-S*-P-D-S252-P-L-P-Sb,c | ||
| HSF1 (transcription factor [NP_005517]) | G-R-P-P-S363-P-P-P-T | 65 |
| c-Myc (transcription factor [AAA20942]) | T-P-P-L-S62-P-S-R-R | 6 |
| R-G-L-C-S71-P-S-Y-V | ||
| p53 (transcription factor [AAA59988]) | P-A-A-P-T81-P-A-A-Pd | 41, 95 |
| NFATc3 (transcription factor [P97305]) | E-S-S-L-S163-P-S*-P-Ab | 57 |
| S-L-S*-P-S165-P-A-S-Sb | ||
| NFATc1a (transcription factor [NP_765978]) | P-A-L-E-S117-P-R-I-E | 56 |
| P-S-C-L-S172-P-A-S-S | ||
| NFATc2 (transcription factor [NP_036472]) | R-I-E-I-T116-P-S-H-E | 248 |
| FOXO4 (transcription factor [P98177]) | K-A-L-G-T447-P-V-L-T*b | 90 |
| T*-P-V-L-T451-P-P-T-Eb,e | ||
| STAT3 (transcription factor [NP_998824]) | D-L-P-M-S727-P-R-T-L | 349 |
| STAT1 (transcription factor [NP_009330]) | L-L-P-M-S727-P-E-E-F | 352 |
| Pax2 (transcription factor [CAA39302]) | Not determinedf | 43 |
| P-S-T-A-S170-P-P-V-S | ||
| L-P-A-L-T296-P-G-L-D | ||
| A-L-L-S-S296-P-Y-Y-Y | ||
| TCFβ1 (transcription factor [NP_002639]) | G-G-E-P-S232-K-K-R-K | 154 |
| R-T-S-F-T242-P-Q-A-I | ||
| Peroxisome proliferator-activated receptor γ1 (nuclear hormone receptor [AAA19971]) | V-E-P-A-S82-P-P-Y-S | 45 |
| Glucocorticoid receptor (nuclear hormone receptor [AAA41203 and NP_000167]) | E-N-L-L-S246-P-L-A-Gg | 37, 266 |
| N-C-L-L-S226-P-L-A-Gg | 147 | |
| Retinoic receptor RARα (nuclear hormone receptor [NP_000955]) | S-Y-T-L-T181-P-E-V-G | 283 |
| P-G-S-C-S445-P-S-L-S | ||
| P-A-T-H-S461-P | ||
| Retinoic receptor RXRα (nuclear hormone receptor [P28700 and NP_002948]) | S-T-L-S-S61-P-I-N-Gh | 2 |
| S-V-I-S-S75-P-M-G-P | ||
| S-V-P-T-T87-P-T-L-G | ||
| L-N-P-S-S265-P-N-D-P | ||
| M-A-A-P-S32-L-H-P-S | 204 | |
| Nur77 (orphan receptor [P12813]) | Not determinedi | 121, 174 |
| P-S-P-S-T145-P-N-F-Q | ||
| Androgen receptor (nuclear hormone receptor [AAA51729]) | S-S-T-T-S650P-T-E-Ej | 109 |
| hnRNP-K (NP_112553) | L-I-S-E-S216-P-I-K-G | 119 |
| I-D-T-W-S353-P-S-E-W | ||
| TIF-IA (Pol I-specific transcription factor [AJ272050]) | Y-V-P-S-T200-P-W-F-L | 211 |
A range of nuclear proteins that are predominantly transcription factors and nuclear hormone receptors have been demonstrated to be substrates for JNK-mediated phosphorylation. This table summarizes these proteins and provides sequence information on the phosphorylation sites identified. For each phosphorylation site, a nine-amino-acid sequence surrounding the residue that is phosphorylated is provided. As shown, multiple sites of phosphorylation have been identified for many nuclear proteins.
In the indicated sequences, S* or T* represents Ser or Thr residues that have also been shown to be phosphorylated by JNKs, showing that JNKs may phosphorylate a number of closely spaced residues. It remains to be determined whether there is any requirement for hierarchical phosphorylation of these substrates by JNKs.
In the Phospho.ELM database of phosphorylation sites (http://phospho.elm.eu.org/), Net is listed as Elk3. However, the site of phosphorylation that is attributed to JNK (i.e., Ser 357) has been demonstrated as a site of phosphorylation for kinases downstream of MKK6 and Ras, not for those downstream of the JNK activator kinases.
Ser34 was shown to be phosphorylated by JNK, but this residue is found in murine p53 and not conserved in human p53 (219).
In the protein sequence, the phosphorylated residues correspond to Thr451 and Thr455.
In the protein sequence, there are two S-P and three T-P motifs. The motifs shown in italics are the best S-P or T-P matches with the heptapeptide consensus (Scansite [329] score, 0.2074).
In the protein sequence, there are 11 S-P and 2 T-P motifs. The motifs shown in italics are the best S-P or T-P matches with the heptapeptide consensus (Scansite [329] score, 0.13827).
In the protein sequence, the phosphorylated residues correspond to Ser648.
The mammalian JNKs are encoded by three distinct genes (Jnk1, Jnk2, and Jnk3) (Fig. 1C). Additional complexity is generated by alternative splicing, which results in up to 10 different protein products varying in size from 46 kDa to 55 kDa (117; for a review, see reference 17).
Specifically, four splice forms arise from the Jnk1 gene, four arise from the Jnk2 gene, and two arise from the Jnk3 gene (Fig. 1C). While JNK1 and JNK2 are expressed in a variety of tissues, JNK3 expression is restricted primarily to the brain, heart, and testes (207, 225). This tissue-specific distribution, particularly for JNK3 expression, has led to the idea that different isoforms may perform different cellular roles. This has been explored further through studies in which the effects of deletions of the Jnk genes alone and in combination have been evaluated (for a review, see reference 30). Later sections of this review explore this isoform specificity further.
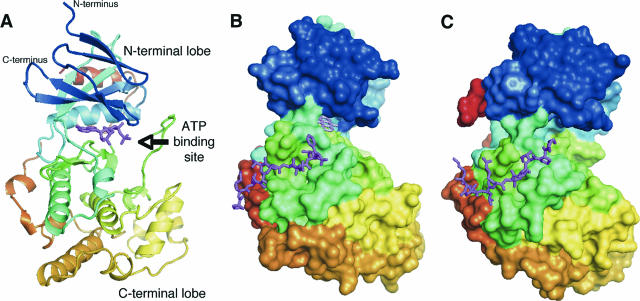
The primary structure of JNK1α1 is illustrated in Fig. 1D, which highlights the conserved features of the protein kinase domains of the JNKs. The subdomain numbering system shown in Fig. 1D denotes conserved sequence regions I to XI, originally defined by Hanks and colleagues for all protein kinases (123). Crystal structures were subsequently determined for JNK3 in the presence of an ATP analogue (327) and several small-molecule inhibitors (273) and for JNK1 in the presence of small-molecule ATP-competitive inhibitors (195, 290) and an inhibitory peptide from the JNK-interacting protein 1 (JIP1), with and without a small-molecule ATP-competitive inhibitor (129). Examples of these structures are shown in Fig. 2.
FIG. 2.
Structures of the JNKs. (A) Crystal structure of JNK3 (PDB accession no. 1JNK) (327). The structure is shown in ribbon representation, colored from the N terminus to the C terminus with colors changing from blue through green and yellow to red. The ATP analogue adenylyl imidodiphosphate is shown in stick representation in magenta. The same coloring scheme is used throughout the figure unless indicated otherwise, and the structures are in approximately the same orientations. (B) Crystal structure of JNK1 (shown in surface representation) in complex with the peptide corresponding to residues 153 to 163 of the substrate and scaffold protein JIP1 (magenta, in stick representation) and the ATP-competitive inhibitor SP600125 (pink, in stick representation) (PDB accession no. 1UKI) (129). (C) Crystal structure of the complex of p38 MAPK (in surface representation) with a peptide corresponding to residues 269 to 280 of the substrate protein MEF2A (magenta, in stick representation) (PDB accession no. 1LEW) (50). The figure was prepared using PyMol (DeLano Scientific LLC).
As expected, the structures of JNK1 and JNK3 are similar to each other and to those of other MAPKs. They have the typical eukaryotic protein kinase fold, as shown in Fig. 2A, comprising two domains or lobes, with an N-terminal domain rich in β-structure (residues 9 to 112 and 347 to 363 in JNK1 and residues 45 to 149 and 379 to 400 in JNK3) and a C-terminal domain rich in α-helices (residues 113 to 337 in JNK1 and residues 150 to 374 in JNK3). The JNK C-terminal domain has an insertion typical of the MAPKs, and this is 12 residues longer in JNKs than in the related MAPKs extracellular signal-regulated kinase 2 (ERK2) and p38. These two domains are connected by two peptide segments, and based on structures of other protein kinases in complex with peptide substrates (34, 140, 171, 198, 335), the peptide substrates for JNK are expected to bind into the groove between the two lobes of JNK. The ATP molecule also binds near the domain interface. All of these structures have been determined for the inactive nonphosphorylated forms of JNK. It is expected that subtle but important changes in structure will accompany activation, as seen for the related MAPK ERK2 (183). The nonphosphorylated JNK structures are inactive due to the misalignment of the catalytic residues accompanying the relative rotation of the two domains and the obstruction of the active site by the “activation loop.” In contrast, the ATP-binding site is well formed in JNK structures.
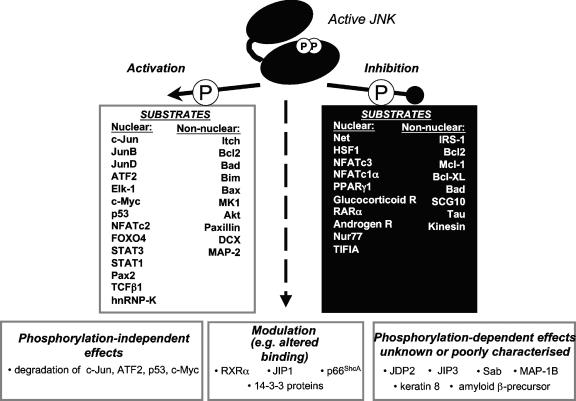
In this review, an overview of the substrates of JNK is presented, beginning with a consideration of the nuclear substrates of JNK that have now been described, in addition to c-Jun. We consider the phosphorylation of 26 nuclear substrates of JNKs (Table 1) and discuss how phosphorylation alters their functions. Many of these nuclear proteins are transcription factors, and therefore their phosphorylation by the JNKs can mediate actions via a direct link to changes in gene expression following the exposure of cells to a range of cytokines and stress stimuli. However, JNK substrates in other cellular compartments have also been described, and the effects of phosphorylation of an additional 26 nonnuclear substrates of JNKs are discussed in turn (see Table 3). These substrates provide a link to a wide range of cellular functions, including cell death and cell movement, as well as allowing for modulation of other signaling events in the cell. We summarize the known effects of phosphorylation on these nuclear and nonnuclear substrates in Fig. 3. This summary shows that JNK-mediated phosphorylation may either enhance or inhibit the activities of its substrates and that, in some cases, the phosphorylation-dependent changes are more complex and involve changes in protein binding and/or localization in the cell. Therefore, the functional effects of phosphorylation following JNK activation must always be specifically tested. Lastly, the determinants of the substrate specificity of JNKs are examined in greater detail, and the effects of JNK-mediated phosphorylation are discussed within the broader context of signal transduction cross talk, integration, and diversification. This analysis reveals the complexities of signal transduction and the new challenges faced in evaluation of signal transduction pathways and their consequent effects.
TABLE 3.
Summary of nonnuclear substrates of JNKs and their phosphorylated sequencesa
| Protein (function [GenBank accession no.]) | Phosphorylation site sequence(s) | Reference(s) |
|---|---|---|
| Regulators of protein turnover | ||
| Itch (E3 ligase [CAI17960]) | S-G-N-N-S199-P-S-L-S | 100 |
| P-P-P-P-T222-P-R-R-P | ||
| S-V-N-G-S232-P-S-A-T | ||
| Adaptor proteins | ||
| IRS-1 (NP_034700) | S-R-T-E-S302-I-T-A-T | 5, 185, 318 |
| I-T-A-T-S307-P-A-S-M | 235 | |
| G-K-P-G-S318-F-R-V-Rc | ||
| JIP1 (Q9R237) | G-F-A-A-S15-P-P-A-Ad | 245, 281 |
| L-H-I-A-S29-P-P-N-F | ||
| A-T-G-D-T103-P-G-A-E | ||
| S-R-S-S-S197-P-L-K-T | ||
| T-G-E-Q-T205-P-P-H-E | ||
| E-I-Y-L-T284-P-V-Q-R | ||
| D-C-L-S-S341-P-E-R-A | ||
| A-S-V-S-S421-P-Y-E-S | ||
| JIP3 | S-A-A-A-T266-P-S-T-T | 161 |
| T-K-S-N-T276-P-T-S-S | ||
| S-A-A-V-T287-P-L-N-E | ||
| p66ShcA (P29353) | E-E-L-P-S36-P-S-A-S | 179 |
| 14-3-3 (cytosolic phosphoprotein binding protein) | ||
| β (AAC14343) | E-I-L-N-S186-P-E-K-Ae | 301 |
| ɛ (CAA79659) | E-I-L-N-S187-P-D-R-Ae | |
| ζ (BAA11751) | E-I-L-N-S184-P-E-K-A | |
| σ (AAF36093) | E-I-A-N-S186-P-E-E-A | |
| Mitochondrial proteins | ||
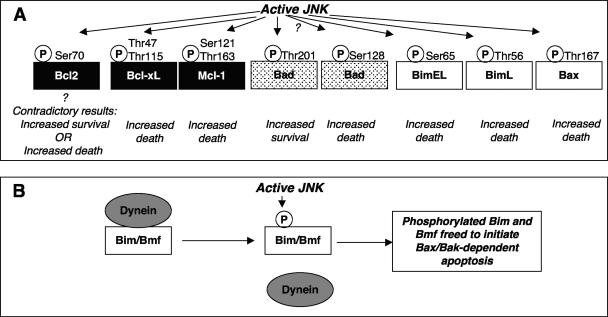
| Bcl2 (antiapoptotic [AAA53662]) | A-A-R-T-S70-P-L-R-P | 76, 331 |
| Bcl-xL (antiapoptotic [CAA80661]) | S-E-M-E-T47-P-S-A-I | 163 |
| Q-L-H-I-T115-P-G-T-A | ||
| Mcl-1 (antiapoptotic [AAF64255]) | D-A-I-M-S121-P-E-E-E | 142 |
| S-L-P-S-T163-P-P-P-A | ||
| Bad (proapoptotic [Q61337]) | K-G-G-S-T201-P-S-Q | 344 |
| E-E-E-L-S128-P-F-Rf | 84 | |
| BimEL(proapoptotic [ACC40029]) | A-P-P-A-S65-P-G-P-F | 258 |
| BimL(proapoptotic [ACC39594]) | P-Q-D-R-S44-P-A-P-M | 186 |
| K-S-T-Q-T56-P-S*-P-Pb | ||
| T-Q-T*-P-S58-P-P-C-Qb | ||
| Bmf (proapoptotic [Q96LC9]) | Not determinedg | 186 |
| L-S-P-A-S77-P-S-Q-G | ||
| Bax (proapoptotic [D47538]) | S-Y-F-G-T167-P-T-W-Q | 166 |
| Sab (BAA25922) | L-D-L-P-S321-P-V-S-L | 63 |
| Protein kinases | ||
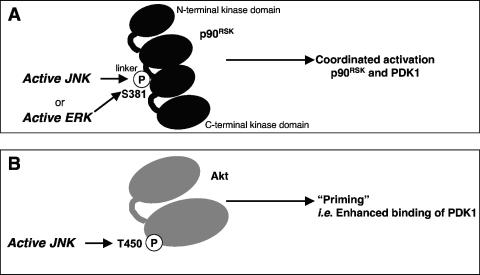
| p90RSK (P18652) | T-P-K-D-S381-P-G-I-P | 350 |
| Akt (P31749) | M-I-T-I-T450-P-P-D-Q | 275 |
| Regulators of cell movement | ||
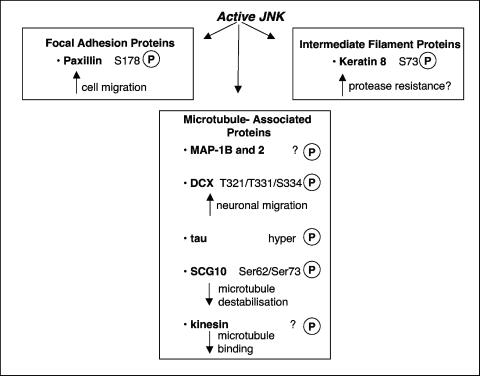
| Paxillin (focal adhesion-associated protein [AAD00648]) | P-G-A-L-S178-P-L-Y-G | 138 |
| Microtubule-associated protein 2 (AAB48098) | Not determinedh | 51 |
| I-T-P-G-T1605-P-P-S-Y | ||
| Microtubule-associated protein 1B (P46821) | Not determinedi | 160 |
| A-S-T-I-S1208-P-P-P-P | ||
| S-P-L-R-S1400-P-P-L-I | ||
| I-S-P-L-T1788-P-R-E-S | ||
| H-S-S-S-S1819-P-P-I-D | ||
| R-D-L-S-T1853-P-G-L-E | ||
| Tau (microtubule-associated protein [NP_005901]) | P-P-A-K-T181-P-P-S-S | 340 |
| S-G-Y-S-S199-P-G-S*-Pb | ||
| G-S*-P-G-S202-P-G-T*-Pb | ||
| G-S*-P-G-T205-P-G-S-Rb | ||
| S-R-S-R-T212-P-S-L-P | ||
| P-S-L-P-T217-P-P-T-R | ||
| I-V-Y-K-S396-P-V-V-S | ||
| S-G-D-T-S404-P-R-H-L | ||
| D-M-V-D-S422-P-Q-L-A | ||
| Amyloid β protein precursor (microtubule-associated protein [AAA36829]) | D-A-A-V-T668-P-E-E-R | 274 |
| SCG10 (stathmin-like protein [AAB36428]) | L-K-P-P-S62-P-I-S-E | 242 |
| T-L-A-S-S73-P-K-K-K | ||
| DCX (microtubule-associated protein [NP_034155]) | S-Q-L-S-T321-P-K-S-K | 105 |
| S-P-I-S-T331-P-T-S*-Pb | ||
| S-T*-P-T-S334-P-G-S-Lb | ||
| Kinesin heavy chain (microtubule-binding protein [P56536]) | Not determinedj | 227 |
| R-F-V-S-S175-P-E-E-V | ||
| Keratin 8 (intermediate filament protein) | Q-S-L-L-S73-P-L-V-L | 127 |
Nonnuclear proteins that have been implicated in a broad range of cellular functions have been demonstrated to be substrates for JNK-mediated phosphorylation. This table summarizes these proteins and provides sequence information on the phosphorylation sites identified. For each phosphorylation site, a nine-amino-acid sequence surrounding the residue that is phosphorylated is provided. As shown, multiple sites of phosphorylation have been identified for some nonnuclear proteins.
In these sequences, S* or T* represents Ser or Thr residues that have also been shown to be phosphorylated by JNKs, showing that JNKs may phosphorylate a number of closely spaced residues. It remains to be determined whether there is any requirement for hierarchical phosphorylation of these substrates by JNKs.
The numbering of the sites refers to the murine IRS-1 sequence. In the human sequence (GenBank accession no. NP_005535), these correspond to the sequences S-R-T-E-S307-I-T-A-T, I-T-A-T-S312-P-A-S-M, and G-K-P-G-S323-F-R-V-R.
The numbering of the sites refers to the human JIP1 sequence.
Phosphorylation sequences in 14-3-3 β and 14-3-3 ɛ are predicted by sequence similarity with 14-3-3 ζ and 14-3-3 σ (301).
JNK phosphorylation of Ser128 of Bad and the role of this in apoptosis have been questioned (347).
In the protein sequence, there are nine S-P and five T-P motifs, and the residues phosphorylated were expected to be similar to those shown in the same study to be phosphorylated in the related protein BimL (186). The motifs shown in italics are the best S-P or T-P matches with the heptapeptide consensus (Scansite [329] score, 0.13827).
In the protein sequence, there are 21 S-P and 23 T-P motifs. The motifs shown in italics are the best S-P or T-P matches with the heptapeptide consensus (Scansite [329] score, 0.13827).
In the protein sequence, there are 53 S-P and 26 T-P motifs. The motifs shown in italics are the best S-P or T-P matches with the heptapeptide consensus (Scansite [329] score, 0.13827).
In the protein sequence, there are 12 S-P and 11 T-P motifs. The motifs shown in italics are the best S-P or T-P matches with the heptapeptide consensus (Scansite [329] score, 0.27655).
FIG. 3.
Summary of the substrates of JNKs discussed in this review. Following its activation by phosphorylation of a specific threonine and tyrosine within its activation loop, JNK can phosphorylate a range of substrates. Phosphorylation can modulate the substrate protein activity in a positive or negative fashion; JNK binding can even modulate the activity in a phosphorylation-independent manner. In some cases, the consequences of phosphorylation by JNK have not yet been defined. The mitochondrial protein Bcl2 is shown as a protein that is both activated and inhibited following its phosphorylation by JNK because currently there is evidence to support either effect (76, 146, 331). Similarly, JNK actions on Bad have been shown to both increase and decrease its activity. In this example, these divergent effects have been attributed to the ability of JNK to phosphorylate distinct sites (84, 344), although the effects of JNK-mediated phosphorylation of Bad Ser128 to increase its proapoptotic actions have been questioned (347).
JNK-MEDIATED PHOSPHORYLATION OF THE ARCHETYPICAL SUBSTRATE c-Jun
Studies on the phosphorylation of c-Jun and related transcription factors have provided many useful insights into the mechanisms of JNK-mediated phosphorylation (117, 152). The phosphorylation sequences in c-Jun conform to the general consensus motif (Pro)-X-Ser/Thr-Pro [(P)-X-S/T-P], as originally defined by substrate phosphorylation studies using the archetypical MAPKs, the ERKs (7, 60, 113). This consensus sequence indicates the ability of MAPKs to phosphorylate either Ser or Thr residues (S/T) within a Pro (P)-containing sequence. JNKs, like other MAPKs, such as the ERKs and the related CDKs, are therefore considered Pro-directed Ser/Thr protein kinases. An explanation of the structural basis for the requirement for a Pro residue immediately following the phosphorylated Ser/Thr has been offered by the structure of the complex of CDK-2 with cyclin and a substrate peptide (34). Specifically, the presence of any amino acid other than Pro in this position would result in an uncompensated hydrogen bond from the nitrogen of the substrate peptide backbone and would therefore not be favored (34). A similar structural explanation is also predicted for the MAPKs, including the JNKs.
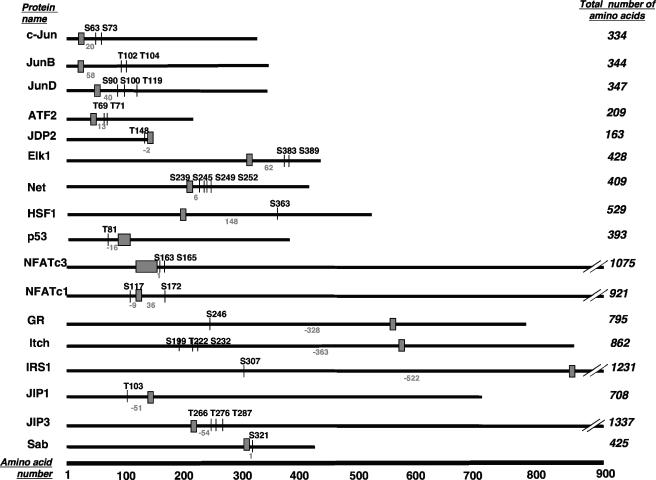
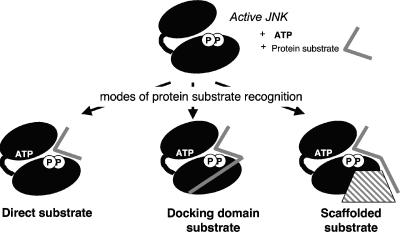
The initial studies on JNK-mediated phosphorylation of c-Jun also revealed a requirement for amino acid sequences, known as the δ domain or the JNK-binding domain (JBD), distant from the amino acids to be phosphorylated (4, 66, 77, 152, 210). The JBD sequence within c-Jun is shown in Table 2, and a general schematic diagram illustrating the relative positions of the phosphorylated residues in relation to the JBDs of c-Jun and other substrates is shown in Fig. 4. These distant targeting domains mediate interactions of other MAPKs with their substrates, upstream activators, phosphatases, and scaffold proteins (295) and thus are more generally termed common docking (CD) domains. The use of docking domains by MAPKs can enhance the efficiency and specificity of substrate phosphorylation (19, 99, 148, 276). Furthermore, small peptides making up the JBD of c-Jun inhibit JNK activity (4). However, as we describe in this review, docking sequences for JNK have not yet been identified for all substrates of JNKs. This raises the possibility that either these substrates are recognized independently of a docking site region or their docking domains do not conform to the sequences currently recognized as forming a JBD. The docking domains have significant implications for the substrate specificity of JNKs, as discussed in later sections of this review.
TABLE 2.
Summary of identified JBDsa
| Protein (function) | Experimentally determined JNK-binding sequenceb | Reference(s) |
|---|---|---|
| Nuclear proteins | ||
| c-Jun (transcription factor) | I33-L-K-Q-S-M-T-L-N-L-A43 | 77 |
| JunB (transcription factor) | K33-L-L-K-P-T-L-A-L-N-L-A44 | 339 |
| JunD (transcription factor) | L50-K-K-D-A-L-T-L-S-L-A60 | 339 |
| ATF2 (transcription factor) | K46-H-K-H-E-M-T-L-K-F-G56 | 118, 197 |
| JDP2 (transcription factor) | G153-N-L-L-E-Q-L-D-K-K163c | 155, 156 |
| Elk-1 (transcription factor) | G311-K-G-R-K-P-D-L-E-L-P321d | 321, 337 |
| Net (transcription factor) | S221-A-K-I-S-S-L-M-L-P-N-A-A233 | 86 |
| HSF1 (transcription factor) | G204-V-K-R-K-I-P-L-M-L-N-D215 | 65 |
| c-Myc (transcription factor) | C171-S-T-S-S-L-Y-L-Q-D-L-S-A-A-A-S-E187e | 6, 246 |
| P53 (transcription factor) | V97-P-S-Q-K-T-Y-H-G-S-Y-G-F-R-L-G-F-L-H-S-G117f | 41, 95 |
| NFATc3 (transcription factor) | P136-E-R-E-F-L-E-R-P-S-R-D-H-L-Y-L-P-L-E-P-S-Y-R-E-S-S-L162 | 57 |
| NFATc1α (transcription factor) | L126-G-L-Y-H-N-N-N-Q-F-F-H-D138 | 56 |
| Glucocorticoid receptor (nuclear hormone receptor) | A574-W-R-I-M-T-L-N-M-L 584g | 37, 266 |
| Regulator of protein turnover | ||
| Itch (E3 ligase) | R595-R-R-L-W-V-I-F-P-G604h | 100 |
| Adaptor proteins | ||
| IRS-1 | R849-L-A-R-P-T-R-L-S-L-G859 | 5, 185, 318 |
| JIP1 | R154-P-K-R-P-T-T-L-N-L-F164 | 245, 281 |
| JIP2 | H134-K-H-R-P-T-T-L-R-L-T144 | 338 |
| JIP3 | R202-K-E-R-P-T-S-L-N-V-F212 | 161 |
| β-Arrestin 2 | L192-M-S-D-R-R-S-L-H-L-E202 | 218 |
| Mitochondrial protein | ||
| Sab (function unknown) | A310-V-V-R-P-G-S-L-D-L-R320 | 322 |
| Regulator of cell movement | ||
| DCX | DC domainsi | 105 |
A range of JNK substrate proteins also interact with JNKs via sequences remote from their phosphorylation sites. This table summarizes these proteins and provides sequence information on the JBDs identified.
The consensus sequence for JBDs has generally been shown to follow the pattern R/K2-3-X1-6-L/I-X-L/I. Residues matching the consensus are shown in bold.
Although JDP2 has a JBD-like sequence, this C-terminal sequence mediates the interaction with JNK.
A Leu323→Ala/Ser324→Ala Elk-1 mutant failed to interact with JNK2 and was compromised in its interaction with JNK1, and thus the Elk-1 interaction site might extend beyond this JBD.
Not determined directly, but 171C-S-T-S-S-L-Y-L-Q-D-L-S-A-A-A-S-E187 is considered a δ domain-like sequence (6, 246).
A peptide sequence corresponding to p53 Val97 to Gly117 prevented complex formation between JNK and p53 (41, 95).
Bruna and colleagues (37) identified this sequence as a potential mediator of the interaction of the glucocorticoid receptor with JNK in the context of the receptor acting as an inhibitor of JNK.
In the database sequence (accession no. CAI17960), this sequence corresponds to residues 533 to 542.
The DC domains correspond to DCX residues 51 to 135 and 178 to 259.
FIG. 4.
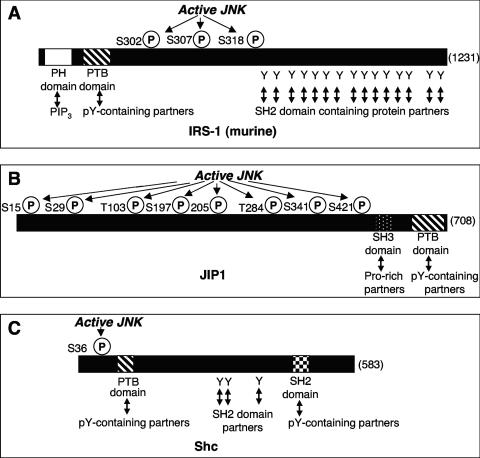
Schematic diagram illustrating the relative positions of the phosphorylated residues in relation to the JBDs of c-Jun and 16 other substrate proteins. At the bottom of the figure, the scale indicates the number of amino acids. Each protein substrate is represented by a solid line, and the total number of amino acids in each protein is indicated on the right. The positions of the amino acids phosphorylated by JNK indicated by the vertical lines are labeled with the residue phosphorylated (i.e., Ser or Thr [S or T] and its number in the sequence). The JNK-binding domains are each denoted by a small box, and the residue numbers are indicated. The distances between the JNK-binding domains and the closest residue phosphorylated are indicated, where positive numbers indicate that the phosphorylation sites lie to the C-terminal side of the JNK-binding domain and negative numbers indicate that the phosphorylation sites lie to the N-terminal side of the JNK-binding domain. This information was derived from the data presented in Tables 1, 2, and 3.
As mentioned in the opening paragraph, current analyses suggest that 518 protein kinases form the human kinome (205). Furthermore, if one-third of intracellular proteins can be phosphorylated, once various protein splice forms are taken into account, this may amount to some 20,000 phosphoproteins (151). A simple calculation would therefore suggest that each protein kinase should, on average, have ∼40 substrates. This calculation does not take into consideration the idea that some proteins will be phosphorylated at multiple sites by different protein kinases or that many different protein kinases may phosphorylate the same site on one substrate. The MAPKs are likely to be consistent with this calculation, with an initial proteomic study identifying 25 phosphoproteins following ERK activation (189). Although not all of these phosphoproteins may be direct substrates of ERK, this does confirm the complexity of signaling events downstream of MAPKs such as the ERKs.
OTHER NUCLEAR SUBSTRATES OF JNK
Early studies on the ERK subfamily of MAPKs suggested that these kinases must be located within the same subcellular compartment as their substrates and demonstrated that these kinases could translocate to compartments such as the nucleus upon exposure of the cells to the appropriate stimulation (114, 187, 300). Similarly, the JNKs have been shown to translocate to the nucleus following cell exposure to agents that lead to activation of the JNKs, including UV irradiation, osmotic shock, and ischemia (47, 158, 223).
Transcription Factors as JNK Substrates
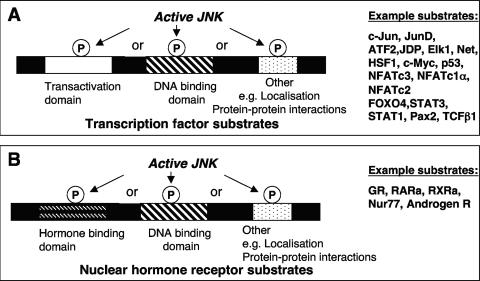
Proteins that act as transcription factors are regulators of gene expression in eukaryotic cells. Typical transcription factor structure includes a transactivation domain together with a DNA-binding domain that recognizes specific DNA elements within the promoters of target genes. Transcription factor activity can be regulated, either positively or negatively, by a number of biochemical processes, including phosphorylation (Fig. 5A).
FIG. 5.
Major classes of known nuclear substrates of JNK. (A) A range of transcription factors have been shown to be JNK substrates. JNK-mediated phosphorylation may be directed towards the transactivation domains, DNA binding domains, or other protein domains, and this alters transcription factor activity. (B) A range of nuclear hormone receptors have also been shown to be JNK substrates.
The Jun family of transcription factors.
Phosphorylation increases the transcriptional activity of c-Jun, as described in the preceding section, as well as that of the related protein JunD (Fig. 3) (339). JunD, although dispensable for development, has been shown to be involved in muscle differentiation (11) and has been implicated in the development of cardiac hypertrophy (263). The JunD phosphorylation and docking site sequences (Tables 1 and 2) are conserved in a shorter splice form of JunD, Δ-JunD, which lacks the N-terminal menin interaction domain, suggesting that both JunD splice forms are under the control of JNK phosphorylation (339). The C-terminal part of JunD is also involved in interactions with ERKs, thus allowing JunD regulation by both the ERK and JNK pathways, and indeed, the N-terminal sites in JunD are phosphorylated by both ERK and JNK (306, 309). The sequences surrounding JunD Ser90 and Ser100 are analogous to sequences surrounding Ser63 and Ser73 of c-Jun (Table 1). Additionally, the sequence surrounding JunD Thr117 shows similarity to the sequences surrounding c-Jun Thr91, which is also phosphorylated by JNK (230). Note that JNK binding to the JunD JBD has been shown to be poor compared with JNK binding to the JBD of c-Jun or JunB (117, 152). This is reinforced by the finding that c-Jun is a more efficient substrate than JunD in vitro for four different JNK isoforms (the jnk1 splice form JNK1α1, the jnk2 splice forms JNK2α2 and JNK2β2, and the the jnk3 splice form JNK3α1) (339). This study suggests that protein kinases from the JNK family exhibit considerable specificities in substrate docking and phosphorylation, even for related transcription factor substrates.
Although the related transcription factor JunB was initially not considered a JNK substrate because it lacks serine residues in the appropriate consensus sequences (Ser74 and Ser84 in murine JunB [accession number NP_032442], within the sequence G71QGS74DTGASLKLAS84TELERL90) (152), subsequent studies showed that JunB is phosphorylated by JNK at two closely spaced threonine residues, Thr102 and Thr104 (Table 1) (1). Like in the case of c-Jun, these phosphorylation sites are C-terminal to a conserved JBD sequence (Table 2 and Fig. 4). Furthermore, this phosphorylation of JunB would appear to potentiate the transcriptional activity of JunB (Fig. 3), as seen when a Thr102→Glu/Thr104→Glu mutant (designed to mimic the JNK-mediated phosphorylation events) showed enhanced ability to synergize with c-Maf in transcriptional activation of the interleukin-4 promoter (1). Thus, this has implicated the JunB protein in T-cell development and in directing Th2 differentiation.
The ATF family of transcription factors.
Together with the transcription factors of the Jun family, the Fos and ATF2 families of bZIP transcription factors also form part of the transcription factor complexes known as the activator protein 1 (AP-1) family (188). Within this transcription factor family, there are additional substrates for JNK. While most evidence supports the presence of a Fos kinase that is not related to JNK (75, 288), ATF2 is recognized as a JNK substrate (118, 197). As seen for JNK-mediated phosphorylation of c-Jun, JNK-mediated phosphorylation of ATF2 is directed to two closely spaced residues, namely, Thr69 and Thr71, in its N-terminal transactivation domain (118, 197) (Tables 1 and 2). In a manner that therefore shows similarity to c-Jun regulation by JNK, the JNK-mediated phosphorylation of ATF2 enhances its transcriptional activity (Fig. 3) (118, 197, 304).
Knowledge of the regions of ATF2 interacting with and phosphorylated by JNK has led to the development of an ATF2-derived protein fragment (ATF250-100). This peptide, when delivered to cells, alters the balance between c-Jun and ATF2 transcriptional activities, leading to the attenuation of ATF2 activity and the induction of c-Jun activity as well as the sensitization of cultured melanoma cells to chemotherapeutic agents (26). These observations, together with recent studies on the substrate-binding characteristics of ERK2 (226), suggest that appropriate substrate-derived peptides will allow a subset of protein kinase substrates to be selectively inhibited. This is a significant advance over the ATP-competitive inhibitors of kinases currently in use. ATP-competitive inhibitors would be expected to inhibit the phosphorylation of all protein substrates for a particular protein kinase and may not be specific for a particular kinase due to the difficulty in discriminating between the conserved ATP-binding sites of various protein kinases (for a review, see reference 92). Thus, a greater understanding of the range of the possible intracellular JNK substrates is critical in the development of new approaches to achieve substrate-selective and specific inhibition of JNK.
There may be additional complex relationships between JNK and the ATF family of transcription factors. While one study has shown the importance of phosphorylation of residues Thr51 and Thr53 in the N-terminal activation of ATFa for the transcriptional activation of this specific transcription factor, it appeared that ATFa was not a direct substrate for JNK2 (74). Instead, the N-terminal domain of ATFa served as a docking site for JNK (29), allowing ATFa-associated partners, such as JunD, to then be phosphorylated by JNK (74). This relationship emphasizes the possibilities of trans-phosphorylation events in the regulation of transcription factor complexes.
JDP2 as a JNK substrate.
JNK can also phosphorylate another binding partner of c-Jun, i.e., Jun dimerization protein 2 (JDP2). JDP2 is a basic leucine zipper transcription factor family member that interacts with c-Jun as well as the transcription factors ATF2 and CCAAT/enhancer-binding protein gamma (156). The site of phosphorylation of JDP2 has been mapped to Thr148 (156) and a JBD identified in subsequent studies (155) (Tables 1 and 2). It is important that although the JDP2 sequence apparently contains a classic JBD consensus sequence within its leucine zipper domain (i.e., K136NEKQHLIYMLNLH149 [residues of the consensus are shown in bold]), the site of interaction was mapped to the JDP2 C-terminal region beyond residue 153 (155). Indeed, a 14-amino-acid fragment derived from the JDP2 sequence (i.e., JDP2150-163), when added to the transcription factor ATF3, which is usually not a JNK substrate, facilitated JNK phosphorylation of ATF3; this sequence alone was therefore sufficient for JNK interaction (155). In contrast to c-Jun-binding partners such as c-Fos or ATF2, JDP2 acts as a repressor at the AP-1 site, and it also inhibits Ras-driven transformation of NIH 3T3 cells and suppresses tumor formation in vivo in a PC3 cell xenograft model (128). The functional consequences of JDP2 phosphorylation by JNK remain to be elucidated (Fig. 4).
Elk-1 as a JNK substrate.
JNK also phosphorylates a number of other transcription factors that do not form part of the AP-1 complex. JNK phosphorylates the Ets domain-containing transcription factor Elk-1 on Ser383 and Ser389 in its C-terminal transactivation domain (Table 1); these same residues are also phosphorylated by the ERK MAPKs (47, 132, 206, 321). This phosphorylation of Elk-1 increases its complex formation with the serum response factor and, in this way, increases transcriptional activity (Fig. 3) (107, 108, 321). The binding of all 10 JNK isoforms (i.e., four JNK1 splice forms, four JNK2 splice forms, and two JNK3 splice forms) to Elk-1 appeared considerably weaker than the binding of these JNK proteins to either c-Jun or ATF2 (117, 337) (see Table 2 for the Elk-1 JBD sequence). Although the exact residues within the JBD required for binding either JNKs or ERKs do differ, p38 MAPK phosphorylation of Elk-1 does not appear to require an intact JBD (337). These differences in specificity determinants can thus play a pivotal role in producing unique nuclear responses following activation of the different MAPK pathways.
The requirement for specific docking domains and subsequent kinase-specific phosphorylation events is further illustrated by the observation that JNK binds to the ternary complex factor Net (also known as Elk-3) via a binding motif (Table 2) that is distinct from that bound by ERK or p38 (86). This JNK-mediated phosphorylation regulates nuclear export of Net and inhibits Net-mediated effects (Fig. 3). The mechanism of transcription factor inhibition can be explained by the actions of JNK to phosphorylate four Ser residues of Net, namely, Ser239, Ser245, Ser249, and Ser252 (Table 1), within the Net nuclear export box, enhancing nuclear export. The docking domain also shows homology to other JBD sequences (Table 2). Interestingly, the Net residues phosphorylated by JNK are distinct from those phosphorylated by ERK or p38, and the actions are distinct from those of JNK-mediated effects to increase the transcriptional activity of the related Ets family transcription factor Elk-1 (Fig. 3). Heat shock factor 1 (HSF-1) is another transcription factor that is also inactivated following its phosphorylation by JNK (65). In this case, transcriptional activity is decreased, rather than inhibition of its actions, requiring its changes in nuclear localization. The site of phosphorylation in HSF-1 appears to be Ser363 (Table 1), one of five Ser/Thr residues within S/T-P motifs. This phosphorylation also depended on an interaction motif showing similarity to the JBD of c-Jun (Table 2) (65). This inhibition by JNK-mediated phosphorylation provides a mechanism, in addition to ERK-mediated phosphorylation or interaction with a range of heat shock proteins, to downregulate the actions of this transcription factor (65).
c-Myc as a JNK substrate.
Other transcription factors have also been investigated as mediators of the nuclear actions of JNK. In c-Myc, Ser61 and Ser71 have been shown to be phosphorylated by JNK1 and, to a lesser extent, by JNK2 and JNK3 (246). This phosphorylation increases c-Myc-mediated apoptosis (Fig. 3) (246). Interestingly, the related transcription factors s-Myc and Max are not JNK substrates (246). The region of c-myc involved in the interaction with JNK was mapped to residues 1 to 262 of c-Myc (246). This was subsequently confirmed in an independent study that blocked the binding of JNK to c-Myc through the use of a peptide corresponding to residues 127 to 189 of c-Myc (6). A δ domain-like region of c-Myc was identified within residues 171 to 187 (6) (Table 2).
p53 as a JNK substrate.
The p53 tumor suppressor protein is another transcription factor that is phosphorylated by many protein kinases, including JNK (3). The interaction between JNK and p53 has been mapped to amino acids 97 to 117 of p53 (Table 2) by the demonstration that a synthetic peptide corresponding to these p53 residues prevented phosphorylation of p53 and its interaction with JNK (3, 94). The overexpression of the JNK pathway upstream kinase MEKK1 (Fig. 1) has been shown to increase p53 stability and transcriptional activity (95), and the p53 residue phosphorylated by JNK was subsequently mapped to Thr81 (41) (Table 1). Phosphorylation of this residue appeared to be critical for p53 stabilization and conferred its transcriptional activity and ability to elicit apoptosis; in the absence of JNK expression or JNK-mediated phosphorylation, p53 was inactive (41). Thus, JNK can be considered an activator of p53 actions (Fig. 3). Interestingly, a JBD-like sequence (I80-F-K-E-Q-G-L-T-L-P-L-Y91, with clear similarity to the I33-L-K-Q-S-M-T-L-N-L-A43 sequence in c-Jun) has also been described for the tumor suppressor protein BRCA2 (221). However, the BRCA2 protein has not been shown to interact with JNK (209), and other protein kinases have been suggested to phosphorylate BRCA2 (220). This emphasizes that all potential JBD-like sequences require experimental validation before any link with JNK-dependent signaling can be suggested. Additional controversy surrounds the role of JNK-mediated phosphorylation of p53, as the sites of phosphorylation are only present in the rat sequence and not conserved in either mouse or human sequences. This raises questions on the importance of these phosphorylation sites in p53 function.
The NFAT family of transcription factors.
Based on the concept that distinct docking domains mediate JNK binding to its substrates, JNK1 has been used as bait in a yeast two-hybrid screen of a mouse embryo cDNA library in a search for novel interacting partners and substrates (57). This screen revealed an interaction of JNK1 with the transcription factor nuclear factor of activated T cells c3′ (NFATc3; also known as NFAT4 or NFATx); the interaction was also confirmed in mammalian cells (57). NFATs are calcium-sensitive transcription factors that have been shown to be critical regulators of T-cell development, and in addition to regulating other differentiation programs, they function in a range of tissues, being involved in skeletal muscle differentiation, cardiac valve development, and osteoclast differentiation (135, 201). The functional utility of the NFATs has been explained by their complex mechanisms of regulation and their ability to integrate calcium signaling with other signaling pathways (for a review, see reference 201). The residues mediating interaction with JNK were mapped to NFATc3 residues 162 to 207, with Ser163 and Ser165 as the sites of phosphorylation (57) (Table 1; Fig. 4). In contrast to the positive effects of JNK in enhancing transcription, as seen in many of the preceding examples of transcription factor substrates of JNK, this JNK-mediated phosphorylation of NFATc3 again resulted in the nuclear exclusion of this transcription factor (57). Thus, the activation of the JNK pathway also antagonizes the actions of NFATc3 (Fig. 3).
JNK also phosphorylates the related NFAT family member NFATc1α on Ser117 and Ser172, and this requires the presence of a JBD within residues 126 to 138 (56) (Tables 1 and 2 and Fig. 4). A comparison of the phosphorylation site sequences suggests that NFATc1α Ser172 is equivalent to NFATc3 Ser165 (Tables 1 and 2). Both NFATc1α phosphorylation sites are close to the domain that interacts with the calcium-dependent phosphatase calcineurin (56). Calcineurin can preferentially dephosphorylate Ser172 in vitro, while the phosphorylation of Ser117 was shown to be critical in regulating the targeting of calcineurin to NFATc1α (56). Thus, phosphorylation of NFATc1α by JNK inhibits the interaction with calcineurin, thus blocking its nuclear entry and providing a molecular mechanism for the observed increased nuclear localization of NFATc1α in the T cells of jnk1−/− mice (83). This antagonism of NFAT signaling by JNK activation is also seen in other systems, such as the heart, where JNK activation negatively regulates NFATc3 activation and inhibition of JNK enhances NFAT signaling, with subsequent enhanced hypertrophic growth (192). These examples again illustrate the importance of JNK in mediating the inhibition of transcriptional events, in addition to its more widely acknowledged role as a positive mediator of signaling.
In contrast to this negative regulation of NFAT signaling, the phosphorylation of a different NFAT transcription family member, NFATc2, by JNK stimulates its transcriptional activity (248) (Fig. 3). The effects of JNK required Thr116 (Table 1) within the docking site for calcineurin in the NFATc2 regulatory domain. No effect of JNK activation on the subcellular localization of NFATc2 was observed (248). Importantly, these different effects of JNK on the different NFAT isoforms highlight the danger of studying the effects of JNK on one member of a transcription factor family and then extrapolating these effects to other closely related members of the same family. Instead, functional testing appears to be required in each case.
The forkhead family of transcription factors.
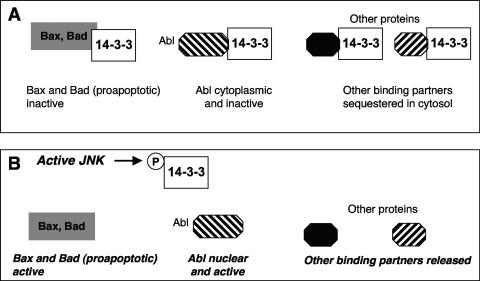
The forkhead family member FOXO4 has also been shown to be phosphorylated following the exposure of cells to TNF-α or oxidative stress in the form of hydrogen peroxide (90). The phosphorylation of the FOXO family has come under increasing attention as an event downstream of activation of the prosurvival protein kinase Akt (for a review, see reference 39). For example, the Akt-mediated phosphorylation of FOXO3a decreases its transcriptional activity because phosphorylated FOXO3a is bound by cytosolic 14-3-3 proteins and thus sequestered in the cytosol. This prevents upregulation of the transcription of enzymes such as catalase and Mn-dependent superoxide dismutase and thus changes the cellular levels of reactive oxygen species (for a review, see reference 40). In contrast, a role for JNKs was suggested more recently, based on the observation that FOXO4 was no longer phosphorylated in cells deficient in both JNK1 and JNK2 (i.e., jnk1−/− jnk2−/− cells) following their exposure to hydrogen peroxide (90). JNK-dependent phosphorylation enhanced FOXO4 transcriptional activity rather than changing its binding to the cytosolic 14-3-3 proteins (90) (Fig. 3). These results reveal a point of cross talk between JNK and other signal transduction pathways. Further points of cross talk will be discussed later in this review.
The positive regulation of FOXO activity in mammalian cells (90) is consistent with studies of the model organism Caenorhabditis elegans (247). In this organism, JNK interacts with and phosphorylates the FOXO homologue DAF-16. Although the phosphorylation sites appeared to be within the N-terminal region of DAF-16 (residues 83 to 307), the residues required for the interaction and for phosphorylation have not yet been identified (247). The significance of this phosphorylation lies in the consequences of DAF-16 regulation. Specifically, the negative regulation of DAF-16, as might result from enhanced signaling from the insulin-like growth factor receptor, has been associated with a shortened life span (for a review, see reference 145). Thus, JNK activation and subsequent DAF-16 phosphorylation and activation resulted in an increased life span, presumably through the upregulation of genes promoting resistance to stress (247).
Similar results with FOXO regulation have also been shown in Drosophila melanogaster, with dfoxo required for JNK-mediated life span extension (313). Thus, in this range of different systems, the FOXO forkhead transcription factors provide a point of convergence in signaling by the insulin-like growth factor and JNK signaling cascades. It will therefore be important to determine which of the mammalian FOXO family members (for a review, see reference 27) are regulated by JNK-mediated phosphorylation. Recent evidence suggests that the JNK pathway is involved in the regulation of the nuclear translocation of FOXO1, with JNK phosphorylation leading to changes in the localization of the transcription factor PDX-1, impairing PDX-1 function, as observed in pancreatic β cells in diabetes (157). However, it is not yet possible to discount a role for JNK as a negative regulator of more membrane-proximal signaling events, such as the phosphorylation of the adaptor protein insulin receptor substrate 1 (IRS-1), as discussed below, which would alter FOXO1 subcellular distribution through the negative impact on Akt signaling. Again, this emphasizes the likely contributions of multiple signaling pathways with extensive opportunities for cross talk and control.
The STAT family of transcription factors.
Other transcription factors are subject to control by multiple phosphorylation events. One example is the signal transducer and activator of transcription (STAT) family, which has been implicated downstream of signaling by both cytokine and growth factor receptors and whose members have been considered critical growth regulators (for a review, see reference 44). The phosphorylation of Tyr705 of STAT3 is mediated by the JAK family of tyrosine kinases, whereas JNK also phosphorylates STAT3 on Ser727 (Table 1) (349). Both phosphorylation events are required for full transcriptional activation of STAT3 (Fig. 3). Similarly, the activation of the JNK pathway downstream of protein kinase C-δ can also result in the phosphorylation and activation of another STAT family member, STAT1 (Fig. 4). This activation requires phosphorylation of Ser727 (352) (Table 1). The involvement of both STAT1 and STAT3 as mediators in a range of diseases, including cancer, inflammatory disease, and ischemia/reperfusion injury (for reviews, see references 255, 284, and 302), warrants further evaluation of the contributions of JNKs to their initiation and development, as JNKs may therefore play critical regulatory roles.
The Pax family of transcription factors.
In addition to the ability to modulate responses to stress through the phosphorylation of a range of transcription factors involved in various aspects of cell growth, as described above, JNKs may also phosphorylate additional transcription factors involved in development. The Pax family of transcription factors is required for the embryonic development of a range of tissues (for a review, see reference 55). Pax2 is required for kidney development as well as for development of the inner ear and the optic cup and has been shown to be a substrate for JNK. Pax2 can be isolated in a complex with the JNK-interacting protein JIP1 (43). Phosphorylation enhances Pax2 transcriptional activity (43) (Fig. 3). Although the phosphorylation site(s) in Pax2 was not identified, it is possible to predict possible phosphorylation sites (Table 1) by using the consensus sequence derived from other JNK substrates (329). It will now be interesting to map the JNK phosphorylation sites in Pax2 and compare these with the predicted sites. In addition, it will be critical to identify possible JNK interaction motifs and to evaluate whether other Pax family members might be regulated by the actions of JNK.
TCFβ1 as a JNK substrate.
Other transcription factors, such as the POU domain-containing protein T-cell factor β1 (TCFβ1), a key regulator during development and lymphocyte activation, also appear to be substrates for JNK, with TCFβ1 being phosphorylated at both the Ser232 and Thr242 residues (Table 1) within its DNA-binding domain (154). This phosphorylation increases the binding of TCFβ1 to DNA and thus likely mediates an increase in transcriptional actions following JNK activation in T cells (154) (Fig. 3). Therefore, in many of the examples discussed thus far, phosphorylation by JNK increases the activities of a range of transcription factor proteins (Fig. 3).
Nuclear Hormone Receptors as JNK Substrates
Nuclear hormone receptors form a specific subset of transcription factor proteins containing a DNA-binding domain which are regulated through direct interaction with families of hydrophobic hormones, such as steroids and the retinoids (Fig. 5B). In addition to the actions of JNK to modulate the activities, localization, or stabilities of the transcription factors described in the preceding section, JNK has also been shown to directly phosphorylate many nuclear hormone receptors. For example, peroxisome proliferator-activated receptor γ1 (PPAR-γ1) is a substrate for JNK (45). PPARs bind to response elements in complex with the retinoic acid receptor and activate transcription in response to a range of endogenous ligands, such as fatty acids and arachidonic acid metabolites, or foreign ligands, such as the antidiabetic drugs thiazolidinediones. JNK phosphorylates Ser82 (Table 1) in the transactivation domain of PPAR-γ1, and this decreases its transcriptional activity (Fig. 3) (45). This phosphorylation may contribute to the development of insulin resistance when adipose tissue releases TNF-α and then signaling via the JNK pathway suppresses PPAR-γ1 activity in vivo.
The glucocorticoid receptor has also been shown to be negatively regulated through its phosphorylation by JNK (266) (Fig. 3). The major site of phosphorylation of the rat glucocorticoid receptor by JNK in vitro was mapped to Ser246 (Table 1). This site was confirmed within cultured cells (266) and was one of four major phosphorylation sites within the N-terminal transcriptional regulatory region (175). Thus, this JNK-mediated inhibition would decrease the actions of glucocorticoids to induce differentiation, regulate gluconeogenesis, and suppress inflammation. Further studies using a form of the human glucocorticoid receptor mutated to prevent phosphorylation by JNK (i.e., Ser226→Ala mutant) suggested that JNK-mediated phosphorylation of the glucocorticoid receptor enhanced nuclear export, apparently by a leptomycin B-sensitive, exportin/CRM1-dependent mechanism (147). This provides an additional mechanism for inhibition of its effects in cells and is similar to the enhancement of nuclear export of the transcription factors Net and NFATc3 described above. The p38 MAPKs were also shown more recently to inhibit glucocorticoid receptor actions, in this case by indirectly targeting the ligand-binding domain (289). This observation demonstrates that there are multiple mechanisms of inhibition of the transcriptional activity of this nuclear hormone receptor.
Conversely, glucocorticoids also inhibit the actions of the JNK pathway (115). This has been attributed to the ability of the glucocorticoid receptor to interact directly with JNK and to inhibit its activity (37). An interaction motif showing similarities to the motif in c-Jun was seen in the glucocorticoid receptor (37) (Table 1). Furthermore, this JBD-like sequence was shown to be required for glucocorticoid-induced nuclear localization of JNK, suggesting a role for the glucocorticoid receptor in shuttling JNK to the nucleus (37). Interestingly, the nuclear translocation of JNK also increased JNK binding to the AP-1-associated response elements in the c-jun gene, and this binding of inactive JNK may maintain repression of AP-1-dependent transcription (37). Clearly, these observations show that all potential JBD-like sequences require experimental validation before any conclusions about their roles in the regulation of JNK-dependent signaling can be drawn.
The retinoic acid receptors RXR and RARα have also been shown to be substrates of JNK, providing one mechanism to explain how stress can inhibit retinoid signaling (2, 181, 283). Interestingly, RXR is a substrate for JNK as well as the dual-specificity kinase MKK4/SEK1, the latter of which is usually considered a JNK activator only (181). MKK4/SEK1-mediated phosphorylation of RXR inhibited retinoid-mediated transcriptional signaling, providing some of the first evidence that MKK4/SEK1 can initiate effects independent of its actions on JNK activation (181). The sites of phosphorylation for MKK4/SEK1 were in domains distinct from those phosphorylated by JNK (181). Although the residues phosphorylated by JNK were not identified, mutation of a single tyrosine in RXR (Tyr249) decreased phosphorylation and abrogated the ability of MKK4/SEK1 to suppress transcriptional activity (181). This is in contrast to the actions of JNKs as Ser/Thr kinases and highlights the possibility that there may be many control points for regulation of retinoid receptor activities.
Initial reports suggested that JNK mediated the phosphorylation of RXRα at residues Ser61, Ser75, Thr87, and Ser265 (Table 1) (2). This phosphorylation did not appear to affect the transactivation properties of either RXRα homodimers or RXRα/RARα heterodimers (2). More recently, the phosphorylation of the three N-terminal residues within the transactivation domain has been shown to be required for the maximal transcriptional activity that results from the cooperation of RXRα and its partner RARγ (106). In addition, the importance of the phosphorylation of Ser265 was also highlighted more recently; this residue lies outside the classic transactivation domain of RXRα in the omega loop of the ligand-binding domain (35). This phosphorylation enhanced the expression of some retinoic acid target genes but decreased the expression of others (35). Thus, JNK-mediated phosphorylation affects RXRα function by modulating its transcriptional effects (Fig. 3). This altered regulation of retinoic acid target genes may thus have important consequences for retinoic acid actions in the cell, as seen for the cooperation of retinoic acid and arsenic trioxide in apoptosis through the JNK-mediated phosphorylation of RXRα (298).
JNK has also been implicated in the inhibition of RXRα transactivation when cells are exposed to stress in the form of arsenic trioxide (204). Mutational analysis has suggested the requirement for Ser32, suggesting this as the novel Ser target for JNK involved in the inhibition of nuclear receptor function (Table 1) (204). The mechanism of this inhibition requires further evaluation, as direct effects on stability, dimer formation, or interaction with DNA have not been observed (204). The JNK-mediated phosphorylation of RARα was recently mapped to residues Thr181, Ser445, and Ser461 (Table 1) (283). This phosphorylation results in the inhibition of RARα through induced proteasomal degradation of RARα (Fig. 3) (283). Specifically, when a RARα mutant lacking these JNK phosphorylation sites was expressed in cells, UV irradiation did not lead to decreases in RARα levels. Conversely, inhibition of JNK in a human lung cancer cell line by the use of the JNK inhibitor SP600125 enhanced RARα levels. This link between the JNK signaling pathway and degradation of specific proteins is explored further in the following section; for example, the E3 ligase Itch has been shown to be a specific substrate for JNK.
Other mechanisms, such as alterations in nuclear export, remain to be investigated, particularly following the observation that the orphan nuclear receptor family member nur77 is phosphorylated in its N terminus by JNK (174). This JNK-mediated phosphorylation of nur77 is involved, in conjunction with phosphorylation by Akt, in the modulation of nur77 functions through regulation of nur77 nuclear export (Fig. 3) (121). The exact residues in nur77 that are phosphorylated by JNK remain to be identified (Table 1). JNK-mediated phosphorylation of Ser650 of the androgen receptor (Table 1) was also recently shown to increase its nuclear export to decrease its transcriptional activity (Fig. 3) (109). Thus, the regulation of subcellular localization by JNK-mediated phosphorylation can be a critical control mechanism in signaling downstream of JNKs.
Additional Nuclear Proteins as JNK Substrates
In addition to the groups of transcription factors and nuclear hormone receptors described as JNK substrates above, other nuclear proteins are phosphorylated by JNK. For example, heterogeneous nuclear ribonucleoprotein K (hnRNP-K) is part of a large family of nuclear RNA-binding proteins and has been implicated in diverse cellular and molecular functions, such as nuclear-cytoplasmic shuttling and RNA transcription and translation (for a review, see reference 31). hnRNP-K was identified as a JNK substrate through a chemical genetic approach that used an ATP analogue and a JNK mutant specifically modified in its ATP-binding pocket to use this ATP analogue (119). Thus, two modifications were made to JNK2 (Met108→Gly and Leu168→Ala) to allow its use of the ATP analogue N6-(2-phenythyl)-ATP. Following expression of this JNK2 mutant in 293T cells and its activation following exposure to UV irradiation, incubation of this kinase in the presence of protein extracts prepared from 293T cells and radiolabeled N6-(2-phenythyl)-ATP allowed the visualization of radiolabeled proteins separated by two-dimensional gel electrophoresis. Tandem nanoflow electrospray mass spectrometry of silver-stained spots that corresponded to phosphorylated proteins identified three peptide sequences, with each corresponding to a peptide from hnRNP-K (119).
Mutational analysis of the hnRNP-K protein has suggested that JNK phosphorylates two sites, Ser216 and Ser353 (Table 1), although a JBD has not been identified (119). JNK phosphorylation of hnRNP-K did not affect inhibition of RNA translation by hnRNP-K (120), but it was shown to enhance the ability of hnRNP-K to drive AP-1-dependent reporter gene expression (Fig. 3) (119). It will be critical to identify how the JNK-mediated phosphorylation of hnRNP-K contributes to its functions in cells, particularly when there are reports that phosphorylation by other protein kinases, such as ERKs and those of the Src family, can regulate hnRNP-K function in translation (31). hnRNP-K has also been shown to function within the DNA damage response pathway, being a target of the HDM2 ubiquitin ligase that is thus stabilized in response to DNA damage stimuli, such as UV irradiation (231). The stabilized hnRNP-K protein can then act as a transcriptional coactivator of the p53 protein (231). It will thus be of considerable interest to evaluate whether phosphorylation controls hnRNP-K activity to allow fine-tuning of DNA damage-induced transcriptional events. Surprisingly, the study identifying hnRNP-K as a JNK substrate appears to be the only study to date to use a chemical genetic approach for the identification of JNK substrates (119). This may reflect the limited availability of the modified ATP analogue, although a recent report has extended this approach with the use of ATP analogue inhibitors and sensitive JNK mutants to dissect the time course of signal transduction of JNKs in primary murine embryonic fibroblasts in response to TNF-α (305).
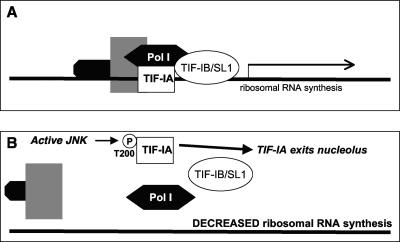
Additional substrates may also help to explain other stress-activated nuclear responses in cells. The exposure of cells to stress decreases the expression of many gene families, including the expression of genes encoding ribosomal proteins and splicing factors (232). The mechanism has been investigated, with inactivation shown to result from phosphorylation of the polymerase I (Pol I)-specific transcription factor TIF-IA by JNK at Thr200 (211) (Table 1). This phosphorylation abrogates complex formation between TIF-IA, Pol I, and the TATA-binding protein-containing factor TIF-IB/SL1 (211) (Fig. 6). The overexpression of the Thr200→Val mutant of TIF-IA that cannot be phosphorylated by JNK was shown to prevent inactivation of TIF-IA and thus lead to Pol I transcription even in the presence of stress (211). Thus, JNK-dependent phosphorylation of TIF-IA following the exposure of cells to stress provides a mechanism to prevent ribosomal synthesis (Fig. 3). This mechanism represents global control of protein translation during stress and may thus act as a protective mechanism for the cell under these situations.
FIG. 6.
The nucleolus as a JNK-responsive stress sensor. (A) rRNA synthesis under normal cellular conditions requires the actions of a protein complex including the Pol I-specific transcription factor TIF-IA, Pol I, and the TATA-binding protein-containing factor TIF1B/SL1. (B) The Pol I-specific transcription factor is phosphorylated by JNK, abrogating complex formation and inhibiting ribosomal synthesis, when JNKs are activated following the exposure of cells to stress.
Taken together, the studies outlined in this and the preceding section highlight the range of nuclear proteins that are JNK substrates. However, active JNK is not restricted to the nucleus and so may have an equally complex range of nonnuclear substrates. For example, the arrestin proteins were recently shown to interact with JNK and allow for the relocalization of JNKs from the nucleus to the cytoplasm (282). In the following sections, we present an overview of nonnuclear JNK substrates that contribute to diverse cellular responses, including protein degradation, signal transduction, apoptotic cell death, and cell movement.
LINKS BETWEEN JNK ACTIVATION AND PROTEIN DEGRADATION
The controlled degradation of proteins provides cells with a robust approach to controlling protein activity. Proteins destined for degradation are thus modified in a highly regulated fashion through their covalent linkage to a conserved 76-amino-acid peptide, ubiquitin (for a review, see reference 256). Indeed, the attachment of multiple ubiquitin molecules, as mediated by a series of ubiquitin-activating enzymes (E1), ubiquitin-conjugating enzymes (E2), and ubiquitin-protein ligases (E3), provides the basis for defining proteins targeted for degradation by the 26S proteasome (256) (Fig. 7). Although the processes of phosphorylation and degradation have been considered separate intracellular events, the possible links between the control of protein ubiquitination and protein phosphorylation are increasingly being recognized (for a review, see reference 101).
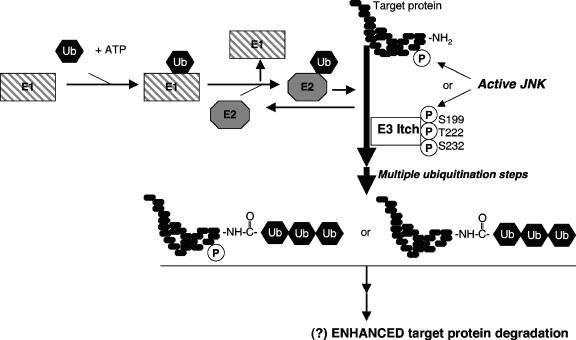
FIG. 7.
Links between JNK and protein degradation. The degradation of proteins follows their covalent modification by polyubiquitination, a multistep reaction requiring the actions of E1, E2, and E3 ligases. The JNK-mediated phosphorylation of the target protein or E3 ligases, such as Itch, can alter the rates of protein degradation. For Itch, the activities are enhanced, but controversy remains on whether all actions of JNKs on protein degradation will result in enhanced degradation.
Studies of T Cells Reveal that the E3 Ligase Itch is a JNK Substrate
There has been a long-standing interest in the roles of JNK in T-cell differentiation and function (82, 83, 265, 334). An evaluation of T cells isolated from mice that express an inactive form of the JNK pathway upstream activator (i.e., Mekk1ΔKD mice) identified a role for JNK-dependent events associated with protein degradation (102). In the T cells of these animals, the increased levels of production of interleukin-2 and the downstream cytokines interleukin-5, interleukin-10, and interleukin-13 were accompanied by increased protein levels (but not mRNA levels) of the transcription factors c-Jun and JunB (102). The enhanced stability of these transcription factors was confirmed in pulse-chase experiments, suggesting that the JNK pathway may assist in accelerating protein turnover in CD4+ T cells and thus may have a role in their polarization into Th1 and Th2 effector cells.
The similarity between the phenotypes of these Mekk1ΔKD T cells and those isolated from animals with disruption of the E3 ligase Itch prompted an evaluation of the functions of JNK (102). Specifically, the E3 ligase Itch was found to be a substrate of JNK with multiple sites of phosphorylation, including Ser199, Ser222, and Ser232 (Table 3). A specific JBD conforming to the general features observed in the JBD of c-Jun was also identified (Table 2) (100), but it should be noted that the hydrophobic residues in the Itch JBD (i.e., Val600 and Phe604) differ from those in many of the other JBDs, where these residues are usually Leu (e.g., in c-Jun [Leu40 and Leu42] or JIP-1 [Leu161 and Leu163]). It will therefore be interesting to compare the exact modes of interaction of JNK with this JBD.
The JNK-mediated phosphorylation of Itch enhances protein degradation (102) (Fig. 3), and this has been attributed to phosphorylation-dependent conformational changes in Itch (100). This mechanism differs from the E3 ligase Fbw7-containing Skp/Cullin/F-box protein complex (SCFFbw7), which is targeted to phosphorylated proteins, such as phosphorylated c-Jun (240). The regulation of Itch activity by JNK phosphorylation has the potential to allow the coordinated regulation of degradation of many different cellular proteins and thus broadens the actions of JNK phosphorylation. Within the context of TNF-α-induced cell death, JNK activation of Itch allows for the degradation of c-FLIP, an inhibitor of caspase-8, and thus the subsequent cleavage of Bid to form tBid (52). Thus, prolonged JNK signaling is proapoptotic. Additional known Itch targets include c-Jun (102) and JunB (91). It will be important now to identify other processes modulated by phosphorylated Itch and determine how these actions integrate with those of the known targets.
JNK Targeting of Transcription Factors for Degradation
In contrast to the studies on Itch-mediated degradation and its enhancement following JNK-mediated phosphorylation of Itch, there remains some disagreement on the roles of JNK in mediating degradation of transcription factor substrates. Specifically, biochemical studies have suggested that the binding of inactive JNK to a number of its transcription factor substrates targets these proteins for degradation. For example, association of inactive JNK with c-Jun enhances c-Jun ubiquitination (96). However, the phosphorylation of c-Jun Ser63 by active JNK protected c-Jun from ubiquitination and increased its half-life (96) rather than enhancing its degradation, as more recently reported (240). JNK binding to promote degradation has also been reported for ATF2 and JunB, whereas JNK-mediated phosphorylation of ATF2 or p53 protected these proteins from degradation (95, 97, 98). In the specific case of c-Jun stability, the de-etiolated 1 protein has been shown to regulate c-Jun levels via the assembly of a multisubunit ubiquitin ligase containing CUL4 (319). The role for JNK in this process remains to be explored fully, as the coexpression of JNK or deletion of the c-Jun JBD did not protect c-Jun from degradation (319).
In contrast, JNK did not associate with Elk-1 or target this transcription factor for degradation (98). It remains to be tested whether some of the differences noted in the effects of JNK on protein stability might have arisen due to the different JNK isoforms tested, particularly in light of the recent report that JNK1 and JNK2 can oppositely regulate p53 levels in cells (291). It is also interesting that a recent study showed that the tyrosine kinase c-Abl was able to promote the proteolytic destruction of damaged DNA-binding proteins and that this was an action independent of the kinase activity of c-Abl (54). It therefore appears that despite the lack of consensus on the effects of JNK-mediated phosphorylation on protein degradation, there may be multiple levels of control by JNK. It will be interesting to see whether JNK regulates the stability of the other non-transcription factor substrates described in the subsequent sections of this review. Furthermore, it will be important to explore whether JNK-mediated phosphorylation can alter the activities of other enzymes involved in posttranslational modifications of proteins, such as recently shown for the Akt-mediated phosphorylation and suppression of methyltransferase activity of EZH2 (48).
JNK PHOSPHORYLATION OF SCAFFOLD AND ADAPTOR PROTEINS
In addition to modulating signaling events within the nucleus and altering signaling events through directed degradation of signaling proteins, Ser and Thr phosphorylation events may regulate membrane-proximal signaling. An important feature of many signaling pathways is their use of additional binding proteins that lack enzymatic activity (for a review, see reference 310). Depending on their exact mode of action in a signaling pathway and, in particular, how many different pathway components they interact with and modulate the actions of, these may be referred to as scaffold or adaptor proteins. Scaffold and adaptor proteins are used not only in the MAPK pathways but also in the regulation of signaling by integrins and G protein-coupled receptors and in metabolic and stress signaling (for reviews, see references 70, 71, 200, 202, 213, 229, 262, 264, 271, and 277). Increasingly, it is recognized that the phosphorylation of these scaffold or adaptor proteins may alter interactions with their binding partners and thus act as critical new ways to modulate intracellular signaling events. Figures 8 and 9 summarize the actions of JNK on four different scaffold molecules that are discussed in further detail in the following sections.
FIG. 8.
Scaffold and adaptor substrates of JNK. Scaffold and adaptor proteins are critical nonenzymatic proteins in signaling pathways that have been proposed to increase the efficiency and specificity of signaling events. The phosphorylation of these proteins, as mediated by JNKs, can alter their functions within signaling pathways. IRS-1 (A), JIP1 (B), and Shc (C) are examples of scaffold and adaptor proteins that are JNK substrates. The sites of phosphorylation by JNK are indicated, together with other interaction domains in these proteins, notably the pleckstrin homology (PH) domain that interacts with phosphatidylinositol 3,4,5-trisphosphate (PIP3), the phosphotyrosine binding (PTB) domains and SH2 domains that interact with tyrosine-phosphorylated (pY) proteins, SH3 domains that interact with proline-rich sequences, and phosphorylated tyrosine residues that themselves may interact with SH2 or PTB domains of other intracellular proteins. The size of each protein is not represented to scale in this figure, but the number of amino acids in each protein is provided in parentheses.
FIG. 9.
JNK-mediated phosphorylation regulates the binding activity of 14-3-3 protein family members. (A) 14-3-3 proteins are a class of adaptor proteins that generally bind their phosphoprotein targets via the consensus motif R-S-X-p(S/T)-X-P. This interaction allows 14-3-3 proteins to act as cytosolic anchors for their binding partners by masking intracellular targeting motifs, such as nuclear localization signals. (B) Any mechanism that would disrupt 14-3-3 protein interactions could thus allow the movement of binding partners to other regions of the cell. Specific examples include the JNK-mediated phosphorylation of 14-3-3 proteins leading to the release of the proapoptotic Bcl2 family members Bax and Bad to allow their movement to the mitochondria and the release of the tyrosine kinase Abl, allowing its translocation to the nucleus. There are likely many other binding partners for 14-3-3 proteins that are affected in this way.
IRS-1: a JNK Substrate That Allows Signal Integration between Stress and Metabolic Events
In the signaling events following activation of the receptors for insulin or insulin-like growth factor, the insulin receptor docking protein known as IRS-1 is phosphorylated at multiple tyrosine residues, and these residues then act to recruit SH2 domain-containing signal transducers, including phosphatidylinositol 3-kinase, Grb2, Shp2, and Crk (for a review, see reference 320) (Fig. 8A). There are also many potential Ser phosphorylation sites within IRS-1, and many different Ser/Thr protein kinases have been shown to phosphorylate IRS-1. These kinases include mammalian target of rapamycin (mTOR), protein kinase C-ζ, and AMP-activated protein kinase as well as JNK (5, 46, 133, 149, 196, 250). These different kinases indicate that insulin signaling can be modulated by a range of growth factors, cytokines, and stresses.
A major site of phosphorylation of IRS-1 by JNK has been mapped to Ser307 (Table 3) and has been proposed to cause insulin insensitivity following increased levels of inflammatory cytokines, such as TNF-α (5) (Table 3). Ser307 is adjacent to the IRS-1 phosphotyrosine-binding domain (Fig. 8A), and its phosphorylation interferes with the interaction of the insulin receptor and IRS-1, thus preventing IRS-1 tyrosine phosphorylation and decreasing insulin action (5). The actions of JNK on IRS-1 require the ability of JNK to interact with IRS-1, and the JNK-binding domain of IRS-1 has been mapped to 1 of 14 sites with L-X-L motifs typical of the JNK-binding domains (as described in the preceding sections for substrates such as c-Jun) (5, 185) (Table 2). Mutation of this JBD therefore not only reduces Ser307 phosphorylation of IRS-1 but also increases insulin-stimulated tyrosine phosphorylation and activation of downstream events, such as the activation of the protein kinase Akt (185). The significance of this JNK-mediated event was seen when these studies were extended to understand the role of JNK signaling in mediating insulin sensitivity in vivo. Specifically, the phosphorylation of Ser307 of IRS-1 was observed in the livers of wild-type mice fed a high-fat diet (which showed signs of insulin resistance) but not in the livers of jnk1-deficient mice also fed this high-fat diet (which did not develop insulin resistance) (134). (The numbering of IRS-1 here comes from the murine IRS-1 sequence [1,231 amino acids in the full-length protein], which differs from the human IRS-1 sequence [NP_005535; 1,244 amino acids in the full-length protein]. Thus, serine 307 of the murine sequence corresponds to serine 312 of the human IRS-1 sequence and is within the conserved sequence TATSPASM [the phosphorylated serine is underlined]. This should not be confused with murine IRS-1 serine 302, which corresponds to human IRS-1 serine 307, within the conserved sequence RTESITAT. The phosphorylation of human IRS-1 serine 307 has been reported to be critical, and insulin signaling and its attenuation of phosphorylation have been linked to insulin resistance in patients with type 2 diabetes [68].) These results are further supported by studies using overexpression of dominant-negative JNK mutants (238). These studies suggest a critical role for the JNK1-mediated phosphorylation of IRS-1 in the development of insulin resistance in diabetes. Thus, JNK inhibition is attracting increasing attention as a strategy in the treatment and prevention of insulin resistance in diabetes (for a review, see reference 136). There may be further benefits to also treating insulin resistance in a diverse range of disorders, including hypertension (285), skeletal muscle disuse atrophy (131), and burn injury (348).
JNK has further been shown to phosphorylate IRS-1 at two additional sites. Ser302 (Table 3) phosphorylation was demonstrated to be important in mediating disruption of binding of IRS-1 to the insulin receptor (318). In addition, Ser318 (Table 3), which was first identified as a novel protein kinase C-ζ-dependent phosphorylation site, has more recently been shown to be a target for both JNK and mTOR (235). Other protein kinases, such as the IκB kinase, may also phosphorylate IRS-1 Ser307 (103, 104, 237). It will be important to evaluate whether the phosphorylation of Ser302, Ser307, and Ser318 of IRS-1 can be mutually exclusive events or whether their combined phosphorylation allows for additive or synergistic effects to inhibit IRS-1. These multiple sites of phosphorylation may allow for greater control over subsequent signaling events.
JNK-Mediated Phosphorylation of Other Adaptor and Scaffold Proteins
In addition to the phosphorylation of IRS-1, as discussed in the preceding section, JNK also phosphorylates other adaptor and scaffold proteins. An interesting observation is the phosphorylation of the JNK scaffold protein JIP1 (245, 281) (Fig. 8B). The JIP1 sequence contains 11 S/T-P sites. Mutagenesis has shown JNK-mediated phosphorylation of multiple sites in JIP1 and that the phosphorylation of JIP1 at Thr103 (Table 3) is necessary for activation of the JNK module, suggesting a possible feedback mechanism for JNK activation which allows operation of the JNK pathway in a switch-like manner (245). JIP1 might also be phosphorylated by kinases other than JNK and facilitate JNK activation; evidence presented from two-dimensional peptide mapping showed JIP1 phosphorylation at multiple sites (245). However, in overexpression studies, a Thr103→Ala mutant form of JIP1 was not phosphorylated upon overexpression of JNK2 and its activation following glucose deprivation (281). The use of this phosphorylation-defective mutant form of JIP1 has implicated JNK-mediated phosphorylation in the dissociation of the protein kinase Akt1 from a JIP1-containing complex following glucose deprivation, thus allowing the activation of Akt1 under these conditions (280). Thus, JNK-mediated phosphorylation of JIP1 provides a critical point in the regulation of prosurvival pathways.
The JBD of JIP1 has been mapped to an L-X-L motif typical of JBDs of substrates such as c-Jun and IRS-1 (80, 338) (Table 2). An 11-amino-acid peptide based on this JIP1 JBD sequence has been characterized as an inhibitor of JNK (16, 18, 80) and has been cocrystallized with JNK1 (129). This structure (Fig. 2B) provides the first insight into how JNK binds its protein substrates through the docking region, which is remote from its active site. Specifically, this structure shows how two conserved hydrophobic residues, together with a proline and an arginine within the JIP-derived peptide, are involved in interactions with JNK (Fig. 2B). The structure agrees with biochemical assays of the ability of this peptide to interact with JNK and act as a substrate-competitive inhibitor through distorting the ATP-binding cleft and inducing relative movement of the two lobes (16, 18, 80). Another available structure of MAPK in complex with a substrate is that of p38 MAPK bound to the peptide derived from the transcription factor MEF2 (Fig. 2C) (50). Although both peptides bind at locations remote from the active site of the kinase, there are sufficient differences between the p38- and JNK-peptide structures to explain the specificity of JIP-derived peptides for binding to JNK (129).
A study examining JIP1 phosphorylation identified 35 sites, 8 of which were Ser/Thr motifs and possible phosphorylation sites for JNK or other MAPKs (67). Interestingly, these phosphorylation sites were present within protein kinase-binding motifs of JIP1, and no modifications were noted in SH3 and PTB domains, which are critical for interactions of JIP1 with proteins such as kinesin and β-amyloid protein (67). This suggests that phosphorylation may further regulate JIP1 interactions with selective groups of binding partners; further experimental evaluation is warranted. Whether other JNK scaffolds, such as plenty of SH2 domains (POSH) (296, 328) or β-arrestin (214), are also substrates for JNK-mediated phosphorylation remains to be evaluated. β-Arrestin 2 has a characterized JBD (Table 2) which is critical for interaction with and activation of JNK3 (218). Interestingly, this motif is not conserved in β-arrestin 1 (218), thus showing distinct mechanisms of interactions with these different arrestin family members. In addition, the JIP family members JIP2 and the more distantly related JIP3 both have JBD sequences (Table 2) (161, 338), with JIP3 being shown to be phosphorylated by JNK at three residues (Thr266, Thr276, and Thr287) (Table 3; Fig. 4) (161). In contrast to the reported effects of JNK-mediated phosphorylation of JIP1 modulating signaling via this scaffolded complex, the phosphorylation of JIP3 by JNK does not appear to contribute to the regulated formation of JNK complexes with JIP3 (Fig. 3) (161). The functional consequences of JNK-mediated phosphorylation of JIP3 therefore require further investigation.
The mammalian Shc proteins represent another important family of adaptor proteins. The Shc proteins are involved in a range of signaling events, including cell survival (for a review, see reference 262) (Fig. 8C). ShcA has three isoforms, and the largest isoform, p66ShcA, is phosphorylated at Ser and Thr residues in response to a range of stimuli (153, 217, 333). Phosphorylation of Ser36 of p66ShcA has been implicated in promoting the cell death response to oxidative stress; an inhibitor of this phosphorylation could therefore provide a novel means for the treatment of diseases associated with oxidative stress. The use of recombinant protein kinases in vitro and of chemical protein kinase inhibitors in cultured cells has implicated the JNKs as major mediators of the phosphorylation of Ser36 of p66ShcA (Table 3) (179). No interaction motif has been reported.
There has also been an increasing interest in the ability of JNKs to phosphorylate 14-3-3 proteins. This family of cytosolic binding proteins has been implicated in many biological processes, including the regulation of cell proliferation and death (71). 14-3-3 proteins generally bind phosphoproteins via the consensus motif R-S-X-p(S/T)-X-P (233). This interaction allows 14-3-3 proteins to act as cytosolic anchors for their binding partners by masking intracellular targeting motifs, such as nuclear localization signals (Fig. 9) (234, 303). Any mechanism that would disrupt 14-3-3 protein interactions could thus allow the movement of binding partners to other regions of the cell.
JNK can phosphorylate the 14-3-3 isoforms ζ and σ at Ser184 and Ser186, respectively (301) (shown in Table 3, alongside predicted JNK phosphorylation sites in 14-3-3 β and 14-3-3 ɛ). Interestingly, the 14-3-3 isoforms γ, η, and θ do not share an equivalent motif (301), suggesting different modes of regulation for different 14-3-3 family members. JNK-mediated phosphorylation of 14-3-3 ζ or σ promotes its dissociation from the proapoptotic Bcl2 family member Bax and its translocation to a mitochondrion, resulting in cell death (301). Conversely, the overexpression of a 14-3-3 mutant that can no longer be phosphorylated by JNK (e.g., the 14-3-3 ζ Ser184→Ala mutant) can inhibit Bax translocation, inhibit cytochrome c release, and prevent cell death (301). Similarly, JNK-mediated phosphorylation of 14-3-3 proteins promoted the release of the proapoptotic Bcl2 family member Bad to antagonize prosurvival signaling initiated by protein kinases such as Akt (286). Interestingly, the site of phosphorylation is adjacent to the ligand-binding groove of 14-3-3 proteins (286), suggesting that binding of other ligands of the 14-3-3 proteins may be modulated by JNK-mediated phosphorylation. Indeed, the use of 35S-labeled cellular proteins showed the release of a subset of bound proteins following 14-3-3 ζ phosphorylation by JNK (286).
The release of other classes of 14-3-3 protein-binding partners has also been demonstrated for the protein tyrosine kinase c-Abl (342). The nucleocytoplasmic shuttling of c-Abl involves three distinct nuclear localization sequences and a nuclear export sequence (254). In response to stress, such as DNA damage or oxidative stress, c-Abl translocates to the nucleus and, importantly, JNK is activated (254). JNK-mediated phosphorylation of the 14-3-3 proteins allows nuclear translocation of c-Abl, presumably because the bound 14-3-3 proteins usually interfere with recognition of the nuclear localization sequences (342; for a review, see reference 341). This important regulatory role for 14-3-3 proteins has been tested specifically with the overexpression of 14-3-3 ζ to attenuate c-Abl translocation in response to DNA damage as well as with the downregulation of 14-3-3 proteins (using small interfering RNAs) to increase nuclear c-Abl (342). The link to phosphorylation by JNK at residue Ser184 was further strengthened by the observation that the 14-3-3 ζ Ser184→Ala mutant protein was more protective than the wild-type 14-3-3 ζ protein in c-Abl-mediated apoptotic events following exposure to either DNA damage or oxidative stress (342).
Studies of 14-3-3 proteins have revealed the power of JNK phosphorylation as a regulatory strategy, particularly when rapid cellular responses are required. Furthermore, the potential to release many different binding partners for their actions in different compartments of the cell, as exemplified by the actions of Bax and Bad being freed to move to the mitochondria or the movement of c-Abl to the nucleus, allows a highly coordinated and rapid response without the requirement for transcriptional events that may take a significantly longer time.
JNK-MEDIATED PHOSPHORYLATION OF MITOCHONDRIAL PROTEINS
Since the initial characterization of JNKs as stress-activated protein kinases, there has been speculation on whether these kinases may be modulators of cell death in response to stress, either by facilitating cell death or by opposing it. Substrates in addition to c-Jun have been proposed to be involved in JNK-mediated apoptosis (64). Thus, in addition to the c-Jun-dependent transcriptional events that could mediate cell survival or cell death (for a review, see reference 9), the actions of JNK to phosphorylate 14-3-3 proteins (see the preceding section) and to facilitate proapoptotic pathways have received increasing interest.
JNK and the activator SEK may colocalize with mitochondria in cardiac myocytes (12), and levels of JNK2 increase in both the nuclei and mitochondria of cells of the PC12 neuronal cell line upon exposure to 6-hydroxydopamine (88). This has led to the suggestion that proteins of the outer mitochondrial membrane may act as JNK substrates. The direct effects of JNK activity on mitochondria have been studied in a cell-free assay, where the addition of purified JNK led to the release of cytochrome c from isolated mitochondria (12). In contrast, dominant-negative mutants of JNK2 prevented apoptotic events, including the release of cytochrome c, and the downstream events, such as cleavage of caspase 3 and poly(ADP)-ribose (88). These results suggest significant roles for JNKs as mediators of apoptotic cell death and for mitochondrial proteins as JNK targets. The regulation of Bcl2 family members is illustrated in Fig. 10 and described in the following section.
FIG. 10.
Mitochondrial substrates of JNK and the regulation of cell death. (A) A number of mitochondrial proteins within the Bcl2 family of apoptosis regulators are direct substrates of JNK phosphorylation. The actions of JNK-mediated phosphorylation to alter the actions of Bcl2 itself remain controversial, and whether the phosphorylation of Ser128 of Bad is JNK mediated has been questioned (?). (B) The link between JNK-mediated phosphorylation of Bim and Bmf proteins and increased cell death has been attributed to the disruption of binding of Bim and Bmf to anchoring proteins, including the dynein and myosin V motor complexes.
JNK-Mediated Phosphorylation of the Bcl2 Family
The Bcl2 family of mitochondrial proteins has increasingly been implicated as regulators of cell death (for reviews, see references 62 and 116), so events that change their activities provide mechanisms to alter the death and survival decisions of the cell. The founding member of this protein family, the antiapoptotic Bcl2 protein, has been shown to be phosphorylated following cell exposure to a variety of stimuli, and JNK has been implicated as the protein kinase in these events (208). The phosphorylation of Ser70 (Table 3) was suggested to inactivate the antiapoptotic function of Bcl2 (331). However, this contradicts earlier suggestions that phosphorylation of this Bcl2 site would enhance its antiapoptotic functions (146) or subsequent studies that showed that Bcl2 phosphorylation at Ser70 was associated with increased cell survival (76). It remains to be evaluated whether this reflects the differences in cell types examined, the type or level of the stimulus, or differences in function upon Bcl2 phosphorylation at sites in addition to Ser70.
Other antiapoptotic Bcl2 family members have also been shown to be substrates for JNK. Specifically, JNK-mediated phosphorylation of Thr47 and Thr115 of Bcl-xL (Table 3) can inactivate the antiapoptotic actions of this protein in response to cellular stresses such as ionizing radiation (Fig. 3) (163). Similarly, JNK-mediated phosphorylation of Ser121 and Thr163 of Mcl-1 (Table 3) in response to cellular stresses such as oxidative stress can inactivate the antiapoptotic actions of this protein (Fig. 3) (142). In contrast, the proapoptotic Bcl2 family member Bad has been shown to be phosphorylated at Thr201 (Table 3), but in this case, phosphorylation suppresses apoptosis by again inactivating the usual actions of this protein (Fig. 3) (344). The latter example explains the requirement for JNK as a mediator of interleukin-3-dependent survival of hematopoietic cells (344). Conversely, JNK phosphorylation of Ser128 of Bad (Table 3) promotes the proapoptotic effects of Bad in primary granule neuron cells (Fig. 3) by antagonizing the actions of growth factors (84). However, it was recently questioned whether Ser128 of Bad is phosphorylated by JNK, and therefore the role of JNK in apoptosis mediated by the Bad protein is not clear (347).
In addition, the proapoptotic protein BimEL has been shown to be a JNK substrate, with phosphorylation of Ser65 (Table 3) potentiating its proapoptotic actions following growth factor withdrawal (Fig. 3) (258). In a separate study, the Bim family members BimL (a splice form of the Bim gene, whose splicing also results in BimEL and BimS) and Bmf were evaluated as JNK substrates (186). BimL was phosphorylated at Thr56 and at least one of Ser44 and Ser58 (Table 3), and Bmf was expected to be phosphorylated at similar sites. The predicted sites for JNK-mediated phosphorylation of Bmf are also shown in Table 3. For both BimL and Bmf, this phosphorylation inhibits their binding to dynein chain proteins that normally sequester these proteins and prevent their proapoptotic activity. Thus, this provides another mechanism to explain the proapoptotic functions of JNK. The JNK- and p38-mediated phosphorylation of the proapoptotic protein Bax has been shown most recently to lead to Bax activation and translocation to the mitochondria (Fig. 3) (166). In this case, the use of specific phosphorylation site mutants has suggested that Bax Thr167 is the target of JNK-mediated phosphorylation (Table 3) (166).
The effects of JNK-mediated phosphorylation of Bcl2 family members should also be considered within the context of JNK-dependent changes in the expression levels of these proteins. For example, increased expression of Bim following arsenite exposure or growth factor withdrawal was shown to be JNK dependent (258, 323). As mentioned previously, Bax translocation to mitochondria can also be modulated by JNK-dependent phosphorylation of 14-3-3 proteins (301). Therefore, JNK acts at multiple levels in the control of mitochondrial Bcl2 family function, and further work is required to comprehensively profile which Bcl2 family members are substrates for JNKs and how phosphorylation affects their functions in apoptotic death.
JNK-Mediated Phosphorylation of Sab
The use of JNK as bait in a yeast two-hybrid screen also identified the SH3 domain-containing mitochondrial protein Sab (also known as SH3BP) as an interacting partner and substrate for JNK (322). Sab was originally identified as a binding partner and negative regulator of Bruton's tyrosine kinase (330). Of two JBD-like sequences, only one was shown to be essential for the interaction with JNK (Table 2). The same motif appears to also mediate interaction with the related MAPK family member SAPK3 (63). In addition, only one of the four Ser residues within an S-P motif were shown to be phosphorylated (Table 3), and this was also the predominant site for phosphorylation by SAPK3 (63). The function of Sab and the way in which it might be altered upon phosphorylation by MAPKs, such as JNK and SAPK3, require further investigation.
REGULATION OF OTHER PROTEIN KINASES THROUGH JNK-MEDIATED PHOSPHORYLATION
Protein kinases, in addition to catalyzing phosphorylation reactions, can also be subjected to control by phosphorylation, in some cases in an autophosphorylation reaction but in other instances in reactions catalyzed by other protein kinases. Such actions of protein kinases to regulate the activities of other kinases are evident in the sequential activation of protein kinases in the MAPK cascade, as shown in Fig. 1A, but there remains the possibility that JNKs phosphorylate additional protein kinase substrates. Two examples are shown in Fig. 11 and described in the following sections.
FIG. 11.
Protein kinases as substrates of JNK. (A) The p90RSK protein kinase consists of two distinct protein kinase domains and a linker domain. Phosphorylation of the linker domain by JNK leads to coordinated phosphorylation and activation of p90RSK and PDK1, which act upstream of the prosurvival kinase Akt. (B) Akt can also be phosphorylated by JNK, and this leads to enhanced binding by the Akt activator PDK1, thus increasing prosurvival Akt actions.
Phosphorylation of MAPK-Activated Protein Kinases
A feature of MAPKs is their ability to phosphorylate a group of substrates that are themselves protein kinases. These substrates are therefore termed the MAPK-activated protein kinases. These protein kinases include the p90 ribosomal S6 kinases (RSK1-4), the mitogen- and stress-activated kinases (MSK1-2), the MAPK-interacting kinases (MNK1-2), and the MAPK-activated protein kinases (MAPKAPK1-5; also now known as MK1-5) (for a review, see reference 268). Interestingly, however, little evidence supports the direct phosphorylation of these protein kinases by JNKs (for a review, see reference 268), with the exception of a report demonstrating p90RSK phosphorylation on Ser381 (also known as RSK2) (Table 1) by both ERK and JNK pathways following exposure of cells to UV irradiation (350). MK1 shows the unusual organization of two kinase domains separated by an ∼100-amino-acid linker, with the N-terminal kinase domain phosphorylating substrates and being controlled by the C-terminal kinase domain and the linker (for a review, see reference 268). Ser381 is within the linker region between the two kinase domains in MK1, and its phosphorylation has been shown to create a docking site for 3-phosphoinositide-dependent protein kinase 1 (PDK1) (93) (Fig. 11A). This then leads to coordinated phosphorylation and activation of both PDK1 and MK1 (93). Thus, phosphorylation of this site can be considered an important rate-limiting step in MK1 activation (Fig. 3); the involvement of JNK in this process is consistent with the activation of MK1 following exposure to stresses such as UV irradiation.
JNK-Mediated Phosphorylation of the Prosurvival Protein Kinase Akt
The protein kinase Akt has been reported to be a JNK substrate (275). Phosphorylation of this protein provides another example of cross talk between the JNK and Akt pathways (in addition to Akt phosphorylation and inhibition of the upstream activators of the JNK pathway [MLK3, ASK, and MKK4/SEK1] [20, 164, 252] or the binding of Akt to the JNK scaffold JIP1 [thereby preventing recruitment of the JNK signaling complexes] [165]). Note that JNK phosphorylates Thr450 of Akt1 (Table 1). This is only one of six candidate JNK phosphorylation sites in Akt. This phosphorylation then primes Akt for phosphorylation and activation by PDK1 (Fig. 11B), thus controlling, in part, the basal activity of Akt, the efficiency of Akt activation after ischemia, and cell survival after ischemia (275). This priming effect shows some similarities to the recent observation that phosphorylation of Ser473 of Akt is also a priming event for subsequent phosphorylation of Akt by PDK1 (270). Both Ser473 and Thr450 are in the hydrophobic tail region of Akt, and thus phosphorylation of these and possibly other residues in this domain of Akt may serve to enhance interactions between Akt and PDK1, thus providing additional links between Akt and other signaling pathways. Further examples of positive regulation of the Akt pathway by JNK remain to be reported, as others have also shown negative regulation of Akt phosphorylation following JNK overexpression (157) and enhancement of Akt phosphorylation by a loss of JNK signaling (287).
REGULATION OF CELL MOVEMENT THROUGH JNK-MEDIATED PHOSPHORYLATION
Cell movement is a complex biological process requiring coordinated changes in the activities of numerous protein complexes, including actin polymerization, adhesion dynamics, changes in cell polarity, and trailing edge retraction (for a review, see reference 308). Critical molecular structures such as the focal adhesions are now seen not only as structural complexes but also as highly sophisticated and regulated signaling networks (for a review, see reference 267). Initial studies on the JNK pathway in Drosophila suggested a role for JNK in cell migration, with the loss of JNK function resulting in failure of dorsal closure in the developing embryo (278). Similarly, in mammalian cells, an upstream kinase in the JNK pathway, MAPK/ERK kinase kinase 1 (MEKK1), was subsequently shown to be essential for cell migration (324, 345). While transcription factor control by JNKs might contribute to this regulation of cell movement (for a review, see reference 249), the observed rapid effects upon JNK inhibition suggested other targets for JNK action (138). A number of focal adhesion, microtubule-associated, and intermediate filament proteins are JNK substrates, and these are summarized in Fig. 12 and described in the following sections.
FIG. 12.
Links between JNK substrates and cell movement. JNK can phosphorylate proteins, such as paxillin, localized within focal adhesions or associated with microtubules or intermediate filaments. These actions can result in changes in cell migration and changes in microtubule stabilization.
JNK-Mediated Phosphorylation of the Focal Adhesion Protein Paxillin
Paxillin has been proposed as a JNK substrate based on its localization in focal adhesions, its function as an adaptor protein involved in cell migration and organization of adhesion, and its Ser phosphorylation following cell exposure to fibronectin or growth factors. In vitro kinase assays showed JNK phosphorylation of paxillin at Ser178 (Table 3), and the relevance of this phosphorylation in cells was confirmed with the use of a Ser178→Ala mutant of paxillin that was not phosphorylated by JNK1 in NBT-II cells (138). The expression of this paxillin mutant also slowed cell migration, as measured in time-lapse single-cell motility assays and monolayers of cells subjected to wound-healing assays (138), suggesting that phosphorylated paxillin enhances cell movement (Fig. 3). Similar results were obtained in wound-healing assays using a variety of cell types, including MDA-MB-231 human breast cancer cells and Chinese hamster ovary K1 cells (138), and JNK-mediated phosphorylation of paxillin was recently shown to be necessary for platelet-derived growth factor-induced chemotaxis of primary foreskin fibroblasts (8). Furthermore, JNK has been shown to localize to the actin-dense membrane ruffles at the cell leading edge following growth factor exposure, suggesting a critical role for JNK in cell migration (8). Taken together, these studies suggest that JNK-mediated phosphorylation of paxillin is a key event in the regulation of migration of many cell types.
In contrast to studies with mammalian cells, studies of Dictyostelium have suggested that JNK may not have any major effect on cell sorting, slug migration, or morphogenesis (38). This conclusion was based on the lack of effects of mutant forms of paxillin that were proposed to disrupt a possible JNK phosphorylation site (38). It should be noted that within the Dictyostelium paxillin sequence, Ser192 had been proposed as a conserved JNK phosphorylation site (38). However, the sequence surrounding this residue (Q-P-A-L-S192-K-A-T-L) varies markedly from the sequence surrounding Ser178 in human paxillin (Table 3). Most notably, it lacks a Pro residue immediately C-terminal to the Ser, and thus the likelihood that this is a JNK phosphorylation site is less likely, though not impossible. The Dictyostelium paxillin sequence (CAA05356) does contain a number of the more favored S/T-P motifs. Whether these are sites for phosphorylation and regulation by JNK or other MAPKs, such as the ERKs, which also modulate cell morphogenesis through interactions with paxillin (144), remains to be evaluated.
JNK Interaction with and Phosphorylation of Microtubule-Associated and Intermediate Filament Proteins
In addition to its nuclear localization, as discussed earlier in this review, JNK localizes in fibroblasts to punctate structures along microtubules (236). Since microtubules are able to affect focal adhesion turnover and are also implicated in aspects of cell migration, the interaction and phosphorylation of microtubules or their associated proteins could be another mechanism for the regulation of cell motility. Indeed, the phosphorylation of microtubule-associated protein 2 appears to be required for binding to microtubules in intact cells (36). JNKs were shown to phosphorylate microtubule-associated protein 2, at least in vitro (177). Microtubule-associated proteins 2 and 1B have been considered in vivo JNK targets only more recently (51, 160), although the exact sites targeted for phosphorylation by JNK remain poorly defined (Table 3).
When the activity of purified JNK1 protein towards crude extracts of mouse brain was evaluated, microtubule-associated protein 2 was shown to be a prominent substrate for the kinase (28). The use of neurons isolated and cultured from jnk1−/− mice showed an increased dendritic arbor number and decreased arbor length. For these cultured cells, the use of cell compartment-specific inhibitors of JNK, namely, the JBD of JIP1 fused to either nuclear exclusion or nuclear localization sequences, also suggested a role for JNK activity outside the nucleus (28). Further experiments will be required to support the proposed pathway leading to effects on dendritic architecture through microtubule-associated protein 2 phosphorylation by JNK. However, JNK1 has now been implicated in the dendritic architecture of the cerebellum and motor cortex (28), in addition to maintaining normal brain axonal growth (51) and contributing to the repression of Wnt (Wnt-4 and Wnt-6) expression and neural differentiation (as recently modeled in embryonic stem cells) (10). It is therefore possible that a complex range of JNK1 substrates regulates these biological processes in vivo.
The microtubule-associated protein tau has also been shown to be a substrate of JNKs (340). The phosphorylation of tau reduces its ability to promote microtubule assembly (194), and hyperphosphorylation inhibits binding to microtubules (32). Thus, JNK-mediated phosphorylation of tau can be viewed as inhibiting the actions of tau (Fig. 3). Nine sites for JNK phosphorylation have been determined (340) (Table 3). This phosphorylation has gained increasing interest due to its correlation with abnormal filamentous inclusions of hyperphosphorylated tau in a range of neurodegenerative diseases, such as Alzheimer's disease (for a review, see reference 184). JNKs are therefore possible targets to prevent hyperphosphorylation of tau, and JNK inhibitors may prove useful in the treatment of tauopathies. Furthermore, the amyloid β protein precursor has also been shown to be phosphorylated by JNK, at Thr668 (Table 3), in a reaction facilitated by the JIP1 scaffold protein (141, 170, 274). Interestingly, the scaffold protein JSAP1/JIP3 has been shown, in addition to JIP1, to cooperate with focal adhesion kinase to regulate JNK and cell migration on fibronectin (292). Thus, there are several layers of complexity and specificity provided by this involvement of specific scaffolds.
The protein known as superior cervical ganglion 10 (SCG10), a stathmin-like family member, is a neuronal growth-associated protein enriched in the growth cones of neurons and is another substrate for JNK (242). SCG10 acts to destabilize microtubules. JNK phosphorylation of SCG10 Ser62 and Ser73 (Table 3) reduces its microtubule-destabilizing activity and provides a means for the control of growth cone microtubule formation following the exposure of neurons to stress (242) (Fig. 3). Although an interaction of JNK with SCG10 comparable to the interaction of JNK with c-Jun has been demonstrated (242) and although SCG10 can be purified alongside other microtubule-destabilizing proteins, such as SCG10-like protein or the stathmin-like proteins RB3 and RB3′, or alongside other JNK-interacting proteins, such as ATF2, β-arrestin, or glutathione S-transferase π (297), no JBD sequence has been reported for the SCG10 protein. While the phosphorylation of Ser73 of SCG10 has been shown following N-methyl d-aspartic acid receptor activation in the rat hippocampus, a link to JNK activation has not been demonstrated directly in this situation (228). However, SCG10 phosphorylation at Ser73 was depleted in the cortexes of brain tissues isolated from jnk1−/− animals, indicating a major role for the JNK1 isoform in mediating SCG10 phosphorylation under physiological conditions and suggesting that this is critical for maintaining neuronal microtubule stability and efficient neurite elongation (297).
The phenotypic changes of lissencephaly in males and abnormal neuronal positioning in females carrying mutations in the X-linked gene DCX (78) have suggested a role for the DCX gene product in neuronal migration. DCX is a microtubule-associated protein that stabilizes microtubules through interactions mediated by the evolutionarily conserved double-cortin (DC) domain (168, 299). A link between neuronal migration and the JNK pathway was initially suggested by increased JNK activity in radially migrating neurons in the cerebral cortex and the ability of a dominant-negative JNK mutant or the JNK inhibitor SP600125 to reduce migration (159). JNK has been shown to phosphorylate the DCX protein (105) (Table 3). The DC domains of DCX are required for JNK-DCX interaction (105), although there is no sequence similarity with the previously recognized docking motifs (295). DCX was also shown to interact with the JNK scaffold protein JIP1, suggesting the possible existence of a large JNK signaling module in the brain (105). DCX mutants lacking the phosphorylation site showed altered neuronal motility by a reduced maximal velocity with prolonged stationary periods (105); JNK-mediated phosphorylation of DCX can therefore be viewed as activating DCX (Fig. 3). The interaction of JIP1 with the motor protein kinesin (307) provides at least one additional point of regulation, localizing phosphorylated DCX to neuron growth cones in a complex with JIP and JNK via the interaction with kinesin (105). More recently, the kinesin heavy chain was shown to be a substrate for JNKs (227). Although the phosphorylation sites are not yet determined (Table 3), this phosphorylation of kinesin inhibits its microtubule-binding activity (Fig. 3); prevention of JNK activity appeared to reverse the suppression of neurite formation by the polyglutamine-expanded androgen receptor protein (227). This suggests that targeting JNK may prove an important therapeutic strategy in neurodegenerative disorders involving polyglutamine-expanded protein.
The intermediate filament protein keratin 8 is also a JNK substrate (127). The phosphorylation of intermediate filament proteins has been suggested to affect their functional and assembly states. Following the exposure of HT-29 cells to anti-Fas receptor antibodies to activate this proapoptotic receptor, the phosphorylation of Ser73 from keratin 8 was observed to show similar kinetics to the activation of JNK (127). Moreover, a comparison of 32P-labeled keratin 8 immunoprecipitated from cells with keratin 8 phosphorylated in vitro by JNK supported the conclusion that JNK phosphorylated this Ser residue (Table 3). Furthermore, JNK and keratin 8 could be isolated in coimmunoprecipitation experiments; however, the residues involved in this interaction were not defined (127). One possible consequence of JNK-mediated phosphorylation of keratin 8 may be protecting the keratin network from cleavage by the proapoptotic proteases, i.e., the caspases (127), although this possibility requires further testing.
GENERAL PRINCIPLES OF JNK SIGNALING VIA PROTEIN PHOSPHORYLATION
JNK Peptide Specificity
The large range of protein substrates of JNK raises a number of interesting questions. First, we discuss the substrate specificity of JNK. Specificity arises from the match between the chemistries of the side chains of a bound peptide substrate and specific pockets, or subsites, which are positioned between the two lobes of the protein kinase (33). Molecular recognition of the peptide sequence surrounding the phosphorylated residue of a substrate is usually referred to as the “peptide specificity” of a protein kinase. The three-dimensional structures of protein kinases with bound peptide substrates (34, 140, 172, 198, 335) have revealed that all three classes of protein kinases bind their peptide substrates in an analogous manner, in an extended conformation and with the same orientation of the substrate peptide chain relative to the protein kinase (173). A recent study has also shown the ability of protein kinase PKR to induce a local unfolding of its substrate (eukaryotic translation initiation factor eIF2), specifically in the region of the phosphorylated serine residue (69).
The distribution of the relative abilities of a protein kinase to phosphorylate different peptide substrates falls sharply after a small number of optimal peptides, and most peptide sequences show negligible phosphorylation (353). However, peptide specificity is not the only element that determines the specificity of phosphorylation in the cell; substrate recruitment plays a critical role in determining substrate preference. Substrate recruitment is any process that can bring a kinase and a substrate in close proximity, thus increasing the effective concentration of the substrate and increasing the chance of forming the enzyme-substrate complex. The docking interactions common in JNK substrates are a classical example of substrate recruitment. The substrate preference is ultimately determined by the combination of peptide specificity and recruitment. Recruitment can have a significant effect on the phosphorylation of an individual substrate, transforming a poor substrate into one that can be effectively phosphorylated.
Inspection of the sequences surrounding JNK phosphorylation sites (Tables 1 and 3) reveals that almost any amino acid is allowed in the positions surrounding the phosphorylated Ser/Thr, with the exception of the position immediately C-terminal to the phosphorylation site, which is restricted to Pro (as discussed above). Tables 1 and 3 reveal four phosphorylated sequences that do not feature a Pro at this position (instead featuring Phe, Leu, Ile, or Lys); in these cases, the lack of Pro and the consequent unfavorable interaction must be compensated for by other mechanisms, such as particularly strong recruitment or specific structural features of the substrate. The lack of apparent peptide specificity at other positions suggests a strong role for substrate recruitment in determining the substrate specificity of JNK.
If we use the substrate sequences in Tables 1 and 3 to construct a consensus sequence for the heptapeptide surrounding the phosphorylated residue, they yield the sequence (S/A/I/G/P/T/L/V)-(L/P/T/S/A)-(L/S/P/T/A/N/G)-(S/T)-(P)-(S/P/T/ E/L/G/A)-(S/L/A/P/R/E/K) (the residues shown are present in >5% of the substrates, and the residues shown in bold are present in >10% of substrates). This consensus sequence is similar to the structure-based optimal phosphorylation sequence prediction by Predikin (33), i.e., (L/I/M/F)-(P/T/A/S/I)-(X)-(S/T)-P-(P/F/L/I/D/E)-(R/K/Q/S/L). All of the heptapeptide substrate-binding residues are conserved among the JNK isoforms; however, analysis of the three-dimensional structure of JNK suggested that surface differences exist among JNK isoforms that may affect substrate binding further N-terminal (four or more residues away) from the phosphorylated residue (327).
To analyze whether the consensus sequence derived above or the Predikin prediction could be used to predict new JNK phosphorylation sites, we scored all heptapeptides surrounding every Ser or Thr residue in the tau protein, using Scansite (329), for their match to the JNK phosphorylation motifs (33). The results indicated that using the consensus motif, the known phosphorylation sites score 0.2228 ± 0.0862 (minimum, 0.1383; maximum, 0.3457 [the lower the value, the closer the match to the motif]), while the sites known to not be phosphorylated score 0.3497 ± 0.0670 (minimum, 0.1382; maximum, 0.4148). Using the motif predicted by Predikin, the corresponding number for known phosphorylation sites is 0.2250 ± 0.0486 (minimum, 0.1306; maximum, 0.2612), while the sites known to not be phosphorylated score 0.2859 ± 0.0502 (minimum, 0.1306; maximum, 0.3266). These results indicate that these motifs discriminate somewhat between phosphorylated and nonphosphorylated sites, but there is substantial overlap between the two distributions, and correspondingly, the probability of predictions is low. One can still use these predictions to find the most likely phosphorylation sites in a protein that is a known JNK substrate (Tables 1 and 3); such predictions can direct mutagenesis experiments to confirm the phosphorylation sites.
Substrate Specificity of JNK Isoforms
In the preceding sections, only limited attention is given to the role of the different JNK isoforms and their substrate preferences. This is largely due to the lack of data, but there are patterns emerging showing that different JNK isoforms, despite their high levels of sequence similarity (reviewed in reference 17), may show substrate preferences. Examples include the preferential interaction of JNK2, rather than JNK1, with the substrate protein DCX (105) and the observation that JNK2 phosphorylates a larger number of sites in tau and that this is correlated with its ability to inhibit microtubule assembly in vitro (340). JNK2 was identified as the ATFa-interacting protein kinase (29), whereas JNK1 is a more efficient Itch kinase than JNK2 (102).
Further clues are emerging with the use of cells lacking one of the JNK isoforms (30). As an example, the use of jnk2-deficient cells has suggested a role for JNK2, rather than JNK1, in the inactivation of TIF-IA and subsequent downregulation of rRNA synthesis (211). There may also be some degree of isoform selectivity in JNK localization, with the report of JNK2 translocation to the mitochondria in a PC12 model of dopaminergic cell death (88). It will be critical to continue to explore possible isoform-specific functions of JNKs within cells and to be able to attribute these characteristics to differences in substrate recognition by these kinases. Because of the high sequence identities of the JNK isoforms and the broad peptide specificity of JNKs, it is clear that a number of subtle mechanisms (including transcriptional regulation, cellular localization, and specific JNK-substrate interactions) must cooperate in the cell to preferentially associate a JNK isoform with a particular substrate.
Interplay of Phosphorylation by JNKs and Other Protein Kinases
Because JNKs act within a broader intracellular signaling network, it is also important to consider whether other kinases phosphorylate the same or additional sites within JNK substrate proteins. This has the potential to allow different kinases to mediate similar effects in cells and to provide a means for signal integration. There are many examples, but the following cases illustrate the possible consequences in our understanding of signal transduction. For the archetypical JNK substrate c-Jun, other protein kinases can also catalyze the phosphorylation of Ser63 and Ser73. For example, the exposure of sympathetic neurons in culture to the DNA-damaging agent etoposide resulted in phosphorylation of both Ser63 and Ser73 and the subsequent upregulation of total c-Jun levels (25). The inhibition of JNK activity or activation could not block either the DNA damage-induced c-Jun phosphorylation or upregulation or the subsequent death of these neurons (25). Instead, pharmacological studies using olomoucine, roscovitine, flavopiridol, or alsterpaullone implicated either the CDKs or a downstream kinase(s) of JNK. Therefore, these kinases, not JNKs, would need to be targeted in further studies preventing neuronal death after DNA damage (25). It will be interesting to see whether inhibitors that target c-Jun itself will be more effective in preventing neuronal cell death and whether such substrate-directed inhibitors will be most effective when targeting the substrate docking sites or the phosphorylation sites themselves.
Other nuclear proteins are also subject to regulation by phosphorylation at the same residues by different kinases. As examples, JNK and p38 can both phosphorylate the same residues of the transcription factor JPD2 (155), and JNK, ERK, and p38 MAPKs can phosphorylate the transcription factor Elk-1 (336). Interestingly, in Drosophila, the homologue of the transcription factor ATF2 is involved in the response to UV irradiation via p38 MAPK but not JNK (269). It is therefore not sufficient to attribute actions to a stress-activated kinase such as JNK without direct confirmation of its involvement. An overlap of kinase functions is also seen in nonnuclear substrates. Thus, the p38 family member SAPK4 (p38-δ) and JNKs share many common phosphorylation sites on the microtubule-associated protein tau, and their actions are comparable in inhibiting microtubule assembly in vitro (340). Similarly, other MAPKs, such as ERK or the p38 family member SAPK3 (p38-γ), glycogen synthase kinase 3, and CDK-5 can all phosphorylate tau (21, 85, 111, 203), and both JNK and p38 appear to phosphorylate the same residue in Bax (166). mTOR and JNK both phosphorylate Ser307 (133) and Ser318 (235) of the adaptor protein IRS-1. The phosphorylation of the same residues by different kinases emphasizes that effective targeting strategies to prevent phosphorylation of critical signaling intermediates, such as IRS-1, will require a better understanding of the role of specific kinases in phosphorylation reactions in vivo and, additionally, that targeting the protein substrate may provide interesting avenues for further investigation.
Many of the proteins phosphorylated by JNKs will also be regulated by phosphorylation at additional sites by other protein kinases. Even the archetypical JNK substrate, c-Jun, is subjected to multisite phosphorylation by different kinases (230), although the consequences of the different phosphorylation sites on Jun function are not currently clear. An interesting recent study (315) has shown that the phosphorylation of c-Jun on Thr293 by glycogen synthase kinase 3 facilitates interaction with Fbw7 ubiquitin ligase in a manner similar to that shown for JNK-mediated phosphorylation (240). Both lead to polyubiquitination of c-Jun and its proteasomal degradation (315). Therefore, there is a critical link between the two signaling pathways through Jun stability and glycogen synthase kinase 3 activity. Importantly, this example also highlights the importance of hierarchical phosphorylation; phosphorylation of Ser243 by a Pro-directed kinase, such as ERK, is required before phosphorylation of Thr293 by glycogen synthase kinase 3 can take place (315). Therefore, the ultimate activation of a specific substrate may require a number of signaling inputs.
For other proteins, studies of multisite phosphorylation have revealed a number of interesting observations about signaling complexity. Tyrosine phosphorylation of the JNK substrate paxillin by c-Src allows its interaction with ERKs, thus providing new areas for cross talk and regulation by additional MAPK family members (144). JNK-mediated phosphorylation of the transcription factor ATF2 at Thr69 following insulin stimulation follows ERK-mediated phosphorylation of ATF2 Thr71 (14). The JNK substrate TIF-IA is phosphorylated at multiple sites by different protein kinases, such as mTOR and ERK, which are required for TIF-IA activity and Pol I transcription (212, 351). This contrasts with the negative regulation of TIF-IA actions by JNK-mediated phosphorylation. Similarly, the JNK substrate Itch is activated by JNK phosphorylation but negatively regulated by tyrosine phosphorylation mediated by Fyn (332). These examples emphasize that proteins can be subjected to both positive and negative regulation by phosphorylation at different sites. Conversely, the transactivation of p53 appears to be augmented by the activities of a number of kinases acting together, and this may help to explain why certain JNK activators, such as interleukin-1, do not stabilize or activate p53 (239).
Complexity of JNK Signaling
The enormous complexity of the response to JNK activation has been illustrated by an evaluation of the JNK-dependent responses of the human breast carcinoma BT474 cell line to genotoxic shock following exposure to cisplatin. JNK has been suggested to regulate DNA repair by transactivation of DNA repair genes (125). Following phosphorylation of ATF2 and c-Jun, a remarkable 269 of 3,083 gene promoters surveyed showed increased binding of these transcription factors (126). Treatment with the JNK inhibitor SP600125 decreased binding of 98% of these genes in the cisplatin profile, confirming the requirement for JNK activity in the response (126). The largest functional group of upregulated genes consisted of 23 DNA repair or repair-associated genes, 13 of which had not been reported previously to be cisplatin sensitive or JNK regulated (126). These 23 genes represent a large proportion of the 89 DNA repair genes represented on the array. Another major group of upregulated genes included the immediate-early regulators of apoptosis (126). Interestingly, the application of SP600125 in the absence of additional cisplatin stimulation showed no major effect, thus suggesting that, at least under the conditions of the study, there was a minimal contribution of JNK to the regulation of these genes. The broader implications of the study include the possibility that JNK/AP-1-mediated resistance to chemotherapy by DNA-damaging drugs may be effectively blocked by inhibition of one or more of the DNA repair genes reported in the study.
A study evaluating JNK-regulated gene expression in rat aortic vascular smooth muscle cells has also revealed a large number of changes, particularly in a range of proteins involved in proinflammatory signaling and extracellular matrix biosynthetic pathways (343). Again, this observation has provided new avenues for evaluation, and in this case, evidence was presented supporting the use of JNK inhibitors as new therapeutic agents in the treatment of abdominal aortic aneurysm (343). It will be interesting to see which transcription factor substrates of JNK are the critical mediators of these responses and whether there may be additional important roles for non-transcription factor substrates.
The substrate repertoire of the JNKs may be increased through the use of proteins such as scaffolds that tether signaling modules. Figure 13 illustrates this general principle alongside the direct recognition of substrates and the use of substrate docking domains in substrate recognition. The use of tethering proteins has been shown for the scaffold protein JNK-associated leucine zipper protein (JLP), as originally identified in the screening of a λgt11 expression library of the mouse myeloid cell line 32Dcl3 with the transcription factor Max (180). In addition to containing two leucine zipper motifs, JLP also showed similarity to the JSAP1/JIP3 scaffold proteins of the JNK pathway, prompting an evaluation of the ability of JLP to interact with other components of the MAPK pathway (180). This analysis revealed that JLP could interact with JNK or p38α MAPK but not with ERK2 (180). JNK and p38α MAPK appeared to interact with two distinct sites on JLP (JLP residues 1 to 110 and JLP residues 160 to 209, respectively), leading to the suggestion that JLP brings JNK and p38α MAPK signaling modules together (180). This was further supported by data showing interactions of JLP with the upstream kinases MKK4 and MEKK3 as well as with the transcription factor substrates c-Myc and Max (180). The scaffolding function of JLP may therefore facilitate specific signaling to c-Myc and Max while also preventing JNK phosphorylation of other substrates (180). With the more recent identification of kinesin light chain 1 as a binding partner for a leucine zipper domain of JLP, it is possible that this motor protein will play a critical role in the spatial regulation of this signaling module (244).
FIG. 13.
Modes of substrate recognition by JNK. Active JNK appears likely to use at least three different modes of substrate recognition. For direct substrate recognition, the amino acids surrounding the residue to be phosphorylated in the substrate protein are recognized at the active site of the kinase. For both docking domain substrates and scaffolded substrates, this requirement for direct substrate recognition at the active site must also be fulfilled, but there are additional sites for substrate recognition. A docking domain on the substrate itself may enhance the interaction with the kinase, and similarly, the interaction with a scaffold protein that can also interact with the kinase will also increase the recognition of the substrate by the kinase.
CONCLUSIONS
In summary, our increased understanding of the substrates of JNK is beginning to highlight the complexities involved in intracellular signal transduction. Over 50 substrates have now been identified for JNKs in the absence of any large-scale or systematic screening strategy. It therefore seems likely that many substrates still remain unknown and that JNKs may phosphorylate more proteins than the estimated average of ∼40 substrates per kinase in humans (151). Improved methods in protein kinase substrate identification (for reviews, see references 24 and 173) should further aid our understanding of how JNKs mediate their complex range of biological actions.
As discussed above, the phosphorylation of a particular substrate by JNK, the identity of the phosphorylation site(s), and the functional consequences of a phosphorylation event remain controversial in a number of cases. In some cases, these proteins remain to be fully validated as JNK substrates. Several criteria need to be met to identify a protein as a physiological protein kinase substrate with reasonable confidence. These criteria include (i) phosphorylation using purified proteins in vitro with significant stoichiometry, (ii) phosphorylation in intact cells in response to physiological stimuli at the same site as that determined in vitro, (iii) stimulation of phosphorylation by constitutively active kinase mutants, and (iv) inhibition of phosphorylation of the substrate by dominant-negative forms of the kinase, by specific inhibitors, or in cells depleted of the kinase (24). Indeed, the variability of sequences phosphorylated by JNK may be overestimated to some degree because of the presence of incorrect phosphorylation sites.
Acknowledgments
We thank Ross Brinkworth, Carl Paulo, and Neil Saunders for helpful discussions.
The work in our laboratories was funded by the National Health and Medical Research Council (NHMRC) and the Australian Research Council (ARC). B.K. is an Australian Research Council (ARC) Federation Fellow and an NHMRC Honorary Research Fellow.
REFERENCES
- 1.Adamson, A. L., D. Darr, E. Holley-Guthrie, R. A. Johnson, A. Mauser, J. Swenson, and S. Kenney. 2000. Epstein-Barr virus immediate-early proteins BZLF1 and BRLF1 activate the ATF2 transcription factor by increasing the levels of phosphorylated p38 and c-Jun N-terminal kinases. J. Virol. 74:1224-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adam-Stitah, S., L. Penna, P. Chambon, and C. Rochette-Egly. 1999. Hyperphosphorylation of the retinoid X receptor alpha by activated c-Jun NH2-terminal kinases. J. Biol. Chem. 274:18932-18941. [DOI] [PubMed] [Google Scholar]
- 3.Adler, V., M. R. Pincus, T. Minamoto, S. Y. Fuchs, M. J. Bluth, P. W. Brandt-Rauf, F. K. Friedman, R. C. Robinson, J. M. Chen, X. W. Wang, C. C. Harris, and Z. Ronai. 1997. Conformation-dependent phosphorylation of p53. Proc. Natl. Acad. Sci. USA 94:1686-1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adler, V., T. Unlap, and A. S. Kraft. 1994. A peptide encoding the c-Jun delta domain inhibits the activity of a c-Jun amino-terminal protein kinase. J. Biol. Chem. 269:11186-11191. [PubMed] [Google Scholar]
- 5.Aguirre, V., T. Uchida, L. Yenush, R. Davis, and M. F. White. 2000. The c-Jun NH2-terminal kinase promotes insulin resistance during association with insulin receptor substrate-1 and phosphorylation of Ser307. J. Biol. Chem. 275:9047-9054. [DOI] [PubMed] [Google Scholar]
- 6.Alarcon-Vargas, D., and Z. Ronai. 2004. c-Jun-NH2 kinase (JNK) contributes to the regulation of c-Myc protein stability. J. Biol. Chem. 279:5008-5016. [DOI] [PubMed] [Google Scholar]
- 7.Alvarez, E., I. C. Northwood, F. A. Gonzalez, D. A. Latour, A. Seth, C. Abate, T. Curran, and R. J. Davis. 1991. Pro-Leu-Ser/Thr-Pro is a consensus primary sequence for substrate protein phosphorylation. Characterization of the phosphorylation of c-Myc and c-Jun proteins by an epidermal growth factor receptor threonine 669 protein kinase. J. Biol. Chem. 266:15277-15285. [PubMed] [Google Scholar]
- 8.Amagasaki, K., H. Kaneto, C. H. Heldin, and J. Lennartsson. 2006. c-Jun N-terminal kinase is necessary for platelet-derived growth factor-mediated chemotaxis in primary fibroblasts. J. Biol. Chem. 281:22173-22179. [DOI] [PubMed] [Google Scholar]
- 9.Ameyar, M., M. Wisniewska, and J. B. Weitzman. 2003. A role for AP-1 in apoptosis: the case for and against. Biochimie 85:747-752. [DOI] [PubMed] [Google Scholar]
- 10.Amura, C. R., L. Marek, R. A. Winn, and L. E. Heasley. 2005. Inhibited neurogenesis in JNK1-deficient embryonic stem cells. Mol. Cell. Biol. 25:10791-10802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andreucci, J. J., D. Grant, D. M. Cox, L. K. Tomc, R. Prywes, D. J. Goldhamer, N. Rodrigues, P. A. Bedard, and J. C. McDermott. 2002. Composition and function of AP-1 transcription complexes during muscle cell differentiation. J. Biol. Chem. 277:16426-16432. [DOI] [PubMed] [Google Scholar]
- 12.Aoki, H., P. M. Kang, J. Hampe, K. Yoshimura, T. Noma, M. Matsuzaki, and S. Izumo. 2002. Direct activation of mitochondrial apoptosis machinery by c-Jun N-terminal kinase in adult cardiac myocytes. J. Biol. Chem. 277:10244-10250. [DOI] [PubMed] [Google Scholar]
- 13.Asehnoune, K., D. Strassheim, S. Mitra, J. Yeol Kim, and E. Abraham. 2005. Involvement of PKCalpha/beta in TLR4 and TLR2 dependent activation of NF-kappaB. Cell Signal. 17:385-394. [DOI] [PubMed] [Google Scholar]
- 14.Baan, B., H. van Dam, G. C. van der Zon, J. A. Maassen, and D. M. Ouwens. 2006. The role of c-Jun N-terminal kinase, p38, and extracellular signal-regulated kinase in insulin-induced Thr69 and Thr71phosphorylation of activating transcription factor 2. Mol. Endocrinol. 20:1786-1795. [DOI] [PubMed] [Google Scholar]
- 15.Bachar, O., M. Adner, R. Uddman, and L. O. Cardell. 2004. Toll-like receptor stimulation induces airway hyper-responsiveness to bradykinin, an effect mediated by JNK and NF-kappa B signaling pathways. Eur. J. Immunol. 34:1196-1207. [DOI] [PubMed] [Google Scholar]
- 16.Barr, R. K., I. Boehm, P. V. Attwood, P. M. Watt, and M. A. Bogoyevitch. 2004. The critical features and the mechanism of inhibition of a kinase interaction motif-based peptide inhibitor of JNK. J. Biol. Chem. 279:36327-36338. [DOI] [PubMed] [Google Scholar]
- 17.Barr, R. K., and M. A. Bogoyevitch. 2001. The c-Jun N-terminal protein kinase family of mitogen-activated protein kinases (JNK MAPKs). Int. J. Biochem. Cell Biol. 33:1047-1063. [DOI] [PubMed] [Google Scholar]
- 18.Barr, R. K., T. S. Kendrick, and M. A. Bogoyevitch. 2002. Identification of the critical features of a small peptide inhibitor of JNK activity. J. Biol. Chem. 277:10987-10997. [DOI] [PubMed] [Google Scholar]
- 19.Barsyte-Lovejoy, D., A. Galanis, A. Clancy, and A. D. Sharrocks. 2004. ERK5 is targeted to myocyte enhancer factor 2A (MEF2A) through a MAPK docking motif. Biochem. J. 381:693-699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barthwal, M. K., P. Sathyanarayana, C. N. Kundu, B. Rana, A. Pradeep, C. Sharma, J. R. Woodgett, and A. Rana. 2003. Negative regulation of mixed lineage kinase 3 by protein kinase B/AKT leads to cell survival. J. Biol. Chem. 278:3897-3902. [DOI] [PubMed] [Google Scholar]
- 21.Baumann, K., E. M. Mandelkow, J. Biernat, H. Piwnica-Worms, and E. Mandelkow. 1993. Abnormal Alzheimer-like phosphorylation of tau-protein by cyclin-dependent kinases cdk2 and cdk5. FEBS Lett. 336:417-424. [DOI] [PubMed] [Google Scholar]
- 22.Behrens, A., W. Jochum, M. Sibilia, and E. F. Wagner. 2000. Oncogenic transformation by ras and fos is mediated by c-Jun N-terminal phosphorylation. Oncogene 19:2657-2663. [DOI] [PubMed] [Google Scholar]
- 23.Behrens, A., M. Sibilia, and E. F. Wagner. 1999. Amino-terminal phosphorylation of c-Jun regulates stress-induced apoptosis and cellular proliferation. Nat. Genet. 21:326-329. [DOI] [PubMed] [Google Scholar]
- 24.Berwick, D. C., and J. M. Tavare. 2004. Identifying protein kinase substrates: hunting for the organ-grinder's monkeys. Trends Biochem. Sci. 29:227-232. [DOI] [PubMed] [Google Scholar]
- 25.Besirli, C. G., and E. M. Johnson. 2003. JNK-independent activation of c-Jun during neuronal apoptosis induced by multiple DNA-damaging agents. J. Biol. Chem. 278:22357-22366. [DOI] [PubMed] [Google Scholar]
- 26.Bhoumik, A., T. G. Huang, V. Ivanov, L. Gangi, R. F. Qiao, S. L. Woo, S. H. Chen, and Z. Ronai. 2002. An ATF2-derived peptide sensitizes melanomas to apoptosis and inhibits their growth and metastasis. J. Clin. Investig. 110:643-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Biggs, W. H., W. K. Cavenee, and K. C. Arden. 2001. Identification and characterization of members of the FKHR (FOX O) subclass of winged-helix transcription factors in the mouse. Mamm. Genome 12:416-425. [DOI] [PubMed] [Google Scholar]
- 28.Bjorkblom, B., N. Ostman, V. Hongisto, V. Komarovski, J. J. Filen, T. A. Nyman, T. Kallunki, M. J. Courtney, and E. T. Coffey. 2005. Constitutively active cytoplasmic c-Jun N-terminal kinase 1 is a dominant regulator of dendritic architecture: role of microtubule-associated protein 2 as an effector. J. Neurosci. 25:6350-6361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bocco, J. L., A. Bahr, J. Goetz, C. Hauss, T. Kallunki, C. Kedinger, and B. Chatton. 1996. In vivo association of ATFa with JNK/SAP kinase activities. Oncogene 12:1971-1980. [PubMed] [Google Scholar]
- 30.Bogoyevitch, M. A. 2006. The isoform-specific functions of the c-Jun N-terminal kinases (JNKs): differences revealed by gene targeting. Bioessays 28:923-934. [DOI] [PubMed] [Google Scholar]
- 31.Bomsztyk, K., O. Denisenko, and J. Ostrowski. 2004. hnRNP K: one protein multiple processes. Bioessays 26:629-638. [DOI] [PubMed] [Google Scholar]
- 32.Bramblett, G. T., M. Goedert, R. Jakes, S. E. Merrick, J. Q. Trojanowski, and V. M. Lee. 1993. Abnormal tau phosphorylation at Ser396 in Alzheimer's disease recapitulates development and contributes to reduced microtubule binding. Neuron 10:1089-1099. [DOI] [PubMed] [Google Scholar]
- 33.Brinkworth, R. I., R. A. Breinl, and B. Kobe. 2003. Structural basis and prediction of substrate specificity in protein serine/threonine kinases. Proc. Natl. Acad. Sci. USA 100:74-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brown, N. R., M. E. Noble, J. A. Endicott, and L. N. Johnson. 1999. The structural basis for specificity of substrate and recruitment peptides for cyclin-dependent kinases. Nat. Cell Biol. 1:438-443. [DOI] [PubMed] [Google Scholar]
- 35.Bruck, N., J. Bastien, G. Bour, A. Tarrade, J. L. Plassat, A. Bauer, S. Adam-Stitah, and C. Rochette-Egly. 2005. Phosphorylation of the retinoid X receptor at the omega loop modulates the expression of retinoic-acid-target genes with a promoter context specificity. Cell Signal. 17:1229-1239. [DOI] [PubMed] [Google Scholar]
- 36.Brugg, B., and A. Matus. 1991. Phosphorylation determines the binding of microtubule-associated protein 2 (MAP2) to microtubules in living cells. J. Cell Biol. 114:735-743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bruna, A., M. Nicolas, A. Munoz, J. M. Kyriakis, and C. Caelles. 2003. Glucocorticoid receptor-JNK interaction mediates inhibition of the JNK pathway by glucocorticoids. EMBO J. 22:6035-6044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bukharova, T., G. Weijer, L. Bosgraaf, D. Dormann, P. J. van Haastert, and C. J. Weijer. 2005. Paxillin is required for cell-substrate adhesion, cell sorting and slug migration during Dictyostelium development. J. Cell Sci. 118:4295-4310. [DOI] [PubMed] [Google Scholar]
- 39.Burgering, B. M., and G. J. Kops. 2002. Cell cycle and death control: long live forkheads. Trends Biochem. Sci. 27:352-360. [DOI] [PubMed] [Google Scholar]
- 40.Burgering, B. M., and R. H. Medema. 2003. Decisions on life and death: FOXO forkhead transcription factors are in command when PKB/Akt is off duty. J. Leukoc. Biol. 73:689-701. [DOI] [PubMed] [Google Scholar]
- 41.Buschmann, T., O. Potapova, A. Bar-Shira, V. N. Ivanov, S. Y. Fuchs, S. Henderson, V. A. Fried, T. Minamoto, D. Alarcon-Vargas, M. R. Pincus, W. A. Gaarde, N. J. Holbrook, Y. Shiloh, and Z. Ronai. 2001. Jun NH2-terminal kinase phosphorylation of p53 on Thr-81 is important for p53 stabilization and transcriptional activities in response to stress. Mol. Cell. Biol. 21:2743-2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Caenepeel, S., G. Charydczak, S. Sudarsanam, T. Hunter, and G. Manning. 2004. The mouse kinome: discovery and comparative genomics of all mouse protein kinases. Proc. Natl. Acad. Sci. USA 101:11707-11712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cai, Y., M. S. Lechner, D. Nihalani, M. J. Prindle, L. B. Holzman, and G. R. Dressler. 2002. Phosphorylation of Pax2 by the c-Jun N-terminal kinase and enhanced Pax2-dependent transcription activation. J. Biol. Chem. 277:1217-1222. [DOI] [PubMed] [Google Scholar]
- 44.Calo, V., M. Migliavacca, V. Bazan, M. Macaluso, M. Buscemi, N. Gebbia, and A. Russo. 2003. STAT proteins: from normal control of cellular events to tumorigenesis. J. Cell Physiol. 197:157-168. [DOI] [PubMed] [Google Scholar]
- 45.Camp, H. S., S. R. Tafuri, and T. Leff. 1999. c-Jun N-terminal kinase phosphorylates peroxisome proliferator-activated receptor-gamma1 and negatively regulates its transcriptional activity. Endocrinology 140:392-397. [DOI] [PubMed] [Google Scholar]
- 46.Carlson, C. J., M. F. White, and C. M. Rondinone. 2004. Mammalian target of rapamycin regulates IRS-1 serine 307 phosphorylation. Biochem. Biophys. Res. Commun. 316:533-539. [DOI] [PubMed] [Google Scholar]
- 47.Cavigelli, M., F. Dolfi, F. X. Claret, and M. Karin. 1995. Induction of c-fos expression through JNK-mediated TCF/Elk-1 phosphorylation. EMBO J. 14:5957-5964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cha, T. L., B. P. Zhou, W. Xia, Y. Wu, C. C. Yang, C. T. Chen, B. Ping, A. P. Otte, and M. C. Hung. 2005. Akt-mediated phosphorylation of EZH2 suppresses methylation of lysine 27 in histone H3. Science 310:306-310. [DOI] [PubMed] [Google Scholar]
- 49.Champion, A., M. Kreis, K. Mockaitis, A. Picaud, and Y. Henry. 2004. Arabidopsis kinome: after the casting. Funct. Integr. Genomics 4:163-187. [DOI] [PubMed] [Google Scholar]
- 50.Chang, C. I., B. E. Xu, R. Akella, M. H. Cobb, and E. J. Goldsmith. 2002. Crystal structures of MAP kinase p38 complexed to the docking sites on its nuclear substrate MEF2A and activator MKK3b. Mol. Cell 9:1241-1249. [DOI] [PubMed] [Google Scholar]
- 51.Chang, L., Y. Jones, M. H. Ellisman, L. S. B. Goldstein, and M. Karin. 2003. JNK1 is required for maintenance of neuronal microtubules and controls phosphorylation of microtubule-associated proteins. Dev. Cell 4:521-533. [DOI] [PubMed] [Google Scholar]
- 52.Chang, L., H. Kamata, G. Solinas, J. L. Luo, S. Maeda, K. Venuprasad, Y.-C. Liu, and M. Karin. 2006. The E3 ubiquitin ligase Itch couples JNK activation to TNFα-induced cell death by inducing c-FLIPL turnover. Cell 124:601-613. [DOI] [PubMed] [Google Scholar]
- 53.Chaussepied, M., D. Lallemand, M. F. Moreau, R. Adamson, R. Hall, and G. Langsley. 1998. Upregulation of Jun and Fos family members and permanent JNK activity lead to constitutive AP-1 activation in Theileria-transformed leukocytes. Mol. Biochem. Parasitol. 94:215-226. [DOI] [PubMed] [Google Scholar]
- 54.Chen, X., J. Zhang, J. Lee, P. S. Lin, J. M. Ford, N. Zheng, and P. Zhou. 2006. A kinase-independent function of c-Abl in promoting proteolytic destruction of damaged DNA binding proteins. Mol. Cell 22:489-499. [DOI] [PubMed] [Google Scholar]
- 55.Chi, N., and J. A. Epstein. 2002. Getting your Pax straight: Pax proteins in development and disease. Trends Genet. 18:41-47. [DOI] [PubMed] [Google Scholar]
- 56.Chow, C. W., C. Dong, R. A. Flavell, and R. J. Davis. 2000. c-Jun NH2-terminal kinase inhibits targeting of the protein phosphatase calcineurin to NFATc1. Mol. Cell. Biol. 20:5227-5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chow, C. W., M. Rincon, J. Cavanagh, M. Dickens, and R. J. Davis. 1997. Nuclear accumulation of NFAT4 opposed by the JNK signaling pathway. Science 278:1638-1641. [DOI] [PubMed] [Google Scholar]
- 58.Clarke, P., S. M. Meintzer, Y. Wang, L. A. Moffitt, S. M. Richardson-Burns, G. L. Johnson, and K. L. Tyler. 2004. JNK regulates the release of proapoptotic mitochondrial factors in reovirus-infected cells. J. Virol. 78:13132-13138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Clarke, P., S. M. Meintzer, C. Widmann, G. L. Johnson, and K. L. Tyler. 2001. Reovirus infection activates JNK and the JNK-dependent transcription factor c-Jun. J. Virol. 75:11275-11283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Clark-Lewis, I., J. S. Sanghera, and S. L. Pelech. 1991. Definition of a consensus sequence for peptide substrate recognition by p44mpk, the meiosis-activated myelin basic protein kinase. J. Biol. Chem. 266:15180-15184. [PubMed] [Google Scholar]
- 61.Cohen, P. 2002. Protein kinases—the major drug targets of the twenty-first century? Nat. Rev. Drug Discov. 1:309-315. [DOI] [PubMed] [Google Scholar]
- 62.Cory, S., and J. M. Adams. 2002. The Bcl2 family: regulators of the cellular life-or-death switch. Nat. Rev. Cancer 2:647-656. [DOI] [PubMed] [Google Scholar]
- 63.Court, N. W., I. Kuo, O. Quigley, and M. A. Bogoyevitch. 2004. Phosphorylation of the mitochondrial protein Sab by stress-activated protein kinase 3. Biochem. Biophys. Res. Commun. 319:130-137. [DOI] [PubMed] [Google Scholar]
- 64.Cuadrado, A., L. Gonzalez, Y. Suarez, T. Martinez, and A. Munoz. 2004. JNK activation is critical for aplidin-induced apoptosis. Oncogene 23:4673-4680. [DOI] [PubMed] [Google Scholar]
- 65.Dai, R., W. Frejtag, B. He, Y. Zhang, and N. F. Mivechi. 2000. c-Jun NH2-terminal kinase targeting and phosphorylation of heat shock factor-1 suppress its transcriptional activity. J. Biol. Chem. 275:18210-18218. [DOI] [PubMed] [Google Scholar]
- 66.Dai, T., E. Rubie, C. C. Franklin, A. Kraft, D. A. F. Gillespie, J. Avruch, J. M. Kyriakis, and J. R. Woodgett. 1995. Stress-activated protein kinases bind directly to the δ domain of c-Jun in resting cells: implications for repression of c-Jun function. Oncogene 10:849-855. [PubMed] [Google Scholar]
- 67.D'Ambrosio, C., S. Arena, G. Fulcoli, M. H. Scheinfeld, D. Zhou, L. D'Adamio, and A. Scaloni. 2006. Hyperphosphorylation of JNK-interacting protein 1, a protein associated with Alzheimer disease. Mol. Cell. Proteomics 5:97-113. [DOI] [PubMed] [Google Scholar]
- 68.Danielsson, A., A. Ost, F. H. Nystrom, and P. Stralfors. 2005. Attenuation of insulin-stimulated insulin receptor substrate-1 serine 307 phosphorylation in insulin resistance of type 2 diabetes. J. Biol. Chem. 280:34389-34392. [DOI] [PubMed] [Google Scholar]
- 69.Dar, A. C., T. E. Dever, and F. Sicheri. 2005. Higher-order substrate recognition of eIF2alpha by the RNA-dependent protein kinase PKR. Cell 122:887-900. [DOI] [PubMed] [Google Scholar]
- 70.Dard, N., and M. Peter. 2006. Scaffold proteins in MAP kinase signaling: more than simple passive activating platforms. Bioessays 28:146-156. [DOI] [PubMed] [Google Scholar]
- 71.Darling, D. L., J. Yingling, and A. Wynshaw-Boris. 2005. Role of 14-3-3 proteins in eukaryotic signaling and development. Curr. Top. Dev. Biol. 68:281-315. [DOI] [PubMed] [Google Scholar]
- 72.David, J. P., K. Sabapathy, O. Hoffmann, M. H. Idarraga, and E. F. Wagner. 2002. JNK1 modulates osteoclastogenesis through both c-Jun phosphorylation-dependent and -independent mechanisms. J. Cell Sci. 115:4317-4325. [DOI] [PubMed] [Google Scholar]
- 73.Davis, R. J. 2000. Signal transduction by the JNK group of MAP kinases. Cell 103:239-252. [DOI] [PubMed] [Google Scholar]
- 74.De Graeve, F., A. Bahr, K. T. Sabapathy, C. Hauss, E. F. Wagner, C. Kedinger, and B. Chatton. 1999. Role of the ATFa/JNK2 complex in Jun activation. Oncogene 18:3491-3500. [DOI] [PubMed] [Google Scholar]
- 75.Deng, T., and M. Karin. 1994. c-Fos transcriptional activity stimulated by H-Ras-activated protein kinase distinct from JNK and ERK. Nature 371:171-175. [DOI] [PubMed] [Google Scholar]
- 76.Deng, X., L. Xiao, W. Lang, F. Gao, P. Ruvolo, and M. Stratford. 2001. Novel role for JNK as a stress-activated Bcl2 kinase. J. Biol. Chem. 276:23681-23688. [DOI] [PubMed] [Google Scholar]
- 77.Dérijard, B., M. Hibi, I.-H. Wu, T. Barrett, B. Su, T. Deng, M. Karin, and R. J. Davis. 1994. JNK1: a protein kinase stimulated by UV light and Ha-Ras that binds and phosphorylates the c-Jun activation domain. Cell 76:1025-1037. [DOI] [PubMed] [Google Scholar]
- 78.des Portes, V., J. M. Pinard, P. Billuart, M. C. Vinet, A. Koulakoff, A. Carrie, A. Gelot, E. Dupuis, J. Motte, Y. Berwald-Netter, M. Catala, A. Kahn, C. Beldjord, and J. Chelly. 1998. A novel CNS gene required for neuronal migration and involved in X-linked subcortical laminar heterotopia and lissencephaly syndrome. Cell 92:51-61. [DOI] [PubMed] [Google Scholar]
- 79.Diao, L., B. Zhang, C. Xuan, S. Sun, K. Yang, Y. Tang, W. Qiao, Q. Chen, Y. Geng, and C. Wang. 2005. Activation of c-Jun N-terminal kinase (JNK) pathway by HSV-1 immediate early protein ICP0. Exp. Cell Res. 308:196-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dickens, M., J. S. Rogers, J. Cavanagh, A. Raitano, Z. Xia, J. R. Halpern, M. E. Greenberg, C. L. Sawyers, and R. J. Davis. 1997. A cytoplasmic inhibitor of the JNK signal transduction pathway. Science 277:693-696. [DOI] [PubMed] [Google Scholar]
- 81.Dong, C., R. J. Davis, and R. A. Flavell. 2002. MAP kinases in the immune response. Annu. Rev. Immunol. 20:55-72. [DOI] [PubMed] [Google Scholar]
- 82.Dong, C., D. D. Yang, C. Tournier, A. J. Whitmarsh, J. Xu, R. J. Davis, and R. A. Flavell. 2000. JNK is required for effector T-cell function but not for T-cell activation. Nature 405:91-94. [DOI] [PubMed] [Google Scholar]
- 83.Dong, C., D. D. Yang, M. Wysk, A. J. Whitmarsh, R. J. Davis, and R. A. Flavell. 1998. Defective T cell differentiation in the absence of Jnk1. Science 282:2092-2095. [DOI] [PubMed] [Google Scholar]
- 84.Donovan, N., E. B. E. Becker, Y. Konishi, and A. Bonni. 2002. JNK phosphorylation and activation of BAD couples the stress-activated signaling pathway to the cell death machinery. J. Biol. Chem. 277:40944-40949. [DOI] [PubMed] [Google Scholar]
- 85.Drewes, G., B. Lichtenberg-Kraag, F. Doring, E. M. Mandelkow, J. Biernat, J. Goris, M. Doree, and E. Mandelkow. 1992. Mitogen activated protein (MAP) kinase transforms tau protein into an Alzheimer-like state. EMBO J. 11:2131-2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ducret, C., S. M. Maira, Y. Lutz, and B. Wasylyk. 2000. The ternary complex factor Net contains two distinct elements that mediate different responses to MAP kinase signalling cascades. Oncogene 19:5063-5072. [DOI] [PubMed] [Google Scholar]
- 87.Eliopoulos, A. G., E. R. Waites, S. M. Blake, C. Davies, P. Murray, and L. S. Young. 2003. TRAF1 is a critical regulator of JNK signaling by the TRAF-binding domain of the Epstein-Barr virus-encoded latent infection membrane protein 1 but not CD40. J. Virol. 77:1316-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Eminel, S., A. Klettner, L. Roemer, T. Herdegen, and V. Waetzig. 2004. JNK2 translocates to the mitochondria and mediates cytochrome c release in PC12 cells in response to 6-hydroxydopamine. J. Biol. Chem. 279:55385-55392. [DOI] [PubMed] [Google Scholar]
- 89.Esen, M., B. Schreiner, V. Jendrossek, F. Lang, K. Fassbender, H. Grassme, and E. Gulbins. 2001. Mechanisms of Staphylococcus aureus induced apoptosis of human endothelial cells. Apoptosis 6:431-439. [DOI] [PubMed] [Google Scholar]
- 90.Essers, M. A., S. Weijzen, A. M. de Vries-Smits, I. Saarloos, N. D. de Ruiter, J. L. Bos, and B. M. Burgering. 2004. FOXO transcription factor activation by oxidative stress mediated by the small GTPase Ral and JNK. EMBO J. 8:4802-4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fang, D., C. Elly, B. Gao, N. Fang, Y. Altman, C. Joazeiro, T. Hunter, N. Copeland, N. Jenkins, and Y. C. Liu. 2002. Dysregulation of T lymphocyte function in itchy mice: a role for Itch in TH2 differentiation. Nat. Immunol. 3:281-287. [DOI] [PubMed] [Google Scholar]
- 92.Fischer, P. M. 2004. The design of drug candidate molecules as selective inhibitors of therapeutically relevant protein kinases. Curr. Med. Chem. 11:1563-1583. [DOI] [PubMed] [Google Scholar]
- 93.Frodin, M., C. J. Jensen, K. Merienne, and S. Gammeltoft. 2000. A phosphoserine-regulated docking site in the protein kinase RSK2 that recruits and activates PDK1. EMBO J. 19:2924-2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fuchs, S. Y., V. Adler, T. Buschmann, Z. Yin, X. Wu, S. N. Jones, and Z. Ronai. 1998. JNK targets p53 ubiquitination and degradation in nonstressed cells. Genes Dev. 12:2658-2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fuchs, S. Y., V. Adler, M. R. Pincus, and Z. Ronai. 1998. MEKK1/JNK signaling stabilizes and activates p53. Proc. Natl. Acad. Sci. USA 95:10541-10546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Fuchs, S. Y., L. Dolan, R. J. Davis, and Z. Ronai. 1996. Phosphorylation-dependent targeting of c-Jun ubiquitination by Jun N-kinase. Oncogene 13:1531-1535. [PubMed] [Google Scholar]
- 97.Fuchs, S. Y., I. Tappin, and Z. Ronai. 2000. Stability of the ATF2 transcription factor is regulated by phosphorylation and dephosphorylation. J. Biol. Chem. 275:12560-12564. [DOI] [PubMed] [Google Scholar]
- 98.Fuchs, S. Y., B. Xie, V. Adler, V. A. Fried, R. J. Davis, and Z. Ronai. 1997. c-Jun NH2-terminal kinases target the ubiquitination of their associated transcription factors. J. Biol. Chem. 272:32163-32168. [DOI] [PubMed] [Google Scholar]
- 99.Galanis, A., S. H. Yang, and A. D. Sharrocks. 2001. Selective targeting of MAPKs to the ETS domain transcription factor SAP-1. J. Biol. Chem. 276:965-973. [DOI] [PubMed] [Google Scholar]
- 100.Gallagher, E., M. Gao, Y. C. Liu, and M. Karin. 2006. Activation of the E3 ubiquitin ligase Itch through a phosphorylation-induced conformational change. Proc. Natl. Acad. Sci. USA 103:1717-1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gao, M., and M. Karin. 2005. Regulating the regulators: control of protein ubiquitination and ubiquitin-like modifications by extracellular stimuli. Mol. Cell 19:581-593. [DOI] [PubMed] [Google Scholar]
- 102.Gao, M., T. Labuda, Y. Xia, E. Gallagher, D. Fang, Y. C. Liu, and M. Karin. 2004. Jun turnover is controlled through JNK-dependent phosphorylation of the E3 ligase Itch. Science 306:271-275. [DOI] [PubMed] [Google Scholar]
- 103.Gao, Z., D. Hwang, F. Bataille, M. Lefevre, D. York, M. J. Quon, and J. Ye. 2002. Serine phosphorylation of insulin receptor substrate 1 by inhibitor kappa B kinase complex. J. Biol. Chem. 277:48115-48121. [DOI] [PubMed] [Google Scholar]
- 104.Gao, Z., X. Zhang, A. Zuberi, D. Hwang, M. J. Quon, M. Lefevre, and J. Ye. 2004. Inhibition of insulin sensitivity by free fatty acids requires activation of multiple serine kinases in 3T3-L1 adipocytes. Mol. Endocrinol. 18:2024-2034. [DOI] [PubMed] [Google Scholar]
- 105.Gdalyahu, A., I. Ghosh, T. Levy, T. Sapir, S. Sapoznik, Y. Fishler, D. Azoulai, and O. Reiner. 2004. DCX, a new mediator of the JNK pathway. EMBO J. 23:823-832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gianni, M., A. Tarrade, E. A. Nigro, E. Garattini, and C. Rochette-Egly. 2003. The AF-1 and AF-2 domains of RAR gamma 2 and RXR alpha cooperate for triggering the transactivation and the degradation of RAR gamma 2/RXR alpha heterodimers. J. Biol. Chem. 278:34458-34466. [DOI] [PubMed] [Google Scholar]
- 107.Gille, H., M. Kortenjann, O. Thomae, C. Moomaw, C. Slaughter, M. H. Cobb, and P. E. Shaw. 1995. ERK phosphorylation potentiates Elk-1-mediated ternary complex formation and transactivation. EMBO J. 14:951-962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gille, H., T. Strahl, and P. E. Shaw. 1995. Activation of ternary complex factor Elk-1 by stress-activated protein kinases. Curr. Biol. 5:1191-1200. [DOI] [PubMed] [Google Scholar]
- 109.Gioeli, D., B. E. Black, V. Gordon, A. Spencer, C. T. Kesler, S. T. Eblen, B. M. Paschal, and M. J. Weber. 2006. Stress kinase signaling regulates androgen receptor phosphorylation, transcription, and localization. Mol. Endocrinol. 20:503-515. [DOI] [PubMed] [Google Scholar]
- 110.Girardin, S. E., R. Tournebize, M. Mavris, A. L. Page, X. Li, G. R. Stark, J. Bertin, P. S. DiStefano, M. Yaniv, P. J. Sansonetti, and D. J. Philpott. 2001. CARD4/Nod1 mediates NF-kappaB and JNK activation by invasive Shigella flexneri. EMBO Rep. 2:736-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Goedert, M., M. Hasegawa, R. Jakes, S. Lawler, A. Cuenda, and P. Cohen. 1997. Phosphorylation of microtubule-associated protein tau by stress-activated protein kinases. FEBS Lett. 409:57-62. [DOI] [PubMed] [Google Scholar]
- 112.Goldberg, J. M., G. Manning, A. Liu, P. Fey, K. E. Pilcher, Y. Xu, and J. L. Smith. 2006. The Dictyostelium kinome—analysis of the protein kinases from a simple model organism. PLoS Genet. 2:e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gonzalez, F. A., D. L. Raden, and R. J. Davis. 1991. Identification of substrate recognition determinants for human ERK1 and ERK2 protein kinases. J. Biol. Chem. 266:22159-22163. [PubMed] [Google Scholar]
- 114.Gonzalez, F. A., A. Seth, D. L. Raden, D. S. Bowman, F. S. Fay, and R. J. Davis. 1993. Serum-induced translocation of mitogen-activated protein kinase to the cell surface ruffling and the nucleus. J. Cell Biol. 122:1089-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Gonzalez, M. V., B. Jimenez, M. T. Berciano, J. M. Gonzalez-Sancho, C. Caelles, M. Lafarga, and A. Munoz. 2000. Glucocorticoids antagonize AP-1 by inhibiting the activation/phosphorylation of JNK without affecting its subcellular distribution. J. Cell Biol. 150:1199-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Green, D. R., and G. Kroemer. 2004. The pathophysiology of mitochondrial cell death. Science 305:626-629. [DOI] [PubMed] [Google Scholar]
- 117.Gupta, S., T. Barrett, A. J. Whitmarsh, J. Cavanagh, H. K. Sluss, B. Dérijard, and R. J. Davis. 1996. Selective interaction of JNK protein kinase isoforms with transcription factors. EMBO J. 15:2760-2770. [PMC free article] [PubMed] [Google Scholar]
- 118.Gupta, S., D. Campbell, B. Dérijard, and R. J. Davis. 1995. Transcription factor ATF2 regulation by the JNK signal transduction pathway. Science 267:389-393. [DOI] [PubMed] [Google Scholar]
- 119.Habelhah, H., K. Shah, L. Huang, A. L. Burlingame, K. M. Shokat, and Z. Ronai. 2001. Identification of new JNK substrate using ATP pocket mutant JNK and a corresponding ATP analogue. J. Biol. Chem. 276:18090-18095. [DOI] [PubMed] [Google Scholar]
- 120.Habelhah, H., K. Shah, L. Huang, A. Ostareck-Lederer, A. L. Burlingame, K. M. Shokat, M. W. Hentze, and Z. Ronai. 2001. ERK phosphorylation drives cytoplasmic accumulation of hnRNP-K and inhibition of mRNA translation. Nat. Cell Biol. 3:325-330. [DOI] [PubMed] [Google Scholar]
- 121.Han, Y. H., X. Cao, B. Lin, F. Lin, S. K. Kolluri, J. Stebbins, J. C. Reed, M. I. Dawson, and X. K. Zhang. 2006. Regulation of Nur77 nuclear export by c-Jun N-terminal kinase and Akt. Oncogene 25:2974-2986. [DOI] [PubMed] [Google Scholar]
- 122.Hanks, S. K., and A. M. Quinn. 1991. Protein kinase catalytic domain sequence database: identification of conserved features of primary structure and classification of family members. Methods Enzymol. 200:38-62. [DOI] [PubMed] [Google Scholar]
- 123.Hanks, S. K., A. M. Quinn, and T. Hunter. 1988. The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science 241:42-52. [DOI] [PubMed] [Google Scholar]
- 124.Hargett, D., T. McLean, and S. L. Bachenheimer. 2005. Herpes simplex virus ICP27 activation of stress kinases JNK and p38. J. Virol. 79:8348-8360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hayakawa, J., C. Depatie, M. Ohmichi, and D. Mercola. 2003. The activation of c-Jun NH2-terminal kinase (JNK) by DNA-damaging agents serves to promote drug resistance via activating transcription factor 2 (ATF2)-dependent enhanced DNA repair. J. Biol. Chem. 278:20582-20592. [DOI] [PubMed] [Google Scholar]
- 126.Hayakawa, J., S. Mittal, Y. Wang, K. S. Korkmaz, E. Adamson, C. English, M. Ohmichi, M. McClelland, and D. Mercola. 2004. Identification of promoters bound by c-Jun/ATF2 during rapid large-scale gene activation following genotoxic stress. Mol. Cell 16:521-535. [DOI] [PubMed] [Google Scholar]
- 127.He, T., A. Stepulak, T. H. Holmstrom, M. B. Omary, and J. E. Eriksson. 2002. The intermediate filament protein keratin 8 is a novel cytoplasmic substrate for c-Jun N-terminal kinase. J. Biol. Chem. 277:10767-10774. [DOI] [PubMed] [Google Scholar]
- 128.Heinrich, R., E. Livne, O. Ben-Izhak, and A. Aronheim. 2004. The c-Jun dimerization protein 2 inhibits cell transformation and acts as a tumor suppressor gene. J. Biol. Chem. 279:5708-5715. [DOI] [PubMed] [Google Scholar]
- 129.Heo, Y. S., S. K. Kim, C. I. Seo, Y. K. Kim, B. J. Sung, H. S. Lee, J. I. Lee, S. Y. Park, J. H. Kim, K. Y. Hwang, Y. L. Hyun, Y. H. Jeon, S. Ro, J. M. Cho, T. G. Lee, and C. H. Yang. 2004. Structural basis for the selective inhibition of JNK1 by the scaffolding protein JIP1 and SP600125. EMBO J. 23:2185-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hidding, U., K. Mielke, V. Waetzig, S. Brecht, U. Hanisch, A. Behrens, E. Wagner, and T. Herdegen. 2002. The c-Jun N-terminal kinases in cerebral microglia: immunological functions in the brain. Biochem. Pharmacol. 64:781-788. [DOI] [PubMed] [Google Scholar]
- 131.Hilder, T. L., J. C. Tou, R. E. Grindeland, C. E. Wade, and L. M. Graves. 2003. Phosphorylation of insulin receptor substrate-1 serine 307 correlates with JNK activity in atrophic skeletal muscle. FEBS Lett. 553:63-67. [DOI] [PubMed] [Google Scholar]
- 132.Hill, C. S., R. Marais, S. John, J. Wynne, S. Dalton, and R. Treisman. 1993. Functional analysis of a growth factor-responsive transcription factor complex. Cell 73:395-406. [DOI] [PubMed] [Google Scholar]
- 133.Hiratani, K., T. Haruta, A. Tani, J. Kawahara, I. Usui, and M. Kobayashi. 2005. Roles of mTOR and JNK in serine phosphorylation, translocation, and degradation of IRS-1. Biochem. Biophys. Res. Commun. 335:836-842. [DOI] [PubMed] [Google Scholar]
- 134.Hirosumi, J., G. Tuncman, L. Chang, C. Z. Gorgun, K. T. Uysal, K. Maeda, M. Karin, and G. S. Hotamisligil. 2002. A central role for JNK in obesity and insulin resistance. Nature 420:333-336. [DOI] [PubMed] [Google Scholar]
- 135.Hogan, P. G., L. Chen, J. Nardone, and A. Rao. 2003. Transcriptional regulation by calcium, calcineurin, and NFAT. Genes Dev. 17:2205-2232. [DOI] [PubMed] [Google Scholar]
- 136.Hotamisligil, G. S. 2005. Role of endoplasmic reticulum stress and c-Jun NH2-terminal kinase pathways in inflammation and origin of obesity and diabetes. Diabetes 54(Suppl. 2):S73-S78. [DOI] [PubMed] [Google Scholar]
- 137.Hrstka, R., J. Stulik, and B. Vojtesek. 2005. The role of MAPK signal pathways during Francisella tularensis LVS infection-induced apoptosis in murine macrophages. Microbes Infect. 7:619-625. [DOI] [PubMed] [Google Scholar]
- 138.Huang, C., Z. Rajfur, C. Borchers, M. D. Schaller, and K. Jacobson. 2003. JNK phosphorylates paxillin and regulates cell migration. Nature 424:219-223. [DOI] [PubMed] [Google Scholar]
- 139.Huang, H., S. B. Petkova, A. W. Cohen, B. Bouzahzah, J. Chan, J. N. Zhou, S. M. Factor, L. M. Weiss, M. Krishnamachary, S. Mukherjee, M. Wittner, R. N. Kitsis, R. G. Pestell, M. P. Lisanti, C. Albanese, and H. B. Tanowitz. 2003. Activation of transcription factors AP-1 and NF-kappa B in murine chagasic myocarditis. Infect. Immun. 71:2859-2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Hubbard, S. R. 1997. Crystal structure of the activated insulin receptor tyrosine kinase in complex with peptide substrate and ATP analog. EMBO J. 16:5572-5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Inomata, H., Y. Nakamura, A. Hayakawa, H. Takata, T. Suzuki, K. Miyazawa, and N. Kitamura. 2003. A scaffold protein JIP-1b enhances amyloid precursor protein phosphorylation by JNK and its association with kinesin light chain 1. J. Biol. Chem. 278:22946-22955. [DOI] [PubMed] [Google Scholar]
- 142.Inoshita, S., K. Takeda, T. Hatai, Y. Terada, M. Sano, J. Hata, A. Umezawa, and H. Ichijo. 2002. Phosphorylation and inactivation of myeloid cell leukemia 1 by JNK in response to oxidative stress. J. Biol. Chem. 277:43730-43734. [DOI] [PubMed] [Google Scholar]
- 143.Iordanov, M. S., J. M. Paranjape, A. Zhou, J. Wong, B. R. Williams, E. F. Meurs, R. H. Silverman, and B. E. Magun. 2000. Activation of p38 mitogen-activated protein kinase and c-Jun NH2-terminal kinase by double-stranded RNA and encephalomyocarditis virus: involvement of RNase L, protein kinase R, and alternative pathways. Mol. Cell. Biol. 20:617-627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ishibe, S., D. Joly, X. Zhu, and L. G. Cantley. 2003. Phosphorylation-dependent paxillin-ERK association mediates hepatocyte growth factor-stimulated epithelial morphogenesis. Mol. Cell 12:1275-1285. [DOI] [PubMed] [Google Scholar]
- 145.Ishii, N., S. Goto, and P. S. Hartman. 2002. Protein oxidation during aging of the nematode Caenorhabditis elegans. Free Radic. Biol. Med. 33:1021-1025. [DOI] [PubMed] [Google Scholar]
- 146.Ito, T., X. Deng, B. Carr, and W. S. May. 1997. Bcl-2 phosphorylation required for anti-apoptosis function. J. Biol. Chem. 272:11671-11673. [DOI] [PubMed] [Google Scholar]
- 147.Itoh, M., M. Adachi, H. Yasui, M. Takekawa, H. Tanaka, and K. Imai. 2002. Nuclear export of glucocorticoid receptor is enhanced by c-Jun N-terminal kinase-mediated phosphorylation. Mol. Endocrinol. 16:2382-2392. [DOI] [PubMed] [Google Scholar]
- 148.Jacobs, D., D. Glossip, H. Xing, A. J. Muslin, and K. Kornfeld. 1999. Multiple docking sites on substrate proteins form a modular system that mediates recognition by ERK MAP kinase. Genes Dev. 13:163-175. [PMC free article] [PubMed] [Google Scholar]
- 149.Jakobsen, S. N., D. G. Hardie, N. Morrice, and H. E. Tornqvist. 2001. 5′-AMP-activated protein kinase phosphorylates IRS-1 on Ser-789 in mouse C2C12 myotubes in response to 5-aminoimidazole-4-carboxamide riboside. J. Biol. Chem. 276:46912-46916. [DOI] [PubMed] [Google Scholar]
- 150.Javelaud, D., J. Laboureau, E. Gabison, F. Verrecchia, and A. Mauviel. 2003. Disruption of basal JNK activity differentially affects key fibroblast functions important for wound healing. J. Biol. Chem. 278:24624-24628. [DOI] [PubMed] [Google Scholar]
- 151.Johnson, S. A., and T. Hunter. 2005. Kinomics: methods for deciphering the kinome. Nat. Methods 2:17-25. [DOI] [PubMed] [Google Scholar]
- 152.Kallunki, T., T. Deng, M. Hibi, and M. Karin. 1996. c-Jun can recruit JNK to phosphorylate dimerization partners via specific docking interactions. Cell 87:929-939. [DOI] [PubMed] [Google Scholar]
- 153.Kao, A. W., S. B. Waters, S. Okada, and J. E. Pessin. 1997. Insulin stimulates the phosphorylation of the 66- and 52-kilodalton Shc isoforms by distinct pathways. Endocrinology 138:2474-2480. [DOI] [PubMed] [Google Scholar]
- 154.Kasibhatla, S., P. Tailor, N. Bonefoy-Berard, T. Mustelin, A. Altman, and A. Fotedar. 1999. Jun kinase phosphorylates and regulates the DNA binding activity of an octamer binding protein, T-cell factor β1. Mol. Cell. Biol. 19:2021-2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Katz, S., and A. Aronheim. 2002. Differential targeting of the stress mitogen-activated protein kinases to the c-Jun dimerization protein 2. Biochem. J. 368:939-945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Katz, S., R. Heinrich, and A. Aronheim. 2001. The AP-1 repressor, JDP2, is a bona fide substrate for the c-Jun N-terminal kinase. FEBS Lett. 506:196-200. [DOI] [PubMed] [Google Scholar]
- 157.Kawamori, D., H. Kaneto, Y. Nakatani, T. A. Matsuoka, M. Matsuhisa, M. Hori, and Y. Yamasaki. 2006. The forkhead transcription factor Foxo1 bridges the JNK pathway and the transcription factor PDX-1 through its intracellular translocation. J. Biol. Chem. 281:1091-1098. [DOI] [PubMed] [Google Scholar]
- 158.Kawasaki, H., T. Moriguchi, S. Matsuda, H. Z. Li, S. Nakamura, S. Shimohama, J. Kimura, Y. Gotoh, and E. Nishida. 1996. Ras-dependent and Ras-independent activation pathways for the stress-activated-protein-kinase cascade. Eur. J. Biochem. 241:315-321. [DOI] [PubMed] [Google Scholar]
- 159.Kawauchi, T., K. Chihama, Y. Nabeshima, and M. Hoshino. 2003. The in vivo roles of STEF/Tiam1, Rac1 and JNK in cortical neuronal migration. EMBO J. 22:4190-4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Kawauchi, T., K. Chihama, Y. V. Nishimura, Y. Nabeshima, and M. Hoshino. 2005. MAP1B phosphorylation is differentially regulated by Cdk5/p35, Cdk5/p25, and JNK. Biochem. Biophys. Res. Commun. 331:50-55. [DOI] [PubMed] [Google Scholar]
- 161.Kelkar, N., S. Gupta, M. Dickens, and R. J. Davis. 2000. Interaction of a mitogen-activated protein kinase signaling module with the neuronal protein JIP3. Mol. Cell. Biol. 20:1030-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Kenzel, S., G. Mancuso, R. Malley, G. Teti, D. T. Golenbock, and P. Henneke. 2006. c-Jun kinase is a critical signaling molecule in a neonatal model of group B streptococcal sepsis. J. Immunol. 176:3181-3188. [DOI] [PubMed] [Google Scholar]
- 163.Kharbanda, S., S. Saxena, K. Yoshida, P. Pandey, M. Kaneki, Q. Wang, K. Cheng, Y. N. Chen, A. Campbell, T. Sudha, Z. M. Yuan, J. Narula, R. Weichselbaum, C. Nalin, and D. Kufe. 2000. Translocation of SAPK/JNK to mitochondria and interaction with Bcl-x(L) in response to DNA damage. J. Biol. Chem. 275:322-327. [DOI] [PubMed] [Google Scholar]
- 164.Kim, A. H., G. Khursigara, X. Sun, T. F. Franke, and M. V. Chao. 2001. Akt phosphorylates and negatively regulates apoptosis signal-regulating kinase 1. Mol. Cell. Biol. 21:893-901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Kim, A. H., H. Yano, H. Cho, D. Meyer, B. Monks, B. Margolis, M. J. Birnbaum, and M. V. Chao. 2002. Akt1 regulates a JNK scaffold during excitotoxic apoptosis. Neuron 35:697-709. [DOI] [PubMed] [Google Scholar]
- 166.Kim, B. J., S. W. Ryu, and B. J. Song. 2006. JNK- and p38 kinase-mediated phosphorylation of Bax leads to its activation and mitochondrial translocation and to apoptosis of human hepatoma HepG2 cells. J. Biol. Chem. 281:21256-21265. [DOI] [PubMed] [Google Scholar]
- 167.Kim, H. T., W. Qiang, N. Liu, V. L. Scofield, P. K. Wong, and G. Stoica. 2005. Up-regulation of astrocyte cyclooxygenase-2, CCAAT/enhancer-binding protein-homology protein, glucose-related protein 78, eukaryotic initiation factor 2 alpha, and c-Jun N-terminal kinase by a neurovirulent murine retrovirus. J. Neurovirol. 11:166-179. [DOI] [PubMed] [Google Scholar]
- 168.Kim, M. H., T. Cierpicki, U. Derewenda, D. Krowarsch, Y. Feng, Y. Devedjiev, Z. Dauter, C. A. Walsh, J. Otlewski, J. H. Bushweller, and Z. S. Derewenda. 2003. The DCX-domain tandems of doublecortin and doublecortin-like kinase. Nat. Struct. Biol. 10:324-333. [DOI] [PubMed] [Google Scholar]
- 169.Kim, S. M., J. H. Park, S. K. Chung, J. Y. Kim, H. Y. Hwang, K. C. Chung, I. Jo, S. I. Park, and J. H. Nam. 2004. Coxsackievirus B3 infection induces cyr61 activation via JNK to mediate cell death. J. Virol. 78:13479-13488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Kimberly, W. T., J. B. Zheng, T. Town, R. A. Flavell, and D. J. Selkoe. 2005. Physiological regulation of the beta-amyloid precursor protein signaling domain by c-Jun N-terminal kinase JNK3 during neuronal differentiation. J. Neurosci. 25:5533-5543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Knighton, D. R., J. H. Zheng, L. F. Ten Eyck, V. A. Ashford, N. H. Xuong, S. S. Taylor, and J. M. Sowadski. 1991. Crystal structure of the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science 253:407-414. [DOI] [PubMed] [Google Scholar]
- 172.Knighton, D. R., J. H. Zheng, L. F. Ten Eyck, N. H. Xuong, S. S. Taylor, and J. M. Sowadski. 1991. Structure of a peptide inhibitor bound to the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science 253:414-420. [DOI] [PubMed] [Google Scholar]
- 173.Kobe, B., T. Kampmann, J. K. Forwood, P. Listwan, and R. I. Brinkworth. 2005. Substrate specificity of protein kinases and computational prediction of substrates. Biochim. Biophys. Acta 1754:200-209. [DOI] [PubMed] [Google Scholar]
- 174.Kolluri, S. K., N. Bruey-Sedano, X. Cao, B. Lin, F. Lin, Y. H. Han, M. I. Dawson, and X. K. Zhang. 2003. Mitogenic effect of orphan receptor TR3 and its regulation by MEKK1 in lung cancer cells. Mol. Cell. Biol. 23:8651-8667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Krstic, M. D., I. Rogatsky, K. R. Yamamoto, and M. J. Garabedian. 1997. Mitogen-activated and cyclin-dependent protein kinases selectively and differentially modulate transcriptional enhancement by the glucocorticoid receptor. Mol. Cell. Biol. 17:3947-3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Kujime, K., S. Hashimoto, Y. Gon, K. Shimizu, and T. Horie. 2000. p38 mitogen-activated protein kinase and c-Jun NH2-terminal kinase regulate RANTES production by influenza virus-infected human bronchial epithelial cells. J. Immunol. 164:3222-3228. [DOI] [PubMed] [Google Scholar]
- 177.Kyriakis, J. M., and J. Avruch. 1990. pp54 microtubule-associated protein 2 kinase. A novel serine/threonine protein kinase regulated by phosphorylation and stimulated by poly-l-lysine. J. Biol. Chem. 265:17355-17363. [PubMed] [Google Scholar]
- 178.Lan, K. P., C. J. Wang, J. D. Hsu, K. M. Chen, S. C. Lai, and H. H. Lee. 2004. Induced eosinophilia and proliferation in Angiostrongylus cantonensis-infected mouse brain are associated with the induction of JAK/STAT1, IAP/NF-kappaB and MEKK1/JNK signals. J. Helminthol. 78:311-317. [DOI] [PubMed] [Google Scholar]
- 179.Le, S., T. J. Connors, and A. C. Maroney. 2001. c-Jun N-terminal kinase specifically phosphorylates p66ShcA at serine 63 in response to ultraviolet radiation. J. Biol. Chem. 276:48332-48336. [DOI] [PubMed] [Google Scholar]
- 180.Lee, C. M., D. Onesime, C. D. Reddy, N. Dhanasekaran, and E. P. Reddy. 2002. JLP: a scaffolding protein that tethers JNK/p38MAPK signaling modules and transcription factors. Proc. Natl. Acad. Sci. USA 99:14189-14194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Lee, H. Y., Y. A. Suh, M. J. Robinson, J. L. Clifford, W. K. Hong, J. R. Woodgett, M. H. Cobb, D. J. Mangelsdorf, and J. M. Kurie. 2000. Stress pathway activation induces phosphorylation of retinoid X receptor. J. Biol. Chem. 275:32193-32199. [DOI] [PubMed] [Google Scholar]
- 182.Lee, S., J. Hong, S. Y. Choi, S. B. Oh, K. Park, J. S. Kim, M. Karin, and S. J. Lee. 2004. CpG oligodeoxynucleotides induce expression of proinflammatory cytokines and chemokines in astrocytes: the role of c-Jun N-terminal kinase in CpG ODN-mediated NF-kappaB activation. J. Neuroimmunol. 153:50-63. [DOI] [PubMed] [Google Scholar]
- 183.Lee, T., A. N. Hoofnagle, K. A. Resing, and N. G. Ahn. 2005. Hydrogen exchange solvent protection by an ATP analogue reveals conformational changes in ERK2 upon activation. J. Mol. Biol. 353:600-612. [DOI] [PubMed] [Google Scholar]
- 184.Lee, V. M., M. Goedert, and J. Q. Trojanowski. 2001. Neurodegenerative tauopathies. Annu. Rev. Neurosci. 24:1121-1159. [DOI] [PubMed] [Google Scholar]
- 185.Lee, Y. H., J. Giraud, R. J. Davis, and M. F. White. 2003. c-Jun N-terminal kinase (JNK) mediates feedback inhibition of the insulin signaling cascade. J. Biol. Chem. 278:2896-2902. [DOI] [PubMed] [Google Scholar]
- 186.Lei, K., and R. J. Davis. 2003. JNK phosphorylation of Bim-related members of the Bcl2 family induces Bax-dependent apoptosis. Proc. Natl. Acad. Sci. USA 100:2432-2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Lenormand, P., C. Sardet, G. Pagès, G. L'Allemain, A. Brunet, and J. Pouysségur. 1993. Growth factors induce nuclear translocation of MAP kinases (p42mapk and p44mapk) but not of their activator MAP kinase kinase (p45mapkk) in fibroblasts. J. Cell Biol. 122:1079-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Leppa, S., and D. Bohmann. 1999. Diverse functions of JNK signaling and c-Jun in stress response and apoptosis. Oncogene 18:6158-6162. [DOI] [PubMed] [Google Scholar]
- 189.Lewis, T. S., J. B. Hunt, L. D. Aveline, K. R. Jonscher, D. F. Louie, J. M. Yeh, T. S. Nahreini, K. A. Resing, and N. G. Ahn. 2000. Identification of novel MAP kinase pathway signaling targets by functional proteomics and mass spectrometry. Mol. Cell 6:1343-1354. [DOI] [PubMed] [Google Scholar]
- 190.Li, B., C. Tournier, R. J. Davis, and R. A. Flavell. 1999. Regulation of IL-4 expression by the transcription factor JunB during T helper cell differentiation. EMBO J. 18:420-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Li, X. D., H. Lankinen, N. Putkuri, O. Vapalahti, and A. Vaheri. 2005. Tula hantavirus triggers proapoptotic signals of ER stress in Vero E6 cells. Virology 333:180-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Liang, Q., O. F. Bueno, B. J. Wilkins, C. Y. Kuan, Y. Xia, and J. D. Molkentin. 2003. c-Jun N-terminal kinases (JNK) antagonize cardiac growth through cross-talk with calcineurin-NFAT signaling. EMBO J. 22:5079-5089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Lin, C., S. G. Zimmer, Z. Lu, R. E. Holland, Q. Dong, and T. M. Chambers. 2001. The involvement of a stress-activated pathway in equine influenza virus-mediated apoptosis. Virology 287:202-213. [DOI] [PubMed] [Google Scholar]
- 194.Lindwall, G., and R. D. Cole. 1984. Phosphorylation affects the ability of tau protein to promote microtubule assembly. J. Biol. Chem. 259:5301-5305. [PubMed] [Google Scholar]
- 195.Liu, M., Z. Xin, J. E. Clampit, S. Wang, R. J. Gum, D. L. Haasch, J. M. Trevillyan, C. Abad-Zapatero, E. H. Fry, H. L. Sham, and G. Liu. 2006. Synthesis and SAR of 1,9-dihydro-9-hydroxypyrazolo[3,4-b]quinolin-4-ones as novel, selective c-Jun N-terminal kinase inhibitors. Bioorg. Med. Chem. Lett. 16:2590-2594. [DOI] [PubMed] [Google Scholar]
- 196.Liu, Y. F., K. Paz, A. Herschkovitz, A. Alt, T. Tennenbaum, S. R. Sampson, M. Ohba, T. Kuroki, D. LeRoith, and Y. Zick. 2001. Insulin stimulates PKCzeta-mediated phosphorylation of insulin receptor substrate-1 (IRS-1). A self-attenuated mechanism to negatively regulate the function of IRS proteins. J. Biol. Chem. 276:14459-14465. [DOI] [PubMed] [Google Scholar]
- 197.Livingstone, C., G. Patel, and N. Jones. 1995. ATF-2 contains a phosphorylation-dependent transcriptional activation domain. EMBO J. 14:1785-1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Lowe, E. D., M. E. Noble, V. T. Skamnaki, N. G. Oikonomakos, D. J. Owen, and L. N. Johnson. 1997. The crystal structure of a phosphorylase kinase peptide substrate complex: kinase substrate recognition. EMBO J. 16:6646-6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Ludwig, S., C. Ehrhardt, E. R. Neumeier, M. Kracht, U. R. Rapp, and S. Pleschka. 2001. Influenza virus-induced AP-1-dependent gene expression requires activation of the JNK signaling pathway. J. Biol. Chem. 276:10990-10998. [PubMed] [Google Scholar]
- 200.Luo, W., and S. C. Lin. 2004. Axin: a master scaffold for multiple signaling pathways. Neurosignals 13:99-113. [DOI] [PubMed] [Google Scholar]
- 201.Macian, F. 2005. NFAT proteins: key regulators of T-cell development and function. Nat. Rev. Immunol. 5:472-484. [DOI] [PubMed] [Google Scholar]
- 202.Malbon, C. C., J. Tao, and H. Y. Wang. 2004. AKAPs (A-kinase anchoring proteins) and molecules that compose their G-protein-coupled receptor signalling complexes. Biochem. J. 379:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Mandelkow, E. M., G. Drewes, J. Biernat, N. Gustke, J. Van Lint, J. R. Vandenheede, and E. Mandelkow. 1992. Glycogen synthase kinase-3 and the Alzheimer-like state of microtubule-associated protein tau. FEBS Lett. 314:315-321. [DOI] [PubMed] [Google Scholar]
- 204.Mann, K. K., A. M. Padovani, Q. Guo, A. L. Colosimo, H. Y. Lee, J. M. Kurie, and W. H. Miller. 2005. Arsenic trioxide inhibits nuclear receptor function via SEK1/JNK-mediated RXRalpha phosphorylation. J. Clin. Investig. 115:2924-2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Manning, G., D. B. Whyte, R. Martinez, T. Hunter, and S. Sudarsanam. 2002. The protein kinase complement of the human genome. Science 298:1912-1934. [DOI] [PubMed] [Google Scholar]
- 206.Marais, R., J. Wynne, and R. Treisman. 1993. The SRF accessory protein Elk-1 contains a growth factor-regulated transcriptional activation domain. Cell 73:381-393. [DOI] [PubMed] [Google Scholar]
- 207.Martin, J. H., A. A. Mohit, and C. A. Miller. 1996. Developmental expression in the mouse nervous system of the p493F12 SAP kinase. Brain Res. Mol. Brain Res. 35:47-57. [DOI] [PubMed] [Google Scholar]
- 208.Maundrell, K., B. Antonsson, E. Magnenat, M. Camps, M. Muda, C. Chabert, C. Gillieron, U. Boschert, E. Vial-Knecht, J. C. Martinou, and S. Arkinstall. 1997. Bcl-2 undergoes phosphorylation by c-Jun N-terminal kinase/stress-activated protein kinases in the presence of the constitutively active GTP-binding protein Rac1. J. Biol. Chem. 272:25238-25242. [DOI] [PubMed] [Google Scholar]
- 209.May, G. H., F. Harris, D. Gillespie, and D. M. Black. 1999. The BRCA2 transactivation domain does not interact with JNK. Genes Chromosomes Cancer 25:407-409. [DOI] [PubMed] [Google Scholar]
- 210.May, G. H. W., K. E. Allen, W. Clark, M. Funk, and D. A. F. Gillespie. 1998. Analysis of the interaction between c-Jun and c-Jun N-terminal kinase in vivo. J. Biol. Chem. 273:33429-33435. [DOI] [PubMed] [Google Scholar]
- 211.Mayer, C., H. Bierhoff, and I. Grummt. 2005. The nucleolus as a stress sensor: JNK2 inactivates the transcription factor TIF-IA and down-regulates rRNA synthesis. Genes Dev. 19:933-941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Mayer, C., J. Zhao, X. Yuan, and I. Grummt. 2004. mTOR-dependent activation of the transcription factor TIF-IA links rRNA synthesis to nutrient availability. Genes Dev. 18:423-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.McCahill, A., J. Warwicker, G. B. Bolger, M. D. Houslay, and S. J. Yarwood. 2002. The RACK1 scaffold protein: a dynamic cog in cell response mechanisms. Mol. Pharmacol. 62:1261-1273. [DOI] [PubMed] [Google Scholar]
- 214.McDonald, P. H., C. W. Chow, W. E. Miler, S. A. Laporte, M. E. Field, F. T. Lin, R. J. Davis, and R. J. Lefkowitz. 2000. β-Arrestin 2: a receptor-regulated MAPK scaffold for the activation of JNK3. Science 290:1574-1577. [DOI] [PubMed] [Google Scholar]
- 215.McLean, T. I., and S. L. Bachenheimer. 1999. Activation of c-Jun N-terminal kinase by herpes simplex virus type 1 enhances viral replication. J. Virol. 73:8415-8426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Meylan, E., K. Burns, K. Hofmann, V. Blancheteau, F. Martinon, M. Kelliher, and J. Tschopp. 2004. RIP1 is an essential mediator of Toll-like receptor 3-induced NF-kappa B activation. Nat. Immunol. 5:503-507. [DOI] [PubMed] [Google Scholar]
- 217.Migliaccio, E., M. Giorgio, S. Mele, G. Pelicci, P. Reboldi, P. P. Pandolfi, L. Lanfrancone, and P. G. Pelicci. 1999. The p66shc adaptor protein controls oxidative stress response and life span in mammals. Nature 402:309-313. [DOI] [PubMed] [Google Scholar]
- 218.Miller, W. E., P. H. McDonald, S. F. Cai, M. E. Field, R. J. Davis, and R. J. Lefkowitz. 2001. Identification of a motif in the carboxyl terminus of beta-arrestin2 responsible for activation of JNK3. J. Biol. Chem. 276:27770-27777. [DOI] [PubMed] [Google Scholar]
- 219.Milne, D. M., L. E. Campbell, D. G. Campbell, and D. W. Meek. 1995. p53 is phosphorylated in vitro and in vivo by an ultraviolet radiation-induced protein kinase characteristic of the c-Jun kinase, JNK1. J. Biol. Chem. 270:5511-5518. [DOI] [PubMed] [Google Scholar]
- 220.Milner, J., F. Fuks, L. Hughes-Davies, and T. Kouzarides. 2000. The BRCA2 activation domain associates with and is phosphorylated by a cellular protein kinase. Oncogene 19:4441-4445. [DOI] [PubMed] [Google Scholar]
- 221.Milner, J., B. Ponder, L. Hughes-Davies, M. Seltmann, and T. Kouzarides. 1997. Transcriptional activation functions in BRCA2. Nature 386:772-773. [DOI] [PubMed] [Google Scholar]
- 222.Minden, A., A. Lin, T. Smeal, B. Dérijard, M. Cobb, R. Davis, and M. Karin. 1994. c-Jun N-terminal phosphorylation correlates with activation of the JNK subgroup but not the ERK subgroup of mitogen-activated protein kinases. Mol. Cell. Biol. 14:6683-6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Mizukami, Y., K. Yoshioka, S. Morimoto, and K.-I. Yoshida. 1997. A novel mechanism of JNK1 activation. Nuclear translocation and activation of JNK1 during ischemia and reperfusion. J. Biol. Chem. 272:16657-16662. [DOI] [PubMed] [Google Scholar]
- 224.Mizutani, T., S. Fukushi, M. Saijo, I. Kurane, and S. Morikawa. 2005. JNK and PI3k/Akt signaling pathways are required for establishing persistent SARS-CoV infection in Vero E6 cells. Biochim. Biophys. Acta 1741:4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Mohit, A. A., J. H. Martin, and C. A. Miller. 1995. p493F12 kinase: a novel MAP kinase expressed in a subset of neurons in the human nervous system. Neuron 14:67-78. [DOI] [PubMed] [Google Scholar]
- 226.Molina, D. M., S. Grewal, and L. Bardwell. 2005. Characterization of an ERK-binding domain in microphthalmia-associated transcription factor and differential inhibition of ERK2-mediated substrate phosphorylation. J. Biol. Chem. 280:42051-42060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Morfini, G., G. Pigino, G. Szebenyi, Y. You, S. Pollema, and S. T. Brady. 2006. JNK mediates pathogenic effects of polyglutamine-expanded androgen receptor on fast axonal transport. Nat. Neurosci. 9:907-916. [DOI] [PubMed] [Google Scholar]
- 228.Morii, H., T. Yamada, I. Nakano, J. M. Coulson, and N. Mori. 2006. Site-specific phosphorylation of SCG10 in neuronal plasticity: role of Ser73 phosphorylation by N-methyl d-aspartic acid receptor activation in rat hippocampus. Neurosci. Lett. 396:241-246. [DOI] [PubMed] [Google Scholar]
- 229.Morrison, D. K., and R. J. Davis. 2003. Regulation of MAP kinase signaling modules by scaffold proteins in mammals. Annu. Rev. Cell Dev. Biol. 19:91-118. [DOI] [PubMed] [Google Scholar]
- 230.Morton, S., R. J. Davis, A. McLaren, and P. Cohen. 2003. A reinvestigation of the multisite phosphorylation of the transcription factor c-Jun. EMBO J. 22:3876-3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Moumen, A., P. Masterson, M. J. O'Connor, and S. P. Jackson. 2005. hnRNP K: an HDM2 target and transcriptional coactivator of p53 in response to DNA damage. Cell 123:1065-1078. [DOI] [PubMed] [Google Scholar]
- 232.Murray, J. I., M. L. Whitfield, N. D. Trinklein, R. M. Myers, P. O. Brown, and D. Botstein. 2004. Diverse and specific gene expression responses to stresses in cultured human cells. Mol. Biol. Cell 5:2361-2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Muslin, A. J., J. W. Tanner, P. M. Allen, and A. S. Shaw. 1996. Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell 84:889-897. [DOI] [PubMed] [Google Scholar]
- 234.Muslin, A. J., and H. Xing. 2000. 14-3-3 proteins: regulation of subcellular localization by molecular interference. Cell Signal. 12:703-709. [DOI] [PubMed] [Google Scholar]
- 235.Mussig, K., H. Fiedler, H. Staiger, C. Weigert, R. Lehmann, E. D. Schleicher, and H. U. Haring. 2005. Insulin-induced stimulation of JNK and the PI 3-kinase/mTOR pathway leads to phosphorylation of serine 318 of IRS-1 in C2C12 myotubes. Biochem. Biophys. Res. Commun. 335:819-825. [DOI] [PubMed] [Google Scholar]
- 236.Nagata, K., A. Puls, C. Futter, P. Aspenstrom, E. Schaefer, T. Nakata, N. Hirokawa, and A. Hall. 1998. The MAP kinase kinase kinase MLK2 co-localizes with activated JNK along microtubules and associates with kinesin superfamily motor KIF3. EMBO J. 17:149-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Nakamori, Y., M. Emoto, N. Fukuda, A. Taguchi, S. Okuya, M. Tajiri, M. Miyagishi, K. Taira, Y. Wada, and Y. Tanizawa. 2006. Myosin motor Myo1c and its receptor NEMO/IKKγ promote TNF-α-induced serine307 phosphorylation of IRS-1. J. Cell Biol. 173:665-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Nakatani, Y., H. Kaneto, D. Kawamori, M. Hatazaki, T. Miyatsuka, T. A. Matsuoka, Y. Kajimoto, M. Matsuhisa, Y. Yamasaki, and M. Hori. 2004. Modulation of the JNK pathway in liver affects insulin resistance status. J. Biol. Chem. 279:45803-45809. [DOI] [PubMed] [Google Scholar]
- 239.Nalca, A., and V. M. Rangnekar. 1998. The G1-phase growth-arresting action of interleukin-1 is independent of p53 and p21/WAF1 function. J. Biol. Chem. 273:30517-30523. [DOI] [PubMed] [Google Scholar]
- 240.Nateri, A. S., L. Riera-Sans, C. Da Costa, and A. Behrens. 2004. The ubiquitin ligase SCFFbw7 antagonizes apoptotic JNK signaling. Science 303:1374-1378. [DOI] [PubMed] [Google Scholar]
- 241.Nateri, A. S., B. Spencer-Dene, and A. Behrens. 2005. Interaction of phosphorylated c-Jun with TCF4 regulates intestinal cancer development. Nature 437:281-285. [DOI] [PubMed] [Google Scholar]
- 242.Neidhart, S., B. Antonsson, C. Gillieron, F. Vilbois, G. Grenningloh, and S. Arkinstall. 2001. c-Jun N-terminal kinase-3 (JNK3)/stress-activated protein kinase-beta (SAPKbeta) binds and phosphorylates the neuronal microtubule regulator SCG10. FEBS Lett. 508:259-264. [DOI] [PubMed] [Google Scholar]
- 243.N′Guessan, P. D., B. Schmeck, A. Ayim, A. C. Hocke, B. Brell, S. Hammerschmidt, S. Rosseau, N. Suttorp, and S. Hippenstiel. 2005. Streptococcus pneumoniae R6x induced p38 MAPK and JNK-mediated caspase-dependent apoptosis in human endothelial cells. Thromb. Haemost. 94:295-303. [DOI] [PubMed] [Google Scholar]
- 244.Nguyen, M. T., H. Satoh, S. Favelyukis, J. L. Babendure, T. Imamura, J. I. Sbodio, J. Zalevsky, B. I. Dahiyat, N. W. Chi, and J. M. Olefsky. 2005. JNK and tumor necrosis factor-α mediate free fatty acid-induced insulin resistance in 3T3-L1 adipocytes. J. Biol. Chem. 280:35361-35371. [DOI] [PubMed] [Google Scholar]
- 245.Nihalani, D., H. N. Wong, and L. B. Holzman. 2003. Recruitment of JNK to JIP1 and JNK-dependent JIP1 phosphorylation regulates JNK module dynamics and activation. J. Biol. Chem. 278:28694-28702. [DOI] [PubMed] [Google Scholar]
- 246.Noguchi, K., C. Kitanaka, H. Yamana, A. Kokubu, T. Mochizuki, and Y. Kuchino. 1999. Regulation of c-Myc through phosphorylation at Ser-62 and Ser-71 by c-Jun N-terminal kinase. J. Biol. Chem. 274:32580-32587. [DOI] [PubMed] [Google Scholar]
- 247.Oh, S. W., A. Mukhopadhyay, N. Svrzikapa, F. Jiang, R. J. Davis, and H. A. Tissenbaum. 2005. JNK regulates lifespan in Caenorhabditis elegans by modulating nuclear translocation of forkhead transcription factor/DAF-16. Proc. Natl. Acad. Sci. USA 102:4494-4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Ortega-Perez, I., E. Cano, F. Were, M. Villar, J. Vazquez, and J. M. Redondo. 2005. c-Jun N-terminal kinase (JNK) positively regulates NFATc2 transactivation through phosphorylation within the N-terminal regulatory domain. J. Biol. Chem. 280:20867-20878. [DOI] [PubMed] [Google Scholar]
- 249.Ozanne, B. W., L. McGarry, H. J. Spence, I. Johnston, J. Winnie, L. Meagher, and G. Stapleton. 2000. Transcriptional regulation of cell invasion: AP-1 regulation of a multigenic invasion programme. Eur. J. Cancer 36:1640-1648. [DOI] [PubMed] [Google Scholar]
- 250.Ozes, O. N., H. Akca, L. D. Mayo, J. A. Gustin, T. Maehama, J. E. Dixon, and D. B. Donner. 2001. A phosphatidylinositol 3-kinase/Akt/mTOR pathway mediates and PTEN antagonizes tumor necrosis factor inhibition of insulin signaling through insulin receptor substrate-1. Proc. Natl. Acad. Sci. USA 98:4640-4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Pan, H., J. Xie, F. Ye, and S. J. Gao. 2006. Modulation of Kaposi's sarcoma-associated herpesvirus infection and replication by MEK/ERK, JNK, and p38 multiple mitogen-activated protein kinase pathways during primary infection. J. Virol. 80:5371-5382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Park, H. S., M. S. Kim, S. H. Huh, J. Park, J. Chung, S. S. Kang, and E. J. Choi. 2002. Akt (protein kinase B) negatively regulates SEK1 by means of protein phosphorylation. J. Biol. Chem. 277:2573-2578. [DOI] [PubMed] [Google Scholar]
- 253.Park, K. J., S. H. Choi, M. S. Koh, D. J. Kim, S. W. Yie, S. Y. Lee, and S. B. Hwang. 2001. Hepatitis C virus core protein potentiates c-Jun N-terminal kinase activation through a signaling complex involving TRADD and TRAF2. Virus Res. 74:89-98. [DOI] [PubMed] [Google Scholar]
- 254.Pendergast, A. M. 2005. Stress and death: breaking up the c-Abl/14-3-3 complex in apoptosis. Nat. Cell Biol. 7:213-214. [DOI] [PubMed] [Google Scholar]
- 255.Pfitzner, E., S. Kliem, D. Baus, and C. M. Litterst. 2004. The role of STATs in inflammation and inflammatory diseases. Curr. Pharm. Des. 10:2839-2850. [DOI] [PubMed] [Google Scholar]
- 256.Pickart, C. M. 2001. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70:503-533. [DOI] [PubMed] [Google Scholar]
- 257.Pulverer, B. J., J. M. Kyriakis, J. Avruch, E. Nikolakaki, and J. R. Woodgett. 1991. Phosphorylation of c-Jun mediated by MAP kinases. Nature 353:670-673. [DOI] [PubMed] [Google Scholar]
- 258.Putcha, G. V., S. Le, S. Frank, C. G. Besirli, K. Clark, B. Chu, S. Alix, R. J. Youle, A. LaMarche, A. C. Maroney, and E. M. Johnson. 2003. JNK-mediated BIM phosphorylation potentiates BAX-dependent apoptosis. Neuron 38:899-914. [DOI] [PubMed] [Google Scholar]
- 259.Qin, J., J. Yao, G. Cui, H. Xiao, T. W. Kim, J. Fraczek, P. Wightman, S. Sato, S. Akira, A. Puel, J. L. Casanova, B. Su, and X. Li. 2006. TLR8-mediated NFkappa B and JNK activation are TAK1-independent and MEKK3-dependent. J. Biol. Chem. 281:21013-21021. [DOI] [PubMed] [Google Scholar]
- 260.Rahaus, M., N. Desloges, and M. H. Wolff. 2004. Replication of varicella-zoster virus is influenced by the levels of JNK/SAPK and p38/MAPK activation. J. Gen. Virol. 85:3529-3540. [DOI] [PubMed] [Google Scholar]
- 261.Rahaus, M., N. Desloges, and M. H. Wolff. 2005. ORF61 protein of varicella-zoster virus influences JNK/SAPK and p38/MAPK phosphorylation. J. Med. Virol. 76:424-433. [DOI] [PubMed] [Google Scholar]
- 262.Ravichandran, K. S. 2001. Signaling via Shc family adapter proteins. Oncogene 20:6322-6330. [DOI] [PubMed] [Google Scholar]
- 263.Ricci, R., U. Eriksson, G. Y. Oudit, R. Eferl, A. Akhmedov, I. Sumara, G. Sumara, Z. Kassiri, J. P. David, L. Bakiri, B. Sasse, M. H. Idarraga, M. Rath, D. Kurz, H. C. Theussl, J. C. Perriard, P. Backx, J. M. Penninger, and E. F. Wagner. 2005. Distinct functions of junD in cardiac hypertrophy and heart failure. Genes Dev. 19:208-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Riedel, H. 2004. Grb10 exceeding the boundaries of a common signaling adapter. Front. Biosci. 9:603-618. [DOI] [PubMed] [Google Scholar]
- 265.Rincon, M., B. Derijard, C. W. Chow, R. J. Davis, and R. A. Flavell. 1997. Reprogramming the signalling requirement for AP-1 (activator protein-1) activation during differentiation of precursor CD4+ T-cells into effector Th1 and Th2 cells. Genes Funct. 1:51-68. [DOI] [PubMed] [Google Scholar]
- 266.Rogatsky, I., S. K. Logan, and M. J. Garabedian. 1998. Antagonism of glucocorticoid receptor transcriptional activation by the c-Jun N-terminal kinase. Proc. Natl. Acad. Sci. USA 95:2050-2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Romer, L. H., K. G. Birukov, and J. G. Garcia. 2006. Focal adhesions: paradigm for a signaling nexus. Circ. Res. 98:606-616. [DOI] [PubMed] [Google Scholar]
- 268.Roux, P. P., and J. Blenis. 2004. ERK and p38 MAPK-activated protein kinases: a family of protein kinases with diverse biological functions. Microbiol. Mol. Biol. Rev. 68:320-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Sano, Y., H. Akimaru, T. Okamura, T. Nagao, M. Okada, and S. Ishii. 2005. Drosophila activating transcription factor-2 is involved in stress response via activation by p38, but not c-Jun NH2-terminal kinase. Mol. Biol. Cell 16:2934-2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Sarbassov, D. D., D. A. Guertin, S. M. Ali, and D. M. Sabatini. 2005. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 307:1098-1101. [DOI] [PubMed] [Google Scholar]
- 271.Sarmay, G., A. Angyal, A. Kertesz, M. Maus, and D. Medgyesi. 2006. The multiple function of Grb2 associated binder (Gab) adaptor/scaffolding protein in immune cell signaling. Immunol. Lett. 104:76-82. [DOI] [PubMed] [Google Scholar]
- 272.Sato, S., H. Sanjo, K. Takeda, J. Ninomiya-Tsuji, M. Yamamoto, T. Kawai, K. Matsumoto, O. Takeuchi, and S. Akira. 2005. Essential function for the kinase TAK1 in innate and adaptive immune responses. Nat. Immunol. 6:1087-1095. [DOI] [PubMed] [Google Scholar]
- 273.Scapin, G., S. B. Patel, J. Lisnock, J. W. Becker, and P. V. LoGrasso. 2003. The structure of JNK3 in complex with small molecule inhibitors: structural basis for potency and selectivity. Chem. Biol. 10:705-712. [DOI] [PubMed] [Google Scholar]
- 274.Scheinfeld, M. H., E. Ghersi, P. Davies, and L. D'Adamio. 2003. Amyloid beta protein precursor is phosphorylated by JNK-1 independent of, yet facilitated by, JNK-interacting protein (JIP)-1. J. Biol. Chem. 278:42058-42063. [DOI] [PubMed] [Google Scholar]
- 275.Shao, Z., K. Bhattacharya, E. Hsich, L. Park, B. Walters, U. Germann, Y. M. Wang, J. Kyriakis, R. Mohanlal, K. Kuida, M. Namchuk, F. Salituro, Y. M. Yao, W. M. Hou, X. Chen, M. Aronovitz, P. N. Tsichlis, S. Bhattacharya, T. Force, and H. Kilter. 2006. c-Jun N-terminal kinases mediate reactivation of Akt and cardiomyocyte survival after hypoxic injury in vitro and in vivo. Circ. Res. 98:111-118. [DOI] [PubMed] [Google Scholar]
- 276.Sharrocks, A. D., S. H. Yang, and A. Galanis. 2000. Docking domains and substrate-specificity determination for MAP kinases. Trends Biochem. Sci. 25:448-453. [DOI] [PubMed] [Google Scholar]
- 277.Shattil, S. J., and P. J. Newman. 2004. Integrins: dynamic scaffolds for adhesion and signaling in platelets. Blood 104:1606-1615. [DOI] [PubMed] [Google Scholar]
- 278.Sluss, H. K., Z. Han, T. Barrett, R. J. Davis, and Y. T. Ip. 1996. A JNK signal transduction pathway that mediates morphogenesis and an immune response in Drosophila. Genes Dev. 10:2745-2758. [DOI] [PubMed] [Google Scholar]
- 279.Smith, W. E., A. V. Kane, S. T. Campbell, D. W. Acheson, B. H. Cochran, and C. M. Thorpe. 2003. Shiga toxin 1 triggers a ribotoxic stress response leading to p38 and JNK activation and induction of apoptosis in intestinal epithelial cells. Infect. Immun. 71:1497-1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Song, J. J., and Y. J. Lee. 2005. Dissociation of Akt1 from its negative regulator JIP1 is mediated through the ASK1-MEK-JNK signal transduction pathway during metabolic oxidative stress: a negative feedback loop. J. Cell Biol. 170:61-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Song, J. J., and Y. J. Lee. 2005. Cross-talk between JIP3 and JIP1 during glucose deprivation: SEK1-JNK2 and Akt1 act as mediators. J. Biol. Chem. 280:26845-26855. [DOI] [PubMed] [Google Scholar]
- 282.Song, X., D. Raman, E. V. Gurevich, S. A. Vishnivetskiy, and V. V. Gurevich. 2006. Visual and both non-visual arrestins in their “inactive” conformation bind JNK3 and MDM2 and relocalize them from the nucleus to the cytoplasm. J. Biol. Chem. 281:21491-21499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Srinivas, H., D. M. Juroske, S. Kalyankrishna, D. D. Cody, R. E. Price, X. C. Xu, R. Narayanan, N. L. Weigel, and J. M. Kurie. 2005. c-Jun N-terminal kinase contributes to aberrant retinoid signaling in lung cancer cells by phosphorylating and inducing proteasomal degradation of retinoic acid receptor alpha. Mol. Cell. Biol. 25:1054-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Stephanou, A. 2004. Role of STAT-1 and STAT-3 in ischaemia/reperfusion injury. J. Cell Mol. Med. 8:519-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Sugita, M., H. Sugita, and M. Kaneki. 2004. Increased insulin receptor substrate 1 serine phosphorylation and stress-activated protein kinase/c-Jun N-terminal kinase activation associated with vascular insulin resistance in spontaneously hypertensive rats. Hypertension 44:484-489. [DOI] [PubMed] [Google Scholar]
- 286.Sunayama, J., F. Tsuruta, N. Masuyama, and Y. Gotoh. 2005. JNK antagonizes Akt-mediated survival signals by phosphorylating 14-3-3. J. Cell Biol. 170:295-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Sunters, A., P. A. Madureira, K. M. Pomeranz, M. Aubert, J. J. Brosens, S. J. Cook, B. M. Burgering, R. C. Coombes, and E. W. Lam. 2006. Paclitaxel-induced nuclear translocation of FOXO3a in breast cancer cells is mediated by c-Jun NH2-terminal kinase and Akt. Cancer Res. 66:212-220. [DOI] [PubMed] [Google Scholar]
- 288.Swanson, K. D., L. K. Taylor, L. Haung, A. L. Burlingame, and G. E. Landreth. 1999. Transcription factor phosphorylation by pp90rsk2. Identification of Fos kinase and NGFI-B kinase I as pp90rsk2. J. Biol. Chem. 274:3385-3395. [DOI] [PubMed] [Google Scholar]
- 289.Szatmary, Z., M. J. Garabedian, and J. Vilcek. 2004. Inhibition of glucocorticoid receptor-mediated transcriptional activation by p38 mitogen-activated protein (MAP) kinase. J. Biol. Chem. 279:43708-43715. [DOI] [PubMed] [Google Scholar]
- 290.Szczepankiewicz, B. G., C. Kosogof, L. T. Nelson, G. Liu, B. Liu, H. Zhao, M. D. Serby, Z. Xin, M. Liu, R. J. Gum, D. L. Haasch, S. Wang, J. E. Clampit, E. F. Johnson, T. H. Lubben, M. A. Stashko, E. T. Olejniczak, C. Sun, S. A. Dorwin, K. Haskins, C. Abad-Zapatero, E. H. Fry, C. W. Hutchins, H. L. Sham, C. M. Rondinone, and J. M. Trevillyan. 2006. Aminopyridine-based c-Jun N-terminal kinase inhibitors with cellular activity and minimal cross-kinase activity. J. Med. Chem. 49:3563-3580. [DOI] [PubMed] [Google Scholar]
- 291.Tafolla, E., S. Wang, B. Wong, J. Leong, and Y. L. Kapila. 2005. JNK1 and JNK2 oppositely regulate p53 in signaling linked to apoptosis triggered by an altered fibronectin matrix: JNK links FAK and p53. J. Biol. Chem. 280:19992-19999. [DOI] [PubMed] [Google Scholar]
- 292.Takino, T., M. Nakada, H. Miyamori, Y. Watanabe, T. Sato, D. Gantulga, K. Yoshioka, K. M. Yamada, and H. Sato. 2005. JSAP1/JIP3 cooperates with focal adhesion kinase to regulate c-Jun N-terminal kinase and cell migration. J. Biol. Chem. 280:37772-37781. [DOI] [PubMed] [Google Scholar]
- 293.Tamanini, A., R. Rolfini, E. Nicolis, P. Melotti, and G. Cabrini. 2003. MAP kinases and NF-kappaB collaborate to induce ICAM-1 gene expression in the early phase of adenovirus infection. Virology 307:228-242. [DOI] [PubMed] [Google Scholar]
- 294.Tanaka, Y., F. Kanai, T. Ichimura, K. Tateishi, Y. Asaoka, B. Guleng, A. Jazag, M. Ohta, J. Imamura, T. Ikenoue, H. Ijichi, T. Kawabe, T. Isobe, and M. Omata. 2006. The hepatitis B virus X protein enhances AP-1 activation through interaction with Jab1. Oncogene 25:633-642. [DOI] [PubMed] [Google Scholar]
- 295.Tanoue, T., M. Adachi, T. Moriguchi, and E. Nishida. 2000. A conserved docking motif in MAP kinases common to substrates, activators and regulators. Nat. Cell Biol. 2:110-116. [DOI] [PubMed] [Google Scholar]
- 296.Tapon, N., K.-I. Nagata, N. Lamarche, and A. Hall. 1998. A new Rac target POSH is an SH3-containing scaffold protein involved in the JNK and NF-kB signalling pathways. EMBO J. 17:1395-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Tararuk, T., N. Ostman, W. Li, B. Bjorkblom, A. Padzik, J. Zdrojewska, V. Hongisto, T. Herdegen, W. Konopka, M. J. Courtney, and E. T. Coffey. 2006. JNK1 phosphorylation of SCG10 determines microtubule dynamics and axodendritic length. J. Cell Biol. 173:265-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Tarrade, A., J. Bastien, N. Bruck, A. Bauer, M. Gianni, and C. Rochette-Egly. 2005. Retinoic acid and arsenic trioxide cooperate for apoptosis through phosphorylated RXR alpha. Oncogene 24:2277-2288. [DOI] [PubMed] [Google Scholar]
- 299.Taylor, K. R., A. K. Holzer, J. F. Bazan, C. A. Walsh, and J. G. Gleeson. 2000. Patient mutations in doublecortin define a repeated tubulin-binding domain. J. Biol. Chem. 275:34442-34450. [DOI] [PubMed] [Google Scholar]
- 300.Traverse, S., and P. Cohen. 1994. Identification of a latent MAP kinase kinase kinase in PC12 cells as B-raf. FEBS Lett. 350:13-18. [DOI] [PubMed] [Google Scholar]
- 301.Tsuruta, F., J. Sunayama, Y. Mori, S. Hattori, S. Shimizu, Y. Tsujimoto, K. Yoshioka, N. Masuyama, and Y. Gotoh. 2004. JNK promotes Bax translocation to mitochondria through phosphorylation of 14-3-3 proteins. EMBO J. 23:1889-1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Turkson, J. 2004. STAT proteins as novel targets for cancer drug discovery. Expert Opin. Ther. Targets 8:409-422. [DOI] [PubMed] [Google Scholar]
- 303.Tzivion, G., and J. Avruch. 2002. 14-3-3 proteins: active cofactors in cellular regulation by serine/threonine phosphorylation. J. Biol. Chem. 277:3061-3064. [DOI] [PubMed] [Google Scholar]
- 304.Van Dam, H., D. Wilhelm, I. Herr, A. Steffen, P. Herrlich, and P. Angel. 1995. ATF-2 is preferentially activated by stress-activated protein kinases to mediate c-Jun induction in response to genotoxic agents. EMBO J. 14:1798-1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Ventura, J. J., A. Hubner, C. Zhang, R. A. Flavell, K. M. Shokat, and R. J. Davis. 2006. Chemical genetic analysis of the time course of signal transduction by JNK. Mol. Cell 21:701-710. [DOI] [PubMed] [Google Scholar]
- 306.Ventura, J. J., N. J. Kennedy, J. A. Lamb, R. A. Flavell, and R. J. Davis. 2003. c-Jun NH2-terminal kinase is essential for the regulation of AP-1 by tumor necrosis factor. Mol. Cell. Biol. 23:2871-2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Verhey, K. J., D. Meyer, R. Deehan, J. Blenis, B. J. Schnapp, T. A. Rapoport, and B. Margolis. 2001. Cargo of kinesin identified as JIP scaffolding proteins and associated signaling molecules. J. Cell Biol. 152:959-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Vicente-Manzanares, M., D. J. Webb, and A. R. Horwitz. 2005. Cell migration at a glance. J. Cell Sci. 118:4917-4919. [DOI] [PubMed] [Google Scholar]
- 309.Vinciguerra, M., A. Vivacqua, G. Fasanella, A. Gallo, C. Cuozzo, A. Morano, M. Maggiolini, and A. M. Musti. 2004. Differential phosphorylation of c-Jun and JunD in response to the epidermal growth factor is determined by the structure of MAPK targeting sequences. J. Biol. Chem. 279:9634-9641. [DOI] [PubMed] [Google Scholar]
- 310.Vondriska, T. M., J. M. Pass, and P. Ping. 2004. Scaffold proteins and assembly of multiprotein signaling complexes. J. Mol. Cell Cardiol. 37:391-397. [DOI] [PubMed] [Google Scholar]
- 311.Wada, T., N. Joza, H. Y. Cheng, T. Sasaki, I. Kozieradzki, K. Bachmaier, T. Katada, M. Schreiber, E. F. Wagner, H. Nishina, and J. M. Penninger. 2004. MKK7 couples stress signalling to G2/M cell-cycle progression and cellular senescence. Nat. Cell Biol. 6:216-226. [DOI] [PubMed] [Google Scholar]
- 312.Wan, J., L. Sun, J. W. Mendoza, Y. L. Chui, D. P. Huang, Z. J. Chen, N. Suzuki, S. Suzuki, W. C. Yeh, S. Akira, K. Matsumoto, Z. G. Liu, and Z. Wu. 2004. Elucidation of the c-Jun N-terminal kinase pathway mediated by Epstein-Barr virus-encoded latent membrane protein 1. Mol. Cell. Biol. 24:192-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Wang, M. C., D. Bohmann, and H. Jasper. 2005. JNK extends life span and limits growth by antagonizing cellular and organism-wide responses to insulin signaling. Cell 121:115-125. [DOI] [PubMed] [Google Scholar]
- 314.Ward, P., L. Equinet, J. Packer, and C. Doerig. 2004. Protein kinases of the human malaria parasite Plasmodium falciparum: the kinome of a divergent eukaryote. BMC Genomics 5:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Wei, W., J. Jin, S. Schlisio, J. W. Harper, and W. G. Kaelin. 2005. The v-Jun point mutation allows c-Jun to escape GSK3-dependent recognition and destruction by the Fbw7 ubiquitin ligase. Cancer Cell 8:25-33. [DOI] [PubMed] [Google Scholar]
- 316.Weiss, C., S. Schneider, E. F. Wagner, X. Zhang, E. Seto, and D. Bohmann. 2003. JNK phosphorylation relieves HDAC3-dependent suppression of the transcriptional activity of c-Jun. EMBO J. 22:3686-3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Wellen, K. E., and G. S. Hotamisligil. 2005. Inflammation, stress, and diabetes. J. Clin. Investig. 115:1111-1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Werner, E. D., J. Lee, L. Hansen, M. Yuan, and S. E. Shoelson. 2004. Insulin resistance due to phosphorylation of insulin receptor substrate-1 at serine 302. J. Biol. Chem. 279:35298-35305. [DOI] [PubMed] [Google Scholar]
- 319.Wertz, I. E., K. M. O'Rourke, Z. Zhang, D. Dornan, D. Arnott, R. J. Deshaies, and V. M. Dixit. 2004. Human de-etiolated-1 regulates c-Jun by assembling a CUL4A ubiquitin ligase. Science 303:1371-1374. [DOI] [PubMed] [Google Scholar]
- 320.White, M. F. 1998. The IRS-signalling system: a network of docking proteins that mediate insulin action. Mol. Cell Biochem. 182:3-11. [PubMed] [Google Scholar]
- 321.Whitmarsh, A. J., P. Shore, A. D. Sharrocks, and R. J. Davis. 1995. Integration of MAP kinase signal transduction pathways at the serum response element. Science 269:403-407. [DOI] [PubMed] [Google Scholar]
- 322.Wiltshire, C., M. Matsushita, S. Tsukada, D. A. F. Gillespie, and G. H. W. May. 2002. A new c-Jun N-terminal kinase (JNK)-interacting protein, Sab (SH3BP5) associates with mitochondria. Biochem. J. 367:577-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Wong, H. K., M. Fricker, A. Wyttenbach, A. Villunger, E. M. Michalak, A. Strasser, and A. M. Tolkovsky. 2005. Mutually exclusive subsets of BH3-only proteins are activated by the p53 and c-Jun N-terminal kinase/c-Jun signaling pathways during cortical neuron apoptosis induced by arsenite. Mol. Cell. Biol. 25:8732-8747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Xia, Y., C. Makris, B. Su, E. Li, J. Yang, G. R. Nemerow, and M. Karin. 2000. MEK kinase 1 is critically required for c-Jun N-terminal kinase activation by proinflammatory stimuli and growth factor-induced cell migration. Proc. Natl. Acad. Sci. USA 97:5243-5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Xiao, J., and J. Chodosh. 2005. JNK regulates MCP-1 expression in adenovirus type 19-infected human corneal fibroblasts. Investig. Ophthalmol. Vis. Sci. 46:3777-3782. [DOI] [PubMed] [Google Scholar]
- 326.Xie, J., H. Pan, S. Yoo, and S. J. Gao. 2005. Kaposi's sarcoma-associated herpesvirus induction of AP-1 and interleukin 6 during primary infection mediated by multiple mitogen-activated protein kinase pathways. J. Virol. 79:15027-15037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Xie, X., Y. Gu, T. Fox, J. T. Coll, M. A. Fleming, W. Markland, P. R. Caron, K. P. Wilson, and M. S. Su. 1998. Crystal structure of JNK3: a kinase implicated in neuronal apoptosis. Structure 6:983-991. [DOI] [PubMed] [Google Scholar]
- 328.Xu, Z., N. V. Kukekov, and L. A. Greene. 2003. POSH acts as a scaffold for a multiprotein complex that mediates JNK activation in apoptosis. EMBO J. 22:252-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Yaffe, M. B., G. G. Leparc, J. Lai, T. Obata, S. Volinia, and L. C. Cantley. 2001. A motif-based profile scanning approach for genome-wide prediction of signaling pathways. Nat. Biotechnol. 19:348-353. [DOI] [PubMed] [Google Scholar]
- 330.Yamadori, T., Y. Baba, M. Matsushita, S. Hashimoto, M. Kurosaki, T. Kurosaki, T. Kishimoto, and S. Tsukada. 1999. Bruton's tyrosine kinase activity is negatively regulated by Sab, the Btk-SH3 domain-binding protein. Proc. Natl. Acad. Sci. USA 96:6341-6346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Yamamoto, K., H. Ichijo, and S. J. Korsmeyer. 1999. BCL-2 is phosphorylated and inactivated by an ASK1/Jun N-terminal protein kinase pathway normally activated at G2/M. Mol. Cell. Biol. 19:8469-8478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Yang, C., W. Zhou, M. S. Jeon, D. Demydenko, Y. Harada, H. Zhou, and Y. C. Liu. 2006. Negative regulation of the E3 ubiquitin ligase Itch via Fyn-mediated tyrosine phosphorylation. Mol. Cell 21:135-141. [DOI] [PubMed] [Google Scholar]
- 333.Yang, C. P., and S. B. Horwitz. 2002. Distinct mechanisms of Taxol-induced serine phosphorylation of the 66-kDa Shc isoform in A549 and RAW 264.7 cells. Biochim. Biophys. Acta 1590:76-83. [DOI] [PubMed] [Google Scholar]
- 334.Yang, D. D., D. Conze, A. J. Whitmarsh, T. Barrett, R. J. Davis, M. Rincon, and R. A. Flavell. 1998. Differentiation of CD4+ cells to Th1 cells requires MAP kinase JNK2. Immunity 9:575-585. [DOI] [PubMed] [Google Scholar]
- 335.Yang, J., P. Cron, V. M. Good, V. Thompson, B. A. Hemmings, and D. Barford. 2002. Crystal structure of an activated Akt/protein kinase B ternary complex with GSK3-peptide and AMP-PNP. Nat. Struct. Biol. 9:940-944. [DOI] [PubMed] [Google Scholar]
- 336.Yang, S.-H., P. R. Yates, A. J. Whitmarsh, R. J. Davis, and A. D. Sharrocks. 1998. The Elk-1 ETS-domain transcription factor contains a mitogen-activated protein kinase targeting motif. Mol. Cell. Biol. 18:710-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Yang, S. H., A. J. Whitmarsh, R. J. Davis, and A. D. Sharrocks. 1998. Differential targeting of MAP kinases to the ETS-domain transcription factor Elk-1. EMBO J. 17:1740-1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Yasuda, J., A. J. Whitmarsh, J. Cavanagh, M. Sharma, and R. J. Davis. 1999. The JIP group of mitogen-activated protein kinase scaffold proteins. Mol. Cell. Biol. 19:7245-7254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Yazgan, O., and C. M. Pfarr. 2002. Regulation of two JunD isoforms by Jun N-terminal kinases. J. Biol. Chem. 277:29710-29718. [DOI] [PubMed] [Google Scholar]
- 340.Yoshida, H., C. J. Hastie, H. McLauchlan, P. Cohen, and M. Goedert. 2004. Phosphorylation of microtubule-associated protein tau by isoforms of c-Jun N-terminal kinase (JNK). J. Neurochem. 90:352-358. [DOI] [PubMed] [Google Scholar]
- 341.Yoshida, K., and Y. Miki. 2005. Enabling death by the Abl tyrosine kinase: mechanisms for nuclear shuttling of c-Abl in response to DNA damage. Cell Cycle 4:777-779. [DOI] [PubMed] [Google Scholar]
- 342.Yoshida, K., T. Yamaguchi, T. Natsume, D. Kufe, and Y. Miki. 2005. JNK phosphorylation of 14-3-3 proteins regulates nuclear targeting of c-Abl in the apoptotic response to DNA damage. Nat. Cell Biol. 7:278-285. [DOI] [PubMed] [Google Scholar]
- 343.Yoshimura, K., H. Aoki, Y. Ikeda, K. Fujii, N. Akiyama, A. Furutani, Y. Hoshii, N. Tanaka, R. Ricci, T. Ishihara, K. Esato, K. Hamano, and M. Matsuzaki. 2005. Regression of abdominal aortic aneurysm by inhibition of c-Jun N-terminal kinase. Nat. Med. 11:1330-1338. [DOI] [PubMed] [Google Scholar]
- 344.Yu, C., Y. Minemoto, J. Zhang, J. Liu, F. Tang, T. N. Bui, J. Xiang, and A. Lin. 2004. JNK suppresses apoptosis via phosphorylation of the proapoptotic Bcl-2 family protein BAD. Mol. Cell 13:329-340. [DOI] [PubMed] [Google Scholar]
- 345.Yujiri, T., M. Ware, C. Widmann, R. Oyer, D. Russell, E. Chan, Y. Zaitsu, P. Clarke, K. Tyler, Y. Oka, G. R. Fanger, P. Henson, and G. L. Johnson. 2000. MEK kinase1 gene disruption alters cell migration and c-Jun NH2-terminal kinase regulation but does not cause a measurable defect in NF-kB activation. Proc. Natl. Acad. Sci. USA 97:7272-7277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Zachos, G., B. Clements, and J. Conner. 1999. Herpes simplex virus type 1 infection stimulates p38/c-Jun N-terminal mitogen-activated protein kinase pathways and activates transcription factor AP-1. J. Biol. Chem. 274:5097-5103. [DOI] [PubMed] [Google Scholar]
- 347.Zhang, J., J. Liu, C. Yu, and A. Lin. 2005. BAD Ser128 is not phosphorylated by c-Jun NH2-terminal kinase for promoting apoptosis. Cancer Res. 65:8372-8378. [DOI] [PubMed] [Google Scholar]
- 348.Zhang, Q., E. A. Carter, B. Y. Ma, M. White, A. J. Fischman, and R. G. Tompkins. 2005. Molecular mechanism(s) of burn-induced insulin resistance in murine skeletal muscle: role of IRS phosphorylation. Life Sci. 77:3068-3077. [DOI] [PubMed] [Google Scholar]
- 349.Zhang, Y., G. Liu, and Z. Dong. 2001. MSK1 and JNKs mediate phosphorylation of STAT3 in UVA-irradiated mouse epidermal JB6 cells. J. Biol. Chem. 276:42534-42542. [DOI] [PubMed] [Google Scholar]
- 350.Zhang, Y., S. Zhong, Z. Dong, N. Chen, A. M. Bode, W. Ma, and Z. Dong. 2001. UVA induces Ser381 phosphorylation of p90RSK/MAPKAP-K1 via ERK and JNK pathways. J. Biol. Chem. 276:14572-14580. [DOI] [PubMed] [Google Scholar]
- 351.Zhao, J., X. Yuan, M. Frodin, and I. Grummt. 2003. ERK-dependent phosphorylation of the transcription initiation factor TIF-IA is required for RNA polymerase I transcription and cell growth. Mol. Cell 11:405-413. [DOI] [PubMed] [Google Scholar]
- 352.Zhao, K. W., D. Li, Q. Zhao, Y. Huang, R. H. Silverman, P. J. Sims, and G. Q. Chen. 2005. Interferon-α-induced expression of phospholipid scramblase 1 through STAT1 requires the sequential activation of protein kinase Cδ and JNK. J. Biol. Chem. 280:42707-42714. [DOI] [PubMed] [Google Scholar]
- 353.Zhu, G., Y. Liu, and S. Shaw. 2005. Protein kinase specificity. A strategic collaboration between kinase peptide specificity and substrate recruitment. Cell Cycle 4:52-56. [DOI] [PubMed] [Google Scholar]