Abstract
Quorum sensing is widely recognized as an efficient mechanism to regulate expression of specific genes responsible for communal behavior in bacteria. Several bacterial phenotypes essential for the successful establishment of symbiotic, pathogenic, or commensal relationships with eukaryotic hosts, including motility, exopolysaccharide production, biofilm formation, and toxin production, are often regulated by quorum sensing. Interestingly, eukaryotes produce quorum-sensing-interfering (QSI) compounds that have a positive or negative influence on the bacterial signaling network. This eukaryotic interference could result in further fine-tuning of bacterial quorum sensing. Furthermore, recent work involving the synthesis of structural homologs to the various quorum-sensing signal molecules has resulted in the development of additional QSI compounds that could be used to control pathogenic bacteria. The creation of transgenic plants that express bacterial quorum-sensing genes is yet another strategy to interfere with bacterial behavior. Further investigation on the manipulation of quorum-sensing systems could provide us with powerful tools against harmful bacteria.
INTRODUCTION
Quorum sensing is widely employed by a variety of gram-positive and gram-negative bacterial species to coordinate communal behavior. It usually involves the regulation of specific genes in response to population density. This coordinated gene expression is achieved by the production, release, and detection of small signal molecules called autoinducers. At low population densities, basal-level expression of an autoinducer synthase gene results in the production of small amounts of autoinducer signal molecules that diffuse out of the cell and are immediately diluted in the surrounding environment. An increase in bacterial population results in the gradual accumulation of autoinducers in and around the cells. The autoinducer specifically activates a transcriptional regulator protein by binding to it. Activated regulators then interact with target DNA sequences and enhance or block the transcription of quorum-sensing-regulated genes, resulting in the synchronous activation of certain phenotypes in a bacterial population (Fig. 1) (41, 44, 109).
FIG. 1.
Schematic representation of bacterial quorum sensing. At low population densities, basal-level production of autoinducer molecules results in the rapid dilution of the autoinducer signals in the surrounding environment. At high population densities, an increase in bacterial number results in accumulation of autoinducers beyond a threshold concentration, leading to the activation of the response regulator proteins, which in turn initiate the quorum-sensing cascade.
Bacteria use quorum sensing to regulate a variety of phenotypes, such as biofilm formation, toxin production, exopolysaccharide production, virulence factor production, and motility, which are essential for the successful establishment of a symbiotic or pathogenic relationship with their respective eukaryotic hosts (83, 101, 111, 118, 134). According to a previous report, quorum sensing is more common in plant-associated Pseudomonas spp. than in free-living soil Pseudomonas spp. (30). This observation suggests that quorum sensing is important in bacterial relationships with eukaryotes.
Molecular cross talk between bacteria and eukaryotes has been described for a variety of symbiotic or pathogenic relationships (27, 75, 129, 143). Recent research has revealed that eukaryotes are capable of interfering with bacterial communication by the production of molecular signals that interact with the bacterial quorum-sensing system (54, 81, 141, 155). Such quorum-sensing-interfering (QSI) compounds have been intensely investigated for their potential as microbial control agents. This review aims to discuss several natural, synthetic and genetic methods of manipulating bacterial quorum sensing. In addition, we summarize information about the various components of the bacterial quorum sensing system, which could be potential targets for modeling QSI compounds.
Quorum Sensing in Gram-Negative Bacteria
Quorum sensing was first described for the luminous marine bacterium Photobacterium fischeri (Vibrio fischeri). In 1970, Kenneth H. Nealson and John W. Hastings of Harvard University observed that these bacteria do not luminesce until they reach a high population density. Based on this observation, they hypothesized that bioluminescence in this organism was probably regulated by molecular messengers that traveled between cells. They called these messengers “autoinducers” to refer to the fact that the autoinduction was in response to one's own culture supernatants, and they predicted that these molecules could enter target cells and activate the expression of the genes responsible for bioluminescence (36, 76, 95, 96). Further research in this field has confirmed their prediction and identified the autoinducer signal made by P. fischeri as N-acyl homoserine lactone (AHL) (28).
P. fischeri is a facultative symbiont of marine fishes and squids. The bacteria live in the light organs of these marine animals and produce luminescence, which helps the animals escape from predators. In return, the bacteria gain nutrients and shelter from their host (26). The bacteria are also capable of a free-living lifestyle, and they alternate between the symbiotic and free-living modes in accordance with the circadian rhythm of the squid (63). Interestingly, bioluminescence is exhibited by the bacteria only when they are in the symbiotic mode of life and not in the free-living state. This regulation of bioluminescence is mediated by quorum sensing. In the free-living state, the bacterial AHL synthase (LuxI) constitutively produces basal amounts of AHLs, which immediately diffuse out of the cell into the surrounding marine environment. Once the bacteria enter the confined space in the light organs of the squid, the AHLs accumulate as a function of population density. At high cell densities or in a confined space, the increasing concentration of AHLs leads to the binding and activation of a specific response regulator called LuxR (53, 56, 114, 152). The activated LuxR then binds to a specific palindromic sequence on the DNA, called the “lux box,” located upstream of the quorum-sensing-regulated genes. LuxR bound to the lux box recruits RNA polymerase, thus resulting in enhanced transcription of the luciferase enzymes and other proteins involved in the production of bioluminescence (29).
An interesting feature of this type of quorum-sensing system is that the autoinducer synthase gene is a target for LuxR. Thus, activation of the quorum-sensing cascade results in increased expression of autoinducer synthase, leading to the production of more AHLs. This acts as a positive feedback loop and significantly amplifies the quorum-sensing effect (21, 32, 33, 37, 70). Similar quorum-sensing networks involving AHLs as autoinducer signals have been described in a large number of gram-negative bacteria, including Agrobacterium spp., Rhizobium spp., Pseudomonas spp., Brucella spp., and Vibrio spp. (34, 41, 52, 139, 150). In addition to AHLs, bacterial quorum sensing is known to involve other types of autoinducer signals, such as autoinducer 2 (AI-2), cyclic dipeptides, and others (20, 49, 138).
QSI Compounds
The coordinated expression of genes involved in virulence, colonization, and symbiosis is crucial in host-bacterial interactions (16, 59, 105, 121). In the case of pathogenic bacteria, early activation of virulence genes, such as those for toxin production, may result in triggering host defense responses and thereby giving the host an early advantage over the bacteria, particularly when their population density is low (80, 155). It would be beneficial for bacterial survival to avoid the production of virulence factors until their numbers could ensure their success. Coordinated expression of these phenotypes could potentially prevent wasting precious energy incurred during the synthesis of compounds that are not required for bacterial growth and survival at that particular time. A similar logic can be applied to symbiotic bacteria, where coordinated expression of specific genes at high population densities will ensure the presence of higher concentrations of the molecular signals required for the establishment of such a relationship with a eukaryotic host.
Eukaryotes, including algae and higher plants, produce several compounds that interfere with bacterial quorum sensing (38, 40, 140). These QSI compounds include molecules that mimic autoinducer structure and/or function and compounds that are antagonistic to the autoinducer molecules (141). In addition, QSI compounds could potentially target other components of the quorum-sensing system, such as interfering with the stability and function of the regulator protein or the autoinducer synthase (54, 82). Production of QSI compounds is not necessarily targeted towards a specific bacterial population. It has been previously observed that exudates from one plant species contain several types of QSI compounds, which vary in their specificity towards different bacterial strains (38, 89, 141). For example, exudates from pea seedlings appear to inhibit quorum sensing in Chromobacterium violaceum but to activate quorum sensing in Pseudomonas and Serratia (141). It is possible that these activities are due to multiple signal molecules present in the same exudates. Thus, by making the QSI compounds, the eukaryotic host may be simultaneously conversing with a variety of different bacterial strains that it encounters in its natural habitat, potentially encouraging the beneficial ones and antagonizing the harmful strains.
KEY PLAYERS IN A QUORUM-SENSING NETWORK
Autoinducers
Autoinducers are usually small molecules that either diffuse freely across the cell membranes or are actively transported out of the cell (1, 53, 104). This discussion will be limited to the quorum-sensing system in gram-negative bacteria. The quorum-sensing systems of gram-positive bacteria have been extensively described elsewhere (79, 134).
Acyl homoserine lactones.
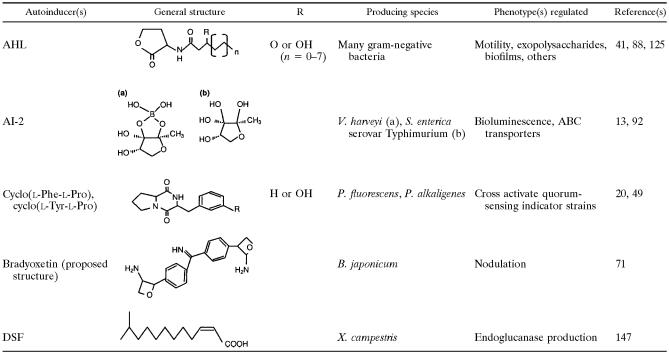
Acyl homoserine lactones are the major group of autoinducer signals in gram-negative bacteria. They have a conserved homoserine lactone (HSL) ring with a variable acyl side chain (Table 1). Based on the length of the acyl groups, AHLs can be broadly classified as short- or long-chain molecules. Short-chain AHLs have 4 to 8 carbon atoms in the acyl moiety, while long-chain AHLs have 10 to 18 carbons. The length and saturation level of the acyl chains coupled to the presence or absence of oxo or hydroxyl substitutions at the C-3 position of the acyl chain provide variation and specificity for quorum-sensing communication in a mixed bacterial population (58, 85, 88, 123, 125).
TABLE 1.
Autoinducer molecules identified in gram-negative bacteria
According to previous reports, a variety of different bacterial strains could make the same AHL, but this AHL may be involved in the regulation of different phenotypes in each strain. For example, 3-oxo-C6-HSL activates bioluminescence in P. fischeri but regulates exopolysaccharide production in Erwinia stewartii (148). This cross talk has facilitated the creation of quorum-sensing indicator strains that can be used to detect the presence of AHLs in a given sample (70, 88, 137, 158).
Autoinducer 2.
AI-2 was first recognized as a quorum-sensing signal in Vibrio harveyi by Bassler et al. (5). Since then, this type of signaling has been discovered in many gram-negative bacteria, such as Salmonella spp., Erwinia spp., and Escherichia spp. AI-2 is described as a global signal molecule for interspecies communication, as it is made by gram-positive as well as gram-negative bacteria (3, 4). The AI-2-type signaling is involved in the regulation of bioluminescence in V. harveyi (6), type III secretion in Escherichia coli O157:H7 (130), the virulence factor VirB in Shigella flexneri (19), and over 400 genes as indicated by microarray analysis conducted with E. coli strains (22, 131). Interestingly, recent work on dental biofilm-forming bacteria shows that AI-2 signaling is required for the formation of mixed-species biofilms (67, 90).
The crystal structure of AI-2 bound to its sensor protein LuxP from V. harveyi identified it as a furanosyl borate diester (Table 1). The proposed structure contains two fused five-member rings containing one boron atom bridging the diester (13). The AI-2 signal molecule isolated from Salmonella enterica serovar Typhimurium is a (2R,4S)-2-methyl-2,3,3,4-tetrahydroxytetrahydrofuran that lacks boron. The two bacterial species recognize two different forms of the AI-2 signal, both derived from the same precursor, dihydroxyl-2,3-pentane dione (92). Yet, it has been previously shown that AI-2 signals cross communicate and activate the V. harveyi indicator strain (135). This leads to the possibility that other gram-positive and gram-negative AI-2 signal molecules could belong to different subsets of dihydroxyl-2,3-pentane dione derivatives.
Cyclic dipeptides.
A new class of autoinducers was recently identified in strains of Pseudomonas based on their ability to activate AHL biosensors. Structural analysis indicated that these new signal molecules were the diketopiperazines (DKPs) cyclo(l-Ala-l-Val) and cyclo(l-Pro-l-Tyr), respectively (see Table 1). Additional DKPs have also been identified in Proteus mirabilis, Citrobacter freundii, Enterobacter agglomerans, Pseudomonas fluorescens, and Pseudomonas alkaligenes. Although the DKPs activate AHL biosensors, they do so at a much higher concentration than AHLs. The induction threshold for DKPs is around 0.3 mM, while that for 3-oxo-C6-HSL is 1 nM for the activation of the LuxR protein (49). The higher concentration of DKPs required for activation may indicate that these compounds may not play a significant role in the natural environment. Some of these DKPs, such as cycloΔ(l-Ala-l-Val), cycloΔ(l-Pro-l-Tyr), and cycloΔ(l-Phe-l-Pro), act as antagonists that actually compete with 3-oxo-C6-HSL for LuxR and inhibit quorum-sensing-regulated genes. Another report of isolation of cyclic dipeptides from Pseudomonas putida suggests that the DKPs are copurified with AHLs and are capable of activating various bacterial AHL biosensors (20). Interestingly, DKPs act as AHL antagonists in some strains and as agonists in others. For example, cyclo(l-Pro-l-Val) antagonized bioluminescence in an E. coli biosensor strain, while it activated violacein pigment production in C. violaceum strain CV026. This ability of DKPs from one bacterial strain to cross-communicate with the quorum-sensing networks of unrelated bacteria adds complexity and diversity to the quorum-sensing language.
Bradyoxetin.
A cell density factor was proposed to be involved in population density-dependent regulation of the nod genes in Bradyrhizobium japonicum, a gram-negative, nitrogen-fixing symbiont of many leguminous plants. The nod genes are repressed at high population densities, and this is mediated by small diffusible signal molecules present in conditioned media of wild-type bacterial cultures (72, 74). The structural elucidation of cell density factor suggests 2-{4-[[4-(3-aminooxetan-2-yl)phenyl](imino)methyl]phenyl}oxetan-3-yl amine, which has been named bradyoxetin (Table 1). The proposed structure of bradyoxetin is similar to that of a siderophore, mugeneic acid. In accordance with this similarity, bradyoxetin appears to be regulated by iron concentrations in the medium. It is maximally produced under iron-deficient conditions and repressed when Fe3+ is provided in excess (71). Cross-linking of quorum sensing with nutrient availability has been observed for other bacterial systems (8). A preliminary analysis suggested that bradyoxetin-like signal molecules were present in the extracts of various Rhizobium strains as well as other species of the α-proteobacterial group (71).
Other types of autoinducers.
In addition to the above-mentioned autoinducers, additional signals have been identified in gram-negative bacteria, including autoinducer (AI-3) in E. coli and diffusible signal factor (DSF) in Xanthomonas campestris (2, 132, 147). The structure of DSF was recently identified as cis-11-methyl-2-dodecenoic acid (Table 1). These signals appear to regulate phenotypes that are normally regulated by quorum sensing. A detailed analysis of the systems involved in the synthesis and regulation of these new autoinducer signals will definitely be a major contribution to our knowledge and understanding of quorum-sensing-dependent communication in bacteria.
Autoinducer Synthases
AHL synthases.
LuxI is the enzyme responsible for the synthesis of AHLs in the quorum-sensing system of V. fischeri (33). The genes for AHL synthases from other bacteria, including species of Rhizobium, Agrobacterium, Pseudomonas, Serratia, Erwinia, and Burkholderia, etc., are homologous to luxI, and their products share conserved residues required for activity (15, 41, 55, 57, 65). This suggests that the mechanisms of action for all LuxI homologs could be similar to each other. The LuxI synthase specifically catalyzes the amide bond formation between S-adenosylmethionine (SAM) and a fatty acyl-acyl carrier protein of a specific chain length. It also catalyzes the formation of the acyl homoserine lactone from the acyl-SAM intermediate (42, 94). The specificity of the AHL synthase to a particular chain length varies depending on the bacterial strain (148). This allows for the variability in the size of the AHLs made by different bacteria and accounts for the production of multiple AHL types by the same bacteria. Such bacterial strains usually have multiple synthases, with each synthase being responsible for the synthesis of a limited range of AHLs (122).
V. harveyi is known to produce AHLs and use them to regulate bioluminescence. The V. harveyi AHL synthase, LuxM, is not homologous to the LuxI of V. fischeri (5). This suggested the presence of additional types of AHL synthases. In accordance with this observation, a second AHL synthase, called AinS, was discovered in V. fischeri (39). This was not homologous to LuxI but appeared to share similarities with LuxM of V. harveyi. The protein was purified, and further studies indicated that this enzyme synthesizes AHLs in a manner similar to that of LuxI; it uses SAM and acyl-acyl carrier protein as substrates and results in AHL production. A unique feature of this synthase is that it can also accept acyl coenzyme A (acyl-CoA) as acyl donor (43). More recent work by Lupp et al. indicate that AinS is part of a hierarchy wherein it precedes the Lux quorum-sensing system in P. fischeri (78).
A third type of AHL synthase, HdtS, was identified in P. fluorescens. When cloned into E. coli, this protein catalyzes the production of 3-OH-C14:1-HSL, C10-HSL, and C6-HSL. This protein is not homologous to LuxI- or LuxM/AinS-type synthases (58).
AI-2 synthase.
LuxS is the synthase responsible for the production of the AI-2 signal molecule in V. harveyi (135). LuxS is an S-ribosylhomocysteinase that catalyzes the cleavage of the thioether linkage of S-ribosylhomocysteine to produce l-homocysteine and 4,5-dihydroxy-2,3-pentanedione. This catalysis is common in both gram-positive and gram-negative bacterial pathways for AI-2 biosynthesis (159). The reaction catalyzed by LuxS is mechanistically unprecedented. Eukaryotic cells contain a similar activity in S-adenosylhomocysteine hydrolase, which utilizes NAD+ to oxidize the 3′-OH group into a ketone intermediate (103, 159).
The X-ray crystal structure of LuxS shows that it exists as a homodimer and that two identical active sites are formed at the dimer interface. Each active site contains a catalytically active metal ion. This metal ion was identified as Zn2+ (46, 66, 120). A more recent study on the crystal structure of LuxS protein suggests that the metal ion in the native protein is Fe2+ and not Zn2+(159). The authors of that paper compared the stability profiles and electrospray ionization-mass spectrometry spectra of LuxS protein containing either Fe2+, Zn2+, or Co2+ as its metal ion. They showed that Fe2+-containing LuxS proteins have stability and Km values identical to those of the native enzyme. The rapid oxidation of Fe2+ by oxygen appears to be responsible for the high degree of instability of the LuxS proteins (159).
Synthases for other types of autoinducers.
In addition to the above-mentioned autoinducer synthases, other enzymes are expected to exist due to the discovery of new autoinducers such as cyclic dipeptides, AI-3, and DSF. The production of cyclic dipeptides by bacterial cells is a novel report (49). This seems to be more common in eukaryotic cells. Although the enzyme responsible for biosynthesis of cyclic dipeptides has not been identified so far, it has been suggested that DKPs can be generated via the nonenzymatic cyclization of linear dipeptides at extremes of temperature and pH (49, 127).
AI-3 activity appears to be dependent on LuxS (132). The structure and the enzyme responsible for AI-3 synthesis remain to be described. The DSF made by X. campestris is dependent on the presence of two genes, rpfF and rpfB. They are predicted to have enoyl-CoA hydratase and long-chain fatty acyl-CoA ligase functions, respectively (2). Additional work on the identification and characterization of the various synthases for these new groups of autoinducer molecules in gram-negative bacteria will be an interesting area for further research.
Quorum-Sensing Regulators
LuxR-type regulators.
Quorum-sensing-dependent gene regulation is mediated by transcriptional regulator proteins that are activated upon binding autoinducer molecules. LuxR is the transcriptional activator in V. fischeri (133). It binds to its cognate AHL, 3-oxo-C6-HSL, via a signal-binding region in the N-terminal domain that spans about two-thirds of the polypeptide chain. The interaction between LuxR and 3-oxo-C6-HSL appears to be very specific (124, 128). The C-terminal domain contains a helix-turn-helix motif and interacts with DNA at a specific site called the “lux box.” The lux box is a 20-bp inverted repeat upstream of the transcription start site of the target gene (23, 29).
LuxR-type proteins are required for AHL-mediated quorum sensing. The crystal structure of TraR, the LuxR homolog in Agrobacterium tumefaciens, shows that the AHL is completely embedded within the protein (146, 156). The crystal structure analysis also revealed that TraR exists as dimers, each binding two molecules of 3-oxo-C8-HSL and one duplex DNA fragment. AHL binding also stabilizes the TraR dimers from degradation by cellular proteases (161, 162).
LuxP/Q-type regulators.
AI-2-type quorum sensing is dependent on the activity of LuxP/Q-type proteins. LuxP is a periplasmic receptor that binds the autoinducer AI-2. LuxP modulates the activity of the sensor kinase LuxQ located on the inner membrane and thereby transduces the AI-2 signaling to the cytoplasm. LuxP belongs to the class of periplasmic binding proteins, which bind diverse ligands by clamping them between two domains. LuxQ is a hybrid two-component sensor kinase consisting of a periplasmic sensor domain and cytoplasmic histidine kinase and response regulatory domains (45, 93).
At low cell densities, LuxQ acts as a kinase and cross phosphorylates histidine residues within the histidine kinase domain. A series of phosphorylation cascades results in the phosphorylation of LuxO. Phospho-LuxO is responsible for the repression of LuxR, the transcriptional activator for luminescence genes in V. harveyi. At high cell densities, AI-2-bound LuxP interacts with LuxQ and converts it to a phosphatase. This leads to the dephosphorylation of LuxQ and initiates a reverse flow of phosphoryl groups through the AI-2 signal pathway, thus derepressing LuxR. LuxR is then free to activate transcription of the luminescence genes (93, 97).
Recent work on the structure of apo-LuxP complexed with the periplasmic domain of LuxQ suggests that LuxP and LuxQ exist in a complex irrespective of the presence of AI-2 in a given environment. However, the binding of AI-2 to LuxP results in conformational changes in the protein structure of LuxP and LuxQ, ultimately resulting in switching the kinase function of LuxQ to a phosphatase activity. The main roles of LuxP/Q are to detect the presence of AI-2 in the environment and to transfer the quorum-sensing information to the cytoplasm, thus regulating gene expression in response to AI-2 concentration (97). In addition to regulation by LuxP/Q proteins, the AI-2 quorum-sensing cascade also involves small RNAs that function as negative regulators (see below).
NEGATIVE REGULATION OF QUORUM SENSING
Antiactivator Proteins
In A. tumefaciens, the activity of the quorum-sensing regulator protein TraR is antagonized by several quorum-sensing antiactivator proteins. TraM is one such protein, encoded by the Ti plasmid in A. tumefaciens and also found in other members of the family Rhizobiaceae (77, 86, 136). The TraR-inhibitory activity of TraM is absolutely required for the normal functioning of the quorum-sensing system in A. tumefaciens. The Tra quorum-sensing system regulates conjugal plasmid transfer and the replication genes in A. tumefaciens, and a TraM mutant is hyperconjugal, resulting in the transfer of the Ti plasmid to recipient cells with high efficiency even at low cell densities (159). Therefore, the main role of TraM is to regulate the activity of the AHL-TraR complex such that it functions effectively only upon reaching a quorum (51, 108).
Crystal structure analysis of TraM has revealed that it forms a highly stable homodimer in solution. Biochemical studies show that TraM forms a heterodimer with TraR and probably binds to the DNA binding loop in the carboxy terminal end of TraR, thereby preventing its interaction with the “tra box” on the DNA. Biochemical studies suggest that the TraM homodimer dissociates in order to form a heterodimer with monomeric TraR, in a 1:1 stoichiometry (12). A second report suggested a multimeric interaction involving two TraR and two TraM dimers (145). The actual mode of binding and inactivation of TraR function by TraM can be confirmed only upon studying the crystal structure of a TraR-TraM complex (12, 110, 145).
Homologs of Transcriptional Regulators
TrlR (TraR-like regulator) is another protein responsible for antagonizing the activity of TraR in A. tumefaciens. The TrlR protein closely resembles the N-terminal sequence of TraR for the first 181 amino acids but is truncated due to a frameshift mutation at codon 182. Due to the similarity of TrlR to the autoinducer binding and dimerization domains of TraR, it is capable of interacting with TraR monomers to form TrlR-TraR heterodimers that are functionally inactive and cannot activate transcription of the target DNA. TrlR binds to one molecule of AHL per monomer and thus can potentially titrate out the amount of AHL available to activate TraR, especially at low population densities when AHL production is at basal levels. Also, TraR not bound to AHL is rapidly degraded by the cellular proteases, and thus TrlR could act in multiple ways to regulate the activity of TraR (11). Interestingly, TrlR expression is greatly enhanced in the presence of mannopine, a type of opine synthesized by the host plant. Mannopine is known to strongly inhibit conjugation in A. tumefaciens (100).
The QscR (quorum-sensing control repressor) protein is a homolog of LasR and RhlR, the transcriptional regulators of Pseudomonas aeruginosa quorum-sensing systems. The main role of QscR appears to be to repress the Las and Rhl quorum-sensing systems in the early growth phase. A QscR mutant expresses lasI and rhlI much earlier than wild-type cells. The mutant also shows premature production of AHLs and is hypervirulent in Drosophila melanogaster compared to wild-type strains (14, 61). It is not clear how QscR regulates the quorum-sensing system. Since it is homologous to the LasR and RhlR proteins, it could potentially bind the promoter region and repress activation of quorum-sensing-regulated genes, or alternatively, it could form inactive heteromultimers with LasR protein in a manner similar to that of TrlR of A. tumefaciens (14). Recent work by Lee et al. shows that this protein directly activates the expression of genes that are indirectly regulated by LasR. This activity requires the presence of AHLs. LasR-specific promoters do not interact with QscR and vice versa. Those authors suggest that in addition to directly activating expression of specific genes, QscR could also interact with the LasR and RhlR proteins and act as a repressor of quorum-sensing-regulated genes (62). Further analysis of this protein and its mechanism of action could result in a better understanding of the function of such LuxR homologs.
AHL-Degrading Enzymes
attM codes for an enzyme that hydrolyzes the lactone ring of 3-oxo-C8-HSL, the cognate AHL made by the Tra quorum-sensing system of A. tumefaciens (154). The expression of attM is repressed by a suppressor encoded by attJ, under logarithmic growth conditions. This allows for the accumulation of AHLs and efficient conjugation in the log phase of bacterial growth. Upon entering the stationary phase, there is a rapid degradation of the AHLs. This correlates with the enhanced expression of attM under stationary-phase conditions. It was recently shown by Zhang et al. that this quormone degradation system is regulated by the stress alarmone signal (p)ppGpp (153). The AttM system of A. tumefaciens helps the bacterium to exit from a quorum-sensing mode upon reaching starvation/stationary-phase conditions. This results in shutting off the energetically expensive conjugation and possibly other such quorum-sensing-regulated phenotypes in the bacterium. Evidence indicates that the AHLs cannot be utilized as a sole source of carbon upon cleavage by the lactonase enzyme. A second enzyme, AiiB, which is homologous to AttM, was identified in A. tumefaciens and was shown to degrade AHLs (10). It has been suggested that AttM could play a role in regulating the AHL turnover (153). A second report by Carlier et al. shows that the attM gene is activated specifically in response to the gamma butyrolactones. In the absence of these inducers, AHLs are not degraded by wild-type bacteria, suggesting that degradation of AHLs may not be the primary function of the enzyme. It is possible that the enzyme evolved for the catabolism of gamma butyrolactones but is able to utilize AHLs as substrates (9).
Recently, AHL acylase and lactonase activities were identified in soil pseudomonads and P. aeruginosa PAO1 strains (50). The AHL-degrading activity was specific towards long-chain AHLs. A soil isolate of Pseudomonas strain PAI-A was able to utilize AHLs for growth. The quorum-sensing strain P. aeruginosa PAO1, which normally produces 3-oxo-C12-HSL and C4-HSL as its quorum-sensing signals, also exhibits the ability to utilize long-chain AHLs for growth. Overexpression of pvdQ, a homolog of the AHL-acylase gene in Ralstonia spp., resulted in the accumulation of C4-HSL but not 3-oxo-C12-HSL molecules in PAO1 when grown in rich media. Interestingly, pvdQ knockout mutants of PAO1 were still able to utilize 3-oxo-C12-HSL for growth, suggesting that additional AHL-degrading enzymes may be present in this bacterium (50). AHL-degrading enzymes identified in various gram-positive as well as gram-negative bacteria are used to control quorum-sensing-regulated bacterial phenotypes (see “Quorum Quenchers” in “USING BACTERIAL COMPONENTS TO MANIPULATE QUORUM SENSING” below).
mRNA-Dependent Regulation
Recent work on the regulation of quorum sensing in V. harveyi and Vibrio cholerae shows that small, regulatory RNAs (sRNAs) are involved in specifically repressing quorum sensing. V. harveyi and V. cholerae have similar quorum-sensing systems based on the AI-1- and AI-2 type autoinducers. At low cell densities, in the absence of the autoinducer molecules, a phosphorylation cascade through the autoinducer sensors and response regulators results in the phosphorylation of LuxO. LuxO phosphate (LuxO-P) is active and negatively regulates the lux system by activating the expression of a repressor. Lenz et al. identified four sRNAs, along with the sRNA binding protein Hfq, that act as repressors of the Lux system in both V. harveyi and V. cholerae (64). They named these sRNA loci qrr1, qrr2, qrr3, and qrr4 (for quorum regulatory rna 1 to 4). These sRNA loci were regulated by LuxO-P and σ54. They are proposed to bind to LuxR and HapR (the LuxR homolog in V. cholerae) mRNAs via the Hfq protein and to enhance degradation of the mRNAs, thereby repressing quorum sensing (64).
The RsmA protein, identified in Erwinia carotovora and P. aeruginosa, is involved in the down regulation of the corresponding quorum-sensing systems. In E. carotovora, the RsmA (repressor of secondary metabolites) is proposed to be an RNA binding protein and reduces the levels of CarI AHL synthase transcripts, thus lowering the amount of AHLs produced (17). More recently, the RsmA protein was identified in P. aeruginosa and shown to down regulate several quorum-sensing-dependent phenotypes, such as protease, elastase, and staphylolytic activities along with production of lectin, HCN, and pyocyanin. Overexpression of RsmA resulted in reduced expression of the AHL synthase genes lasI and rhlI. This suggests that RsmA may down regulate quorum sensing by repression of AHL synthesis (106). The mechanism of RsmA-mediated repression in P. aeruginosa is not clear and needs further investigation.
EUKARYOTIC INTERFERENCE IN BACTERIAL QUORUM SENSING
Quorum-Sensing Cross Talk between A. tumefaciens and Its Host Plant
A. tumefaciens causes crown gall disease in a variety of plants, resulting in large tumors in the crown of the plant. The Ti (tumor-inducing) plasmid of the bacterium contains genes responsible for virulence and harbors the ability to transfer a fragment of its DNA (T-DNA) into the nuclear genome of the host plant. This bacterial DNA then directs the plant cells to synthesize specific carbohydrates called opines which are readily metabolized by Agrobacterium around the crown gall tumors (160). The Ti plasmid is transferred between bacterial cells by conjugation, and this process is regulated by the Tra quorum-sensing system. In addition to the regulation by bacterial proteins discussed above, TraR is expressed only in the presence of plant-produced opines. This ensures that conjugal transfer of the Ti plasmid occurs only in the vicinity of host tumors (Fig. 2) (98, 99, 107).
FIG. 2.
Quorum-sensing cross talk between A. tumefaciens and its host plant. The transfer and integration of bacterial T-DNA into plant cells result in tumorogenesis, leading to crown gall disease in plants. The tumor cells directed by the bacterial DNA produce and release opines, which are metabolized by the A. tumefaciens present in the soil around the plant roots. The conjugal transfer of the Ti plasmid among the rapidly proliferating A. tumefaciens in soil is regulated by bacterial quorum sensing as well as by plant-produced opines.
There are at least eight chemical families of opines (98). Opines are utilized by bacteria as a source of carbon and energy. A subset of the compounds are termed conjugal opines, and they serve as Ti plasmid conjugal transfer-inducing signals. Interestingly, there is variation in the opine-mediated regulation of TraR expression. There are at least two different types of regulation, based on the type of Ti plasmid present: the nopaline type and the octopine type. The nopaline-type Ti plasmid regulation involves a repressor protein, AccR. TraR, in this case, is encoded by part of a five-gene arc operon that is normally repressed by AccR. The presence of conjugal opines, for example, agrocinopine (Table 2), relieves the repression by directly binding AccR, resulting in the transcription of the TraR gene. In the case of octopine-type Ti plasmid regulation, TraR is encoded by the last gene of a 14-gene operon involved in the uptake and catabolism of octopine. Expression of TraR is dependent on OccR, an octopine-responsive transcriptional activator belonging to the LysR family. Therefore, in the presence of octopines, traR is expressed, leading to the onset of a quorum-sensing cascade, which eventually culminates in conjugal plasmid transfer (99).
TABLE 2.
QSI compounds identified from eukaryotes
In addition to the above-described regulation, there appears to be an additional level of QSI activity by the host plant. A third type of opine, mannopine, is involved in activating the transcription of trlR, which is an antiactivator of TraR (100). This multilevel quorum-sensing cross talk between A. tumefaciens and its host plant suggests the possibility that similar intricate QSI signaling could exist in other plant-bacterial interactions.
Furanones: Structural Mimics
Halogenated furanones are naturally produced by the Australian red alga Delisea pulchra and are known to have strong inhibitory activity against fouling organisms and herbivores. Interestingly, the furanones have structural similarity to AHLs (Table 2). Previous research has shown that furanones are capable of interfering with the quorum-sensing behavior of several bacterial strains. One such bacterium is Serratia liquefaciens, which regulates swarming motility via AHL-dependent quorum sensing. The Swr quorum-sensing system of S. liquefaciens comprises the response regulator SwrR and the synthase SwrI, responsible for the production of C4-HSL and C6-HSL (18, 68).
The Swr quorum-sensing system is thought to work in a manner similar to that of the Lux system in V. fischeri. Biochemical studies on the effect of specific halogenated furanones on LuxR protein overexpressed in E. coli indicate that the furanones are capable of interfering with AHL-LuxR interactions. Although furanones bind LuxR, the complex appears to be unstable. Binding of furanone to LuxR renders it highly unstable and accelerates its turnover rate. This results in the rapid disruption of the quorum-sensing-mediated gene regulation. Interestingly, addition of AHLs to the protein prior to introduction of furanones results in protecting the LuxR protein against furanone-promoted degradation. The mode of action of furanones and the nature of the interaction with the LuxR protein remain to be elucidated (81, 82).
Interestingly, recent work by Ren et al. shows that (5Z)-4-bromo-5-bromomethylene-3-butyl-2(5H)-furanone, naturally made by D. pulchra, inhibits AI-2-dependent quorum sensing in E. coli. They showed that the furanone completely inhibits swarming motility in E. coli and greatly inhibits biofilm formation in this strain. They also analyzed the effect of the furanone on AI-1 and AI-2 indicator strains of V. harveyi and found that luminescence was greatly inhibited in both reporter strains. This luminescence suppression was reversible when excess concentrations of either autoinducer signal were present (115).
A microarray analysis comparing the global gene expressions of E. coli K-12 and DH5α (which lacks an AI-2 system), along with the gene expression profile of E. coli K-12 grown in the presence or absence of the furanone, identified 166 genes that were differentially expressed by AI-2 and 90 genes that were differentially expressed by furanones. More importantly, 44 genes, most of which are involved in chemotaxis, motility, and flagellar biosynthesis, were activated by AI-2 but repressed by furanones, suggesting that furanones inhibited AI-2-dependent signaling. The genes luxS and pfs, which code for AI-2-synthesizing enzymes, and rbsB (a homolog of luxP, which codes for AI-2 receptor protein in V. harveyi) were not repressed by furanones (116). Further research in evaluating the molecular and structural details involved in this activity will greatly enhance our understanding of the QSI compounds.
l-Canavanine as a Quorum-Sensing Inhibitor
l-Canavanine is an arginine analog found exclusively in the seeds of legumes. It has been reported to be as abundant as up to 5% (dry weight) of some leguminous seeds (149). In addition to serving as a nitrogen source for the germinating seedlings, l-canavanine is also known to serve as an allelopathic substance by inhibiting the growth of certain bacteria and phytophagous insects (119). Handelsman and her group showed that canavanine exuded from alfalfa seeds has the potential to affect the population biology of Bacillus cereus (31). l-Canavanine is incorporated in place of l-arginine into nascent protein chains during synthesis, resulting in altered protein structure and function and eventually leading to death of the targeted cell (7, 119). In our recent work, we identified l-canavanine as a QSI compound and showed that the QSI activity is independent of its effect on bacterial growth (Table 2) (54).
Our initial screen for QSI compounds from alfalfa (Medicago sativa) exudates detected several signal molecules that appeared to specifically affect quorum sensing of different bacterial strains. Some compounds had quorum-sensing-inhibitory effects, while others seemed to activate quorum sensing (unpublished results). We identified the structure of one of the quorum-sensing-inhibitory compounds that was capable of inhibiting violacein production in the indicator strain Chromobacterium violaceum CV026. The production of violacein pigment, resulting in purple colonies, is a quorum-sensing-regulated phenotype in C. violaceum. CV026 is an AHL synthase mutant and hence cannot produce its own AHLs, but in the presence of externally provided AHLs, it produces violacein pigment. l-Canavanine inhibited the violacein production phenotype in the presence of synthetic AHLs (54).
Since we isolated l-canavanine from seeds of M. sativa, we analyzed its effect on the quorum-sensing system of its bacterial symbiont, Sinorhizobium meliloti. Two quorum-sensing systems, Sin and Tra, have been described for S. meliloti strains (84). The Sin quorum-sensing system is known to regulate the expression of over 200 genes (48). One subset of these genes code for the production of the symbiotically important exopolysaccharide EPS II (83). We showed that l-canavanine inhibited the expression and production of EPS II. We also showed that a gene that is not normally regulated by quorum sensing suffers a negligible inhibition of gene expression compared to that of a quorum-sensing-regulated gene (1.5-fold versus 20-fold, respectively). Increasing the AHL concentration did not titrate out the QSI activity of l-canavanine, but addition of 10-fold excess amounts of l-arginine was sufficient to antagonize its QSI activity, suggesting that l-canavanine was probably interfering with protein structure and function (54).
Human Hormones Interfere with Bacterial Quorum Sensing
A recent study on enterohemorrhagic Escherichia coli (EHEC) showed that human hormones cross communicated with the bacterial quorum-sensing system (132). The LuxS-dependent quorum-sensing system in EHEC controls several phenotypes, including pathogenicity and type III secretion, which eventually results in virulence on HeLa epithelial cells (126). Interestingly, type III secretion and virulence phenotypes were restored in a luxS mutant by supplementing it with the human hormones epinephrine and norepinephrine (Table 2). Other intestinal hormones were tested and found to be inactive in this assay. In addition, treating the cells with antagonists to adrenergic receptors (the hormonal receptors in human cell membranes) blocked the cross-activation of a luxS mutant by the human hormones. Sperandio et al. also suggested the presence of a third type of autoinducer, AI-3, that is dependent on the luxS system. They proposed that the pathogenic genes in EHEC are regulated by AI-3/epinephrine-norepinephrine signals and this might influence the infection process by coordinating the appropriate time and environment for the onset of virulence (132).
Other QSI Compounds
An interesting report by Teplitski et al. described the discovery of several unidentified compounds from higher plants, which were termed AHL-mimic compounds, as they appeared to activate the expression of several quorum-sensing-regulated genes (141). That study analyzed the presence of different AHL-mimic activities in a variety of higher plants and found that the nature of these activities varied depending on the plant species. Pea (Pisum sativum) seedlings and their exudates contained compounds that inhibited violacein production, a quorum-sensing-regulated phenotype in C. violaceum. Addition of excess AHLs after exposure to pea seedlings did not reverse this phenotype; however, exposure to AHLs prior to the addition of pea seedling exudates partially prevented the violacein inhibition phenotype. Also, other quorum-sensing-regulated phenotypes in C. violaceum, such as protease and exochitinase activities, were substantially inhibited in the presence of pea seedling exudates. In addition, the pea seedling exudates were capable of activating several quorum-sensing-regulated phenotypes in different bacterial indicator strains that were based on AHL receptor proteins such as LuxR, AhyR (from Aeromonas hydrophila), or LasR (from P. aeruginosa) (141).
Another study on Medicago truncatula indicated that it produces at least 15 to 20 compounds that are capable of specifically activating or inhibiting quorum-sensing-regulated behavior in different bacterial strains. In that study, young seedlings and seedling exudates were sequentially extracted with ethyl acetate, methanol, and 50% methanol. Each extract was then separated by high-pressure liquid chromatography, and the fractions were tested for activity with several bacterial reporter stains. The M. truncatula extracts activated reporter strains based on the quorum-sensing regulator proteins CepR (P. putida), LasR (P. aeruginosa), and LuxR (V. fischeri) and V. harveyi BB170, which responds to the AI-2 quorum-sensing signal. In addition, several compounds inhibited gene expression regulated by CviR (C. violaceum), LasR, AhyR, and LuxR. The authors also observed that there were time-dependent changes in the secretion of QSI compounds during seedling germination and development (38).
Chlamydomonas reinhardtii, a unicellular soil-fresh water alga, was also found to secrete substances that interfered with bacterial quorum sensing. More than a dozen unidentified compounds were isolated that were capable of activating LasR or CepR but not LuxR, AhyR, or CviR quorum-sensing reporter strains. Interestingly, the ethyl acetate extracts but not the methanol extracts of the C. reinhardtii culture filtrates had the above activities. This is different from previous observations in M. truncatula and other plants, where the mimic activities appeared to prefer more-polar solvents. A partially purified LasR-stimulatory compound from C. reinhardtii extracts was used to analyze its effect on the overall proteome of S. meliloti. It was concluded that a major portion of quorum-sensing-regulated proteins were also affected by the LasR-stimulatory quorum-sensing mimic compound (140).
Screening for QSI compounds is greatly facilitated by the recent creation of bacterial strains designed to detect the presence of quorum-sensing-inhibitory compounds (89, 113). Rasmussen et al. constructed bacterial indicator strains to detect the presence of quorum-sensing-inhibitory compounds in a given sample. They designed two types of quorum-sensing inhibitor selector (QSIS) systems. The first type had a lethal gene fused to a LuxR-regulated promoter. Therefore, in the presence of AHL signal molecules, the lethal gene is expressed, resulting in death of the indicator strains. The presence of a quorum-sensing-inhibitory compound results in neutralizing the activity of AHL, and therefore the lethal gene is not expressed, allowing the growth of the bacteria on the test plates. The second type of QSIS system employed an antibiotic resistance gene that is controlled by a repressor. The repressor in turn is controlled by LuxR. In the presence of AHL, the repressor prevents the expression of the antibiotic resistance. Therefore, the indicator strain is unable to grow in the presence of the antibiotic and AHLs, unless it encounters a quorum-sensing-inhibitory compound that will allow the expression of the antibiotic resistance gene (113).
Both of the above-mentioned QSIS systems have a visible phenotype, such as a lacZ expression or a green fluorescent protein (GFP) marker that facilitates the quantitation of gene induction and bacterial growth. In addition, Rasmussen et al. identified QSI compounds by placing various plant extracts in a well on an agar plate containing a lawn of the QSIS strain. The extracts diffuse out of the well, forming a gradient, thereby allowing simultaneous testing of various concentrations of the QSI compounds. Another advantage of this system is that growth-inhibitory compounds are not detected by this assay, thereby eliminating potential false positives. Rasmussen et al. used the above-described QSIS system to identify QSI activity in garlic extract as well as a synthetic compound, 4-nitro-pyridine-N-oxide. They further observed that these compounds enhanced susceptibility of P. aeruginosa biofilms to the antibiotic tobramycin. They also showed that garlic extract and 4-nitro-pyridine-N-oxide lowered the pathogenicity of P. aeruginosa wild-type strain PAO1 in a Caenorhabditis elegans nematode model (113).
In another study, the QSIS system was used to identify quorum-sensing inhibitors produced by the fungi belonging to the genus Penicillium. Extracts of 50 members of the genus Penicillium were screened, and several of them were found to have QSI activity. Two of the compounds were identified as penicillic acid and patulin (Table 2). These compounds were found to down regulate the expression of several quorum-sensing-regulated genes, as determined by a DNA microarray of P. aeruginosa. Patulin was found to accelerate LuxR turnover in a manner similar to that of the furanone C-30. Patulin also enhanced the susceptibility of PAO1 biofilms to tobramycin and promoted faster clearing of P. aeruginosa in mouse lungs (112).
The above description of the rapidly increasing numbers of QSI compounds isolated from eukaryotes holds promise for the advancement of therapeutic medicine and pharmacology. In addition, the production of multiple QSI compounds by a single eukaryote that can effectively cripple the virulence of a variety of bacterial strains reflects on the complexity of coevolution and suggests the existence of intricate molecular signaling in eukaryote-bacterial relationships.
USING BACTERIAL COMPONENTS TO MANIPULATE QUORUM SENSING
Quorum Quenchers
Several AHL-degrading enzymes identified in various bacteria have the potential to be used as quorum quenchers. Dong et al. initially identified AiiA from Bacillus species and showed that this enzyme inactivates the AHL signal and attenuates virulence when expressed in Erwinia carotovora (25). A second report showed that a soil isolate of Variovorax paradoxus not only breaks down AHLs but is capable of utilizing them as the sole source of carbon and nitrogen (60). Another study on quorum quenching isolated more than 20 bacteria belonging to the Bacillus cereus group which were capable of enzymatic inactivation of AHLs. Further genetic analyses revealed that the enzymes responsible for AHL inactivation were homologs of AiiA from Bacillus species strain 240B1. This enzyme is an AHL lactonase, known to act by hydrolyzing the lactone bond in the AHL (24).
More recent work on the isolation of bacteria capable of degrading C6-HSL identified 25 isolates, which were further classified into six groups based on their genomic profiles. The bacteria were identified as members of the genera Pseudomonas, Comamonas, Variovorax, and Rhodococcus. The strains varied in their specificity for the acyl chain lengths of the AHLs. Interestingly, the degradation properties of the various strains depended on the nutritional medium conditions. Based on these screens, one of the isolates, Rhodococcus erythropolis strain W2, was chosen for further analysis, as it had a broad degradation spectrum. This strain was capable of inhibiting quorum-sensing-regulated phenotypes in C. violaceum and A. tumefaciens and reduced the pathogenicity of Pectobacterium carotovorum subsp. carotovorum in potato tubers (144).
A second class of quorum-quenching enzymes was identified in Ralstonia strain XJ12B. The acylase AiiD isolated from this strain is a 794-amino-acid polypeptide that is capable of hydrolyzing the AHL amide, releasing homoserine lactone and the corresponding fatty acid. Expression of aiiD in P. aeruginosa PAO1 decreased its ability to swarm, produce elastase and pyocyanin, and paralyze nematodes, all of which are quorum-sensing-regulated phenotypes in this bacterium. Interestingly, the AHL acylase activity was found to be present mainly in the cell debris after lysis, suggesting that it may be membrane or periplasm associated. The enzyme, when incubated with 3-oxo-C8-HSL, 3-oxo-C10-HSL, and 3-oxo-C12-HSL, completely inactivated all three signals within a few hours (50). P. aeruginosa PAO1 and a soil Pseudomonas strain, PAI-A, are reported to degrade AHLs, with specificity for long-acyl-chain AHLs. P. aeruginosa pvdQ, which was identified as a homolog of the AHL acylase gene from Ralstonia, was cloned into E. coli and shown to degrade 3-oxo-C12-HSL, cleaving the long acyl chain away to produce homoserine lactones. Interestingly, a pvdQ mutant of P. aeruginosa was still able to utilize AHLs for growth, indicating that other enzymes may contribute to the AHL assimilation phenotype (50).
Three-dimensional structure analysis of the AHL lactonase from Bacillus thuringiensis shows that it has two major β-sheets in a parallel formation at the center of the structure and that this is surrounded by six α-helices. The crystals contain one AHL lactonase subunit per asymmetric unit, which suggests that AHL lactonase is a monomer. The active site contains two zinc ions located 3.3 Å apart in a bridged dinuclear site. Interestingly, the active-site cavity is branched and extends deep into the enzyme with the two zinc ions and the potential catalytic residues, all located on the ridge between the two branches. It has been suggested that this branched active site may contribute to the specificity of lactonase for acyl chain lengths and C-3 substitutions of AHL substrates (69).
Overall, the AHL-dependent quorum quenchers can be broadly classified into AHL lactonases and AHL acylases. It is possible that a single bacterium can possess both types of enzymes, thereby facilitating its metabolism of AHLs. Interestingly, enzymes that degrade AHLs have been identified in bacteria that produce AHLs, and these enzymes could potentially play a role in regulating quorum sensing for, e.g., P. aeruginosa PAO1 (pvdQ) and A. tumefaciens (attM and aiiB) (10, 50). On the other hand, non-AHL-producing bacteria make AHL-degrading enzymes, which probably evolved to utilize this commonly found molecule as a carbon and nitrogen source. It is also possible that quorum quenching is used as a defense mechanism against antibiotic-producing bacteria in the ecological niche. The degradation and inactivation of AHLs also achieve a rapid depletion of the quorum-sensing signals, thereby ensuring that the AHL concentration in the environment is a true indication of the bacterial population density.
Transgenic Plants
Quorum sensing appears to be crucial for plant-bacterial interactions, be they pathogenesis or symbiosis (41, 73, 150). A timely activation of specific phenotypes responsible for interaction with the host plant ensures a successful establishment of the bacterial population on the host. A premature activation of these bacterial phenotypes could trigger early defense responses and therefore may be deleterious to the bacterial population (80, 155). This idea has been exploited to create transgenic plants encoding bacterial AHL synthases, such that the plants are now capable of producing AHL signal molecules. Fray et al. cloned the yenI AHL synthase from Yersinia enterocolitica and targeted it to the chloroplasts of tobacco plants to create transgenics that produced 3-oxo-C6-HSL and C6-HSL (35). Older senescent plant tissue accumulated AHLs to a greater extent than younger tissue. Interestingly, the viral promoter under control of which the yenI gene was cloned is believed to be most active in the younger tissues. This suggested that AHL may not be readily broken down in the plant tissue but may continue to accumulate over time. The presence of AHLs was detected not only in green tissue containing the chloroplasts but also in organs such as roots containing undifferentiated plastids, indicating that either the root plastids are capable of synthesizing AHLs or the AHLs are transported to the root system. The AHLs produced by the transgenic tobacco plants activated quorum-sensing-related phenotypes in AHL synthase mutants of Pseudomonas aureofaciens 30-84 and E. carotovora strains (35).
Another report, involving the cloning of the expI gene from E. carotovora, which is responsible for the synthesis of 3-oxo-C6-HSL, into tobacco, showed that the transgenic plants produced the active signal molecule and exhibited enhanced resistance to infection by wild-type E. carotovora. 3-oxo-C6-HSL is involved in the regulation of extracellular plant cell wall-degrading enzyme (PCWDE) production in E. carotovora. These enzymes release pectic fragments from the plant cell wall that trigger the activation of plant defense. Therefore, it is beneficial to the bacterium to produce PCWDEs only upon reaching a quorum, such that they can overcome the plant defense response. Creation of transgenic plants producing AHL signal molecules sends a false message to the bacterium about its population density, eventually leading to premature production of PCWDEs. Mäe et al. showed that the ability of wild-type E. carotovora to cause disease symptoms (tissue maceration) was reduced to about half in transgenic plants compared to plants harboring only the vector. Increasing the bacterial inoculum size by fourfold or more resulted in overwhelming the enhanced resistance in the transgenic plants. Addition of synthetic AHLs to wild-type plants led to reduced maceration symptoms when challenged with wild-type E. carotovora strains (80). More recent work involving transgenic potato plants containing the yenI gene shows increased susceptibility to soft rot infections by Erwinia strains. It was shown that the degree of susceptibility of transgenic potato plants to soft rot varied depending on the tissue tested and on the strain of E. carotovora. It has been suggested by those authors that coordinated production of PCWDEs at a high bacterial population density may be a response to nutrient limitation rather than a mechanism to overcome plant defenses (142). The contrasting results of cloning AHL synthase into tobacco and potato plants, with one showing increased resistance and the other enhanced susceptibility to the pathogens belonging to the same genus, Erwinia, show that further research on evaluating the mechanism of action along with elucidating the details of bacterial pathogenesis is necessary.
Cloning of a bacterial AHL lactonase enzyme (AiiA from Bacillus sp. strain 2401B) into tobacco and potato plants by Dong et al. illustrates a potentially effective method to control bacterial infections (24). Soluble protein extracted from transgenic tobacco leaves and potato tubers inactivated 3-oxo-C6-HSL activity in vitro. When challenged with the pathogen E. carotovora, the transgenic plants did not develop any symptoms with a low population density of inoculum (600 CFU) and exhibited much smaller maceration areas than controls with a high population density of inoculum (60,000 CFU). The initiated maceration seen with a high-population-density inoculum on transgenic plants showed significantly delayed development of symptoms upon further incubation. That study shows that the enzymatic inactivation of AHL by cloning quorum-quenching genes could be an effective approach to control bacterial infections (24).
Synthetic Analogs for Quorum-Sensing Autoinducers
A variety of structural analogs for different AHL molecules have been studied for their effects on the quorum-sensing system of the related bacterial strain. A study on the analogs of the 3-oxo-C8-HSL shows that the nature of the agonistic or antagonistic activity strongly depends on the expression of the TraR protein (157). Overexpression of the response regulator, as seen in the case of several reporter strains, results in recognition of most AHL analogs as agonists. In that particular study, wild-type levels of TraR identified 3-oxo-C7-HSL, 3-oxo-C11-HSL, and 3-oxo-C12-HSL as agonists. Most of the 33 compounds tested in that study inhibited AHL-dependent gene expression. C8-HSL, 3-oxo-C6-HSL, C7-HSL, C10-HSL, and 3-OH-C9-HSL were identified as the most effective antagonists. Overall, the agonistic or antagonistic activity, each requiring binding to TraR, is effective only when the acyl chain lengths are closer to that of the cognate AHL, 3-oxo-C8-HSL. When TraR was overexpressed, the bacterium was more sensitive to low concentrations of 3-oxo-C8-HSL than its parent strain. In addition, this strain identified 29 of the 33 tested compounds as agonists. Interestingly, compounds with four carbons in their acyl chains were not recognized even when TraR was overexpressed (157). Similar studies conducted with synthetic analogs for cognate AHLs of other quorum-sensing systems indicated that the homoserine lactone ring was very important for biological activity, while the nature of the acyl chains was not as critical for binding the response regulator (117).
Another study aimed at synthesizing analogs to 3-oxo-C6-HSL and C6-HSL with either ramified, cycloalkyl, or aryl substituents at the C-4 position of the acyl chain (117). Those authors concluded that the inducing activity is retained if one branching is introduced at the C-5 position of the acyl chain. The best antagonists were compounds with a phenyl group or a phenyl group bearing a heteroatom in the para position. Naphthyl and biphenyl compounds showed no activity, probably due to steric hindrance. They also observed that the 3-oxo group, which is important for the inducing activity, also favors the antagonistic activity of phenyl derivatives. Overall, the secondary alkyl derivatives had agonistic activity, while the aryl and tertiary alkyl derivatives had antagonistic activity when tested with an E. coli-based luminescent biosensor strain containing a plasmid with the lux genes from V. fischeri (117).
Olsen et al. synthesized AHL analogs based on the strategy that modifying the conserved lactone head group of AHLs will probably result in antagonistic molecules with a broader application range. Based on their observations, they concluded that the analogs with C-4 substitutions on the lactone ring were weak activators, implying that this part of the molecule is crucial for recognition by LuxR, whereas substitution at the third position on the lactone ring resulted in activators of LuxR. Two compounds containing carbamate lactones were identified as inhibitors, although they appeared to be less efficient than a furanone (102).
Several furanone-based structural analogs have also been synthesized and analyzed for their QSI activity. A recent study reported the isolation of two natural products from a marine sponge and a Pseudomonas sp. that were structurally similar to furanones. These compounds, isocladospolide and acaterin, were used as templates for further modifications, and the resulting compounds were tested against LuxR-based E. coli biosensor strains. The 5H-furan-2-ones substituted with short alkyl chains were in general more antagonistic than the longer-alkyl-chain counterparts. Also, a substitution at the C-3 or C-5 position of the alkyl chain resulted in the most active antagonists (47).
Another interesting study using synthetic furanones tested on mouse lungs infected with different bacterial strains indicated that the QSI effect of furanones was functional in vivo and helped to enhance clearance of P. aeruginosa from infected lungs (151). Synthetic furanones were initially tested on mouse lungs infected with E. coli strains carrying luxR-PluxI-gfp-based quorum-sensing sensors. AHL-dependent GFP expression was completely inhibited by intravenous injection of the active furanones. This inhibition was overcome by providing excess amounts of AHLs. This suggested that the synthetic furanones were transported via the blood to the lungs, entered the lung tissues, and inhibited 3-oxo-C6-HSL-dependent gene expression in the bacteria. The death of mice inoculated with wild-type P. aeruginosa PAO1 was significantly delayed when they were treated with synthetic furanone, although the furanone could not prevent the death of the mice. In addition, there were fewer CFU of the bacterium on the furanone-treated lung surface compared to untreated controls. This suggests that the ability of the bacteria to colonize may be greatly reduced owing to inhibition of quorum sensing or because the bacterial clearance from lungs was enhanced due to furanones (151).
A C. violaceum-based screen for QSI compounds used natural and chemically synthesized furanones and aimed at identifying compounds that either inhibited or enhanced quorum-sensing-dependent behavior (87). The authors designed a microtiter dish-based assay that would differentiate between compounds that affect growth, activate quorum sensing in C. violaceum strain CV026, inhibit violacein formation induced by the cognate AHL, or enhance violacein formation in the presence of AHL. They found that some furanones were toxic at a higher concentration but inhibited quorum sensing at a much lower nontoxic concentration. Several compounds were found to enhance quorum sensing at suboptimal C6-HSL concentrations but were antagonistic when optimal concentrations of C6-HSL were present. This study reflects on the variability of the activity of a given compound based on its concentration as well as the availability of quorum-sensing-activating AHLs (87).
CONCLUSION AND PERSPECTIVES
Quorum-sensing-interfering compounds either produced naturally by certain eukaryotic hosts, synthesized by chemical methods, or produced by creating transgenic plants all have either a positive or a negative effect on the expression of bacterial phenotypes regulated by quorum sensing.
Synthetic analogs that function as QSI compounds and either enhance or inhibit quorum-sensing behavior could have potential clinical applications. Two issues appear to be worth considering while evaluating synthetic compounds for QSI activity. First, as shown by Zhu et al., overexpression of the response regulator gives false reactions, as in the case of TraR, which, when overexpressed, detected several antagonistic compounds as agonists (157). Hence, wild-type levels of response regulator should be used for the screening. Second, these compounds should probably be analyzed over a range of concentrations, as a compound acting as an antagonist at higher concentrations may actually function as an agonist when provided at a lower concentration (87).
Creation of transgenic plants by cloning AHL synthases or lactonases holds promise for preventing plant diseases (24). At the same time, a more thorough testing of the effects on the competing ecological flora needs to be considered. Production of AHLs or lactonases may enable the plant to ward off potential attacks, but since this also targets the quorum-sensing systems of the other potentially beneficial flora in soil, it may lead to other repercussions. In addition, in the case of transgenic plants belonging to the edible group, additional caution needs to be exercised, as AHLs seem to persist even in older tissues (35). Strategies to inactivate the transgenic enzymes and their accumulated products before marketing might be worth investigating. One final consideration is that the general suppression of all the AHL-dependent bacteria may result in an unbalanced encouragement of bacteria that use other types of autoinducers for quorum sensing, some of which may actually be pathogenic to the host.
Several host plants have been shown to produce compounds that influence the quorum-sensing-regulated behavior of associated bacteria. It is possible that bacteria have developed mechanisms to counteract the eukaryotic interference. This could explain the presence of multiple types of autoinducer signals made by a single bacterium to regulate common phenotypes (91, 93). Most of the QSI compounds identified so far appear to be targeted towards AHL-dependent quorum sensing. An investigation of QSI compounds directed towards other type of autoinducer molecules could provide us with potential tools against bacteria using multiple types of autoinducers to regulate a single phenotype, as seen in the case of V. harveyi, P. aeruginosa, and others (49, 93).
In conclusion, the study of bacterial quorum sensing has suggested several ideal targets for drug design as well as agricultural and industrial applications. In addition, eukaryotes seem to have evolved efficient mechanisms to manipulate bacterial quorum sensing and thereby protect themselves from pathogenic attack. Parallel to this, certain bacteria have evolved mechanisms to fine-tune gene regulation by incorporating eukaryotic signal molecules in the superregulation of their quorum-sensing behavior. This eukaryote-bacterial cross talk could be exploited to model manipulative techniques that interfere with bacterial quorum sensing. Further investigation on the quorum-sensing systems and eukaryote-bacterial cross talk involving autoinducer signal molecules other than AHLs could reveal additional mechanisms for messing with bacterial quorum sensing.
Acknowledgments
We thank the members of our laboratory for helpful discussions and critical reading of the manuscript.
The work in our laboratory was supported by the Texas Advanced Research Program under grant 0009741-0022-2001 to J.E.G., by National Science Foundation grant MCB-9733532 to J.E.G., and by National Institutes of Health grant 1R01GM069925 to J.E.G.
REFERENCES
- 1.Aendekerk, S., B. Ghysels, P. Cornelis, and C. Baysse. 2002. Characterization of a new efflux pump, MexGHI-OpmD, from Pseudomonas aeruginosa that confers resistance to vanadium. Microbiology 148:2371-2381. [DOI] [PubMed] [Google Scholar]
- 2.Barber, C. E., J. L. Tang, J. X. Feng, M. Q. Pan, T. J. Wilson, H. Slater, J. M. Dow, P. Williams, and M. J. Daniels. 1997. A novel regulatory system required for pathogenicity of Xanthomonas campestris is mediated by a small diffusible signal molecule. Mol. Microbiol. 24:555-566. [DOI] [PubMed] [Google Scholar]
- 3.Bassler, B. L. 1999. How bacteria talk to each other: regulation of gene expression by quorum sensing. Curr. Opin. Microbiol. 2:582-587. [DOI] [PubMed] [Google Scholar]
- 4.Bassler, B. L. 2002. Small talk. Cell-to-cell communication in bacteria. Cell 109:421-424. [DOI] [PubMed] [Google Scholar]
- 5.Bassler, B. L., M. Wright, R. E. Showalter, and M. R. Silverman. 1993. Intercellular signaling in Vibrio harveyi: sequence and function of genes regulating expression of luminescence. Mol. Microbiol. 9:773-786. [DOI] [PubMed] [Google Scholar]
- 6.Bassler, B. L., M. Wright, and M. R. Silverman. 1994. Multiple signalling systems controlling expression of luminescence in Vibrio harveyi: sequence and function of genes encoding a second sensory pathway. Mol. Microbiol. 13:273-286. [DOI] [PubMed] [Google Scholar]
- 7.Bence, A. K., and P. A. Crooks. 2003. The mechanism of l-canavanine cytotoxicity: arginyl tRNA synthetase as a novel target for anticancer drug discovery. J. Enzyme Inhib. Med. Chem. 18:383-394. [DOI] [PubMed] [Google Scholar]
- 8.Bollinger, N., D. J. Hassett, B. H. Iglewski, J. W. Costerton, and T. R. McDermott. 2001. Gene expression in Pseudomonas aeruginosa: evidence of iron override effects on quorum sensing and biofilm-specific gene regulation. J. Bacteriol. 183:1990-1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carlier, A., R. Chevrot, Y. Dessaux, and D. Faure. 2004. The assimilation of gamma-butyrolactone in Agrobacterium tumefaciens C58 interferes with the accumulation of the N-acyl-homoserine lactone signal. Mol. Plant-Microbe Interact. 17:951-957. [DOI] [PubMed] [Google Scholar]
- 10.Carlier, A., S. Uroz, B. Smadja, R. Fray, X. Latour, Y. Dessaux, and D. Faure. 2003. The Ti plasmid of Agrobacterium tumefaciens harbors an attM-paralogous gene, aiiB, also encoding N-acylhomoserine lactonase activity. Appl. Environ. Microbiol. 69:4989-49893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chai, Y., J. Zhu, and S. C. Winans. 2001. TrlR, a defective TraR-like protein of Agrobacterium tumefaciens, blocks TraR function in vitro by forming inactive TrlR:TraR dimers. Mol. Microbiol. 40:414-421. [DOI] [PubMed] [Google Scholar]
- 12.Chen, G., J. W. Malenkos, M. R. Cha, C. Fuqua, and L. Chen. 2004. Quorum-sensing antiactivator TraM forms a dimer that dissociates to inhibit TraR. Mol. Microbiol. 52:1641-1651. [DOI] [PubMed] [Google Scholar]
- 13.Chen, X., S. Schauder, N. Potier, A. Van Dorsselaer, I. Pelczer, B. L. Bassler, and F. M. Hughson. 2002. Structural identification of a bacterial quorum-sensing signal containing boron. Nature 415:545-549. [DOI] [PubMed] [Google Scholar]
- 14.Chugani, S. A., M. Whiteley, K. M. Lee, D. D'Argenio, C. Manoil, and E. P. Greenberg. 2001. QscR, a modulator of quorum-sensing signal synthesis and virulence in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 98:2752-2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Costa, J. M., and J. E. Loper. 1997. EcbI and EcbR: homologs of LuxI and LuxR affecting antibiotic and exoenzyme production by Erwinia carotovora subsp. betavasculorum. Can. J. Microbiol. 43:1164-1171. [DOI] [PubMed] [Google Scholar]
- 16.Crespi, M., and S. Galvez. 2000. Molecular mechanisms in root nodule development. J. Plant Growth Regul. 19:155-166. [DOI] [PubMed] [Google Scholar]
- 17.Cui, Y., A. Chatterjee, Y. Liu, C. K. Dumenyo, and A. K. Chatterjee. 1995. Identification of a global repressor gene, rsmA, of Erwinia carotovora subsp. carotovora that controls extracellular enzymes, N-(3-oxohexanoyl)-l-homoserine lactone, and pathogenicity in soft-rotting Erwinia spp. J. Bacteriol. 177:5108-5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Daniels, R., J. Vanderleyden, and J. Michiels. 2004. Quorum sensing and swarming migration in bacteria. FEMS Microbiol. Rev. 28:261-289. [DOI] [PubMed] [Google Scholar]
- 19.Day, W. A., Jr., and A. T. Maurelli. 2001. Shigella flexneri LuxS quorum-sensing system modulates virB expression but is not essential for virulence. Infect. Immun. 69:15-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Degrassi, G., C. Aguilar, M. Bosco, S. Zahariev, S. Pongor, and V. Venturi. 2002. Plant growth-promoting Pseudomonas putida WCS358 produces and secretes four cyclic dipeptides: cross-talk with quorum sensing bacterial sensors. Curr. Microbiol. 45:250-254. [DOI] [PubMed] [Google Scholar]
- 21.de Kievit, T. R., and B. H. Iglewski. 2000. Bacterial quorum sensing in pathogenic relationships. Infect. Immun. 68:4839-4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DeLisa, M. P., C. F. Wu, L. Wang, J. J. Valdes, and W. E. Bentley. 2001. DNA microarray-based identification of genes controlled by autoinducer 2-stimulated quorum sensing in Escherichia coli. J. Bacteriol. 183:5239-5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Devine, J. H., G. S. Shadel, and T. O. Baldwin. 1989. Identification of the operator of the lux regulon from the Vibrio fischeri strain ATCC7744. Proc. Natl. Acad. Sci. USA 86:5688-5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dong, Y. H., L. H. Wang, J. L. Xu, H. B. Zhang, X. F. Zhang, and L. H. Zhang. 2001. Quenching quorum-sensing-dependent bacterial infection by an N-acyl homoserine lactonase. Nature 411:813-817. [DOI] [PubMed] [Google Scholar]
- 25.Dong, Y. H., J. L. Xu, X. Z. Li, and L. H. Zhang. 2000. AiiA, an enzyme that inactivates the acylhomoserine lactone quorum-sensing signal and attenuates the virulence of Erwinia carotovora. Proc. Natl. Acad. Sci. USA 97:3526-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dunlap, P. V. 1999. Quorum regulation of luminescence in Vibrio fischeri. J. Mol. Microbiol. Biotechnol. 1:5-12. [PubMed] [Google Scholar]
- 27.Dunn, A. K., and J. Handelsman. 2002. Toward an understanding of microbial communities through analysis of communication networks. Antonie Leeuwenhoek 81:565-574. [DOI] [PubMed] [Google Scholar]
- 28.Eberhard, A., A. L. Burlingame, C. Eberhard, G. L. Kenyon, K. H. Nealson, and N. J. Oppenheimer. 1981. Structural identification of autoinducer of Photobacterium fischeri luciferase. Biochemistry 20:2444-2449. [DOI] [PubMed] [Google Scholar]
- 29.Egland, K. A., and E. P. Greenberg. 1999. Quorum sensing in Vibrio fischeri: elements of the luxl promoter. Mol. Microbiol. 31:1197-1204. [DOI] [PubMed] [Google Scholar]
- 30.Elasri, M., S. Delorme, P. Lemanceau, G. Stewart, B. Laue, E. Glickmann, P. M. Oger, and Y. Dessaux. 2001. Acyl-homoserine lactone production is more common among plant-associated Pseudomonas spp. than among soilborne Pseudomonas spp. Appl. Environ. Microbiol. 67:1198-1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Emmert, E. A. B., J. L. Milner, J. C. Lee, K. L. Pulvermacher, H. A. Olivares, J. Clardy, and J. Handelsman. 1998. Effect of canavanine from alfalfa seeds on the population biology of Bacillus cereus. Appl. Environ. Microbiol. 64:4683-4688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Engebrecht, J., K. Nealson, and M. Silverman. 1983. Bacterial bioluminescence: isolation and genetic analysis of functions from Vibrio fischeri. Cell 32:773-781. [DOI] [PubMed] [Google Scholar]
- 33.Engebrecht, J., and M. Silverman. 1984. Identification of genes and gene products necessary for bacterial bioluminescence. Proc. Natl. Acad. Sci. USA 81:4154-4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Farrand, S. K., Y. Qin, and P. Oger. 2002. Quorum-sensing system of Agrobacterium plasmids: analysis and utility. Methods Enzymol. 358:452-484. [DOI] [PubMed] [Google Scholar]
- 35.Fray, R. G., J. P. Throup, M. Daykin, A. Wallace, P. Williams, G. S. Stewart, and D. Grierson. 1999. Plants genetically modified to produce N-acylhomoserine lactones communicate with bacteria. Nat. Biotechnol. 17:1017-1020. [DOI] [PubMed] [Google Scholar]
- 36.Fuqua, C., M. R. Parsek, and E. P. Greenberg. 2001. Regulation of gene expression by cell-to-cell communication: acyl-homoserine lactone quorum sensing. Annu. Rev. Genet. 35:439-468. [DOI] [PubMed] [Google Scholar]
- 37.Fuqua, C., and S. C. Winans. 1996. Conserved cis-acting promoter elements are required for density-dependent transcription of Agrobacterium tumefaciens conjugal transfer genes. J. Bacteriol. 178:435-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gao, M., M. Teplitski, J. B. Robinson, and W. D. Bauer. 2003. Production of substances by Medicago truncatula that affect bacterial quorum sensing. Mol. Plant-Microbe Interact. 16:827-834. [DOI] [PubMed] [Google Scholar]
- 39.Gilson, L., A. Kuo, and P. V. Dunlap. 1995. AinS and a new family of autoinducer synthesis proteins. J. Bacteriol. 177:6946-6951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Givskov, M., R. de Nys, M. Manefield, L. Gram, R. Maximilien, L. Eberl, S. Molin, P. D. Steinberg, and S. Kjelleberg. 1996. Eukaryotic interference with homoserine lactone-mediated prokaryotic signalling. J. Bacteriol. 178:6618-6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.González, J. E., and M. M. Marketon. 2003. Quorum sensing in nitrogen-fixing rhizobia. Microbiol. Mol. Biol. Rev. 67:574-592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hanzelka, B. L., and E. P. Greenberg. 1996. Quorum sensing in Vibrio fischeri: evidence that S-adenosylmethionine is the amino acid substrate for autoinducer synthesis. J. Bacteriol. 178:5291-5294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hanzelka, B. L., M. R. Parsek, D. L. Val, P. V. Dunlap, J. E. Cronan, Jr., and E. P. Greenberg. 1999. Acylhomoserine lactone synthase activity of the Vibrio fischeri AinS protein. J. Bacteriol. 181:5766-5770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Henke, J. M., and B. L. Bassler. 2004. Bacterial social engagements. Trends Cell Biol. 14:648-656. [DOI] [PubMed] [Google Scholar]
- 45.Henke, J. M., and B. L. Bassler. 2004. Three parallel quorum-sensing systems regulate gene expression in Vibrio harveyi. J. Bacteriol. 186:6902-6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hilgers, M. T., and M. L. Ludwig. 2001. Crystal structure of the quorum-sensing protein LuxS reveals a catalytic metal site. Proc. Natl. Acad. Sci. USA 98:11169-11174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hjelmgaard, T., T. Persson, T. B. Rasmussen, M. Givskov, and J. Nielsen. 2003. Synthesis of furanone-based natural product analogues with quorum sensing antagonist activity. Bioorg. Med. Chem. 11:3261-3271. [DOI] [PubMed] [Google Scholar]
- 48.Hoang, H. H., A. Becker, and J. E. González. 2004. The LuxR homolog ExpR, in combination with the Sin quorum-sensing system, plays a central role in Sinorhizobium meliloti gene expression. J. Bacteriol. 186:5460-5472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Holden, M. T., S. Ram Chhabra, R. de Nys, P. Stead, N. J. Bainton, P. J. Hill, M. Manefield, N. Kumar, M. Labatte, D. England, S. Rice, M. Givskov, G. P. Salmond, G. S. Stewart, B. W. Bycroft, S. Kjelleberg, and P. Williams. 1999. Quorum-sensing cross talk: isolation and chemical characterization of cyclic dipeptides from Pseudomonas aeruginosa and other Gram-negative bacteria. Mol. Microbiol. 33:1254-1266. [DOI] [PubMed] [Google Scholar]
- 50.Huang, J. J., J. I. Han, L. H. Zhang, and J. R. Leadbetter. 2003. Utilization of acyl-homoserine lactone quorum signals for growth by a soil pseudomonad and Pseudomonas aeruginosa PAO1. Appl. Environ. Microbiol. 69:5941-5949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hwang, I., A. J. Smyth, Z. Q. Luo, and S. K. Farrand. 1999. Modulating quorum sensing by antiactivation: TraM interacts with TraR to inhibit activation of Ti plasmid conjugal transfer genes. Mol. Microbiol. 34:282-294. [DOI] [PubMed] [Google Scholar]
- 52.Juhas, M., L. Eberl, and B. Tummler. 2005. Quorum sensing: the power of cooperation in the world of Pseudomonas. Environ. Microbiol. 7:459-471. [DOI] [PubMed] [Google Scholar]
- 53.Kaplan, H. B., and E. P. Greenberg. 1985. Diffusion of autoinducer is involved in regulation of the Vibrio fischeri luminescence system. J. Bacteriol. 163:1210-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Keshavan, N. D., P. K. Chowdhary, D. C. Haines, and J. E. González. 2005. l-Canavanine made by Medicago sativa interferes with quorum sensing in Sinorhizobium meliloti. J. Bacteriol. 187:8427-8436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Khan, S. R., D. V. Mavrodi, G. J. Jog, H. Suga, L. S. Thomashow, and S. K. Farrand. 2005. Activation of the phz operon of Pseudomonas fluorescens 2-79 requires the LuxR homolog PhzR, N-(3-OH-hexanoyl)-l-homoserine lactone produced by the LuxI homolog PhzI, and a cis-acting phz box. J. Bacteriol. 187:6517-6527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Koch, B., T. Liljefors, T. Persson, J. Nielsen, S. Kjelleberg, and M. Givskov. 2005. The LuxR receptor: the sites of interaction with quorum-sensing signals and inhibitors. Microbiology 151:3589-3602. [DOI] [PubMed] [Google Scholar]
- 57.Labbate, M., S. Y. Queck, K. S. Koh, S. A. Rice, M. Givskov, and S. Kjelleberg. 2004. Quorum sensing-controlled biofilm development in Serratia liquefaciens MG1. J. Bacteriol. 186:692-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Laue, B. E., Y. Jiang, S. R. Chhabra, S. Jacob, G. S. Stewart, A. Hardman, J. A. Downie, F. O'Gara, and P. Williams. 2000. The biocontrol strain Pseudomonas fluorescens F113 produces the Rhizobium small bacteriocin, N-(3-hydroxy-7-cis-tetradecenoyl)homoserine lactone, via HdtS, a putative novel N-acylhomoserine lactone synthase. Microbiology 146:2469-2480. [DOI] [PubMed] [Google Scholar]
- 59.Lawhon, S. D., J. G. Frye, M. Suyemoto, S. Porwollik, M. McClelland, and C. Altier. 2003. Global regulation by CsrA in Salmonella typhimurium. Mol. Microbiol. 48:1633-1645. [DOI] [PubMed] [Google Scholar]
- 60.Leadbetter, J. R., and E. P. Greenberg. 2000. Metabolism of acyl-homoserine lactone quorum-sensing signals by Variovorax paradoxus. J. Bacteriol. 182:6921-6926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ledgham, F., I. Ventre, C. Soscia, M. Foglino, J. N. Sturgis, and A. Lazdunski. 2003. Interactions of the quorum sensing regulator QscR: interaction with itself and the other regulators of Pseudomonas aeruginosa LasR and RhlR. Mol. Microbiol. 48:199-210. [DOI] [PubMed] [Google Scholar]
- 62.Lee, J. H., Y. Lequette, and E. P. Greenberg. 2006. Activity of purified QscR, a Pseudomonas aeruginosa orphan quorum-sensing transcription factor. Mol. Microbiol. 59:602-609. [DOI] [PubMed] [Google Scholar]
- 63.Lee, K. H., and E. G. Ruby. 1994. Effect of the squid host on the abundance and distribution of symbiotic Vibrio fischeri in nature. Appl. Environ. Microbiol. 60:1565-1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lenz, D. H., K. C. Mok, B. N. Lilley, R. V. Kulkarni, N. S. Wingreen, and B. L. Bassler. 2004. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell 118:69-82. [DOI] [PubMed] [Google Scholar]
- 65.Lewenza, S., B. Conway, E. P. Greenberg, and P. A. Sokol. 1999. Quorum sensing in Burkholderia cepacia: identification of the LuxRI homologs CepRI. J. Bacteriol. 181:748-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lewis, H. A., E. B. Furlong, B. Laubert, G. A. Eroshkina, Y. Batiyenko, J. M. Adams, M. G. Bergseid, C. D. Marsh, T. S. Peat, W. E. Sanderson, J. M. Sauder, and S. G. Buchanan. 2001. A structural genomics approach to the study of quorum sensing: crystal structures of three LuxS orthologs. Structure 9:527-537. [DOI] [PubMed] [Google Scholar]
- 67.Li, Y. H., N. Tang, M. B. Aspiras, P. C. Lau, J. H. Lee, R. P. Ellen, and D. G. Cvitkovitch. 2002. A quorum-sensing signaling system essential for genetic competence in Streptococcus mutans is involved in biofilm formation. J. Bacteriol. 184:2699-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lindum, P. W., U. Anthoni, C. Christophersen, L. Eberl, S. Molin, and M. Givskov. 1998. N-Acyl-l-homoserine lactone autoinducers control production of an extracellular lipopeptide biosurfactant required for swarming motility of Serratia liquefaciens MG1. J. Bacteriol. 180:6384-6388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu, D., B. W. Lepore, G. A. Petsko, P. W. Thomas, E. M. Stone, W. Fast, and D. Ringe. 2005. Three-dimensional structure of the quorum-quenching N-acyl homoserine lactone hydrolase from Bacillus thuringiensis. Proc. Natl. Acad. Sci. USA 102:11882-11887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Llamas, I., N. Keshavan, and J. E. González. 2004. Use of Sinorhizobium meliloti as an indicator for specific detection of long-chain N-acyl homoserine lactones. Appl. Environ. Microbiol. 70:3715-3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Loh, J., R. W. Carlson, W. S. York, and G. Stacey. 2002. Bradyoxetin, a unique chemical signal involved in symbiotic gene regulation. Proc. Natl. Acad. Sci. USA 99:14446-14451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Loh, J., D. P. Lohar, B. Andersen, and G. Stacey. 2002. A two-component regulator mediates population-density-dependent expression of the Bradyrhizobium japonicum nodulation genes. J. Bacteriol. 184:1759-1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Loh, J., E. A. Pierson, L. S. Pierson III, G. Stacey, and A. Chatterjee. 2002. Quorum sensing in plant-associated bacteria. Curr. Opin. Plant Biol. 5:285-290. [DOI] [PubMed] [Google Scholar]
- 74.Loh, J. T., J. P. Yuen-Tsai, M. G. Stacey, D. Lohar, A. Welborn, and G. Stacey. 2001. Population density-dependent regulation of the Bradyrhizobium japonicum nodulation genes. Mol. Microbiol. 42:37-46. [DOI] [PubMed] [Google Scholar]
- 75.Long, S. R. 1996. Rhizobium symbiosis: nod factors in perspective. Plant Cell 8:1885-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Losick, R., and D. Kaiser. 1997. Why and how bacteria communicate. Sci. Am. 276:68-73. [DOI] [PubMed] [Google Scholar]
- 77.Luo, Z. Q., Y. Qin, and S. K. Farrand. 2000. The antiactivator TraM interferes with the autoinducer-dependent binding of TraR to DNA by interacting with the C-terminal region of the quorum-sensing activator. J. Biol. Chem. 275:7713-7722. [DOI] [PubMed] [Google Scholar]
- 78.Lupp, C., M. Urbanowski, E. P. Greenberg, and E. G. Ruby. 2003. The Vibrio fischeri quorum-sensing systems ain and lux sequentially induce luminescence gene expression and are important for persistence in the squid host. Mol. Microbiol. 50:319-331. [DOI] [PubMed] [Google Scholar]
- 79.Lyon, G. J., and R. P. Novick. 2004. Peptide signaling in Staphylococcus aureus and other Gram-positive bacteria. Peptides 25:1389-1403. [DOI] [PubMed] [Google Scholar]
- 80.Mäe, A., M. Montesano, V. Koiv, and E. T. Palva. 2001. Transgenic plants producing the bacterial pheromone N-acyl-homoserine lactone exhibit enhanced resistance to the bacterial phytopathogen Erwinia carotovora. Mol. Plant-Microbe Interact. 14:1035-1042. [DOI] [PubMed] [Google Scholar]
- 81.Manefield, M., R. de Nys, N. Kumar, R. Read, M. Givskov, P. Steinberg, and S. Kjelleberg. 1999. Evidence that halogenated furanones from Delisea pulchra inhibit acylated homoserine lactone (AHL)-mediated gene expression by displacing the AHL signal from its receptor protein. Microbiology 145:283-291. [DOI] [PubMed] [Google Scholar]
- 82.Manefield, M., T. B. Rasmussen, M. Henzter, J. B. Andersen, P. Steinberg, S. Kjelleberg, and M. Givskov. 2002. Halogenated furanones inhibit quorum sensing through accelerated LuxR turnover. Microbiology 148:1119-1127. [DOI] [PubMed] [Google Scholar]
- 83.Marketon, M. M., S. A. Glenn, A. Eberhard, and J. E. González. 2003. Quorum sensing controls exopolysaccharide production in Sinorhizobium meliloti. J. Bacteriol. 185:325-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Marketon, M. M., and J. E. González. 2002. Identification of two quorum-sensing systems in Sinorhizobium meliloti. J. Bacteriol. 184:3466-3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Marketon, M. M., M. R. Gronquist, A. Eberhard, and J. E. González. 2002. Characterization of the Sinorhizobium meliloti sinR/sinI locus and the production of novel N-acyl homoserine lactones. J. Bacteriol. 184:5686-5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Marketon, M. M., I. Llamas, M. R. Gronquist, A. Eberhard, and J. E. González. 2006. Unpublished data.
- 87.Martinelli, D., G. Grossmann, U. Sequin, H. Brandl, and R. Bachofen. 2004. Effects of natural and chemically synthesized furanones on quorum sensing in Chromobacterium violaceum. BMC Microbiol. 4:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.McClean, K. H., M. K. Winson, L. Fish, A. Taylor, S. R. Chhabra, M. Camara, M. Daykin, J. H. Lamb, S. Swift, B. W. Bycroft, G. S. Stewart, and P. Williams. 1997. Quorum sensing and Chromobacterium violaceum: exploitation of violacein production and inhibition for the detection of N-acyl homoserine lactones. Microbiology 143:3703-3711. [DOI] [PubMed] [Google Scholar]
- 89.McLean, R. J., L. S. Pierson III, and C. Fuqua. 2004. A simple screening protocol for the identification of quorum signal antagonists. J. Microbiol. Methods 58:351-360. [DOI] [PubMed] [Google Scholar]
- 90.McNab, R., S. K. Ford, A. El-Sabaeny, B. Barbieri, G. S. Cook, and R. J. Lamont. 2003. LuxS-based signaling in Streptococcus gordonii: autoinducer 2 controls carbohydrate metabolism and biofilm formation with Porphyromonas gingivalis. J. Bacteriol. 185:274-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Miller, M. B., K. Skorupski, D. H. Lenz, R. K. Taylor, and B. L. Bassler. 2002. Parallel quorum sensing systems converge to regulate virulence in Vibrio cholerae. Cell 110:303-314. [DOI] [PubMed] [Google Scholar]
- 92.Miller, S. T., K. B. Xavier, S. R. Campagna, M. E. Taga, M. F. Semmelhack, B. L. Bassler, and F. M. Hughson. 2004. Salmonella typhimurium recognizes a chemically distinct form of the bacterial quorum-sensing signal AI-2. Mol. Cell 15:677-687. [DOI] [PubMed] [Google Scholar]
- 93.Mok, K. C., N. S. Wingreen, and B. L. Bassler. 2003. Vibrio harveyi quorum sensing: a coincidence detector for two autoinducers controls gene expression. EMBO J. 22:870-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Moré, M. I., L. D. Finger, J. L. Stryker, A. C. Fuqua, A. Eberhard, and S. C. Winans. 1996. Enzymatic synthesis of a quorum-sensing autoinducer through use of defined substrates. Science 272:1655-1658. [DOI] [PubMed] [Google Scholar]
- 95.Nealson, K. H., and J. W. Hastings. 1979. Bacterial bioluminescence: its control and ecological significance. Microbiol. Rev. 43:496-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nealson, K. H., T. Platt, and J. W. Hastings. 1970. Cellular control of the synthesis and activity of the bacterial luminescent system. J. Bacteriol. 104:313-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Neiditch, M. B., M. J. Federle, S. T. Miller, B. L. Bassler, and F. M. Hughson. 2005. Regulation of LuxPQ receptor activity by the quorum-sensing signal autoinducer-2. Mol. Cell 18:507-518. [DOI] [PubMed] [Google Scholar]
- 98.Oger, P., and S. K. Farrand. 2001. Co-evolution of the agrocinopine opines and the agrocinopine-mediated control of TraR, the quorum-sensing activator of the Ti plasmid conjugation system. Mol. Microbiol. 41:1173-1185. [DOI] [PubMed] [Google Scholar]
- 99.Oger, P., and S. K. Farrand. 2002. Two opines control conjugal transfer of an Agrobacterium plasmid by regulating expression of separate copies of the quorum-sensing activator gene traR. J. Bacteriol. 184:1121-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Oger, P., K. S. Kim, R. L. Sackett, K. R. Piper, and S. K. Farrand. 1998. Octopine-type Ti plasmids code for a mannopine-inducible dominant-negative allele of traR, the quorum-sensing activator that regulates Ti plasmid conjugal transfer. Mol. Microbiol. 27:277-288. [DOI] [PubMed] [Google Scholar]
- 101.Ohtani, K., H. Hayashi, and T. Shimizu. 2002. The luxS gene is involved in cell-cell signalling for toxin production in Clostridium perfringens. Mol. Microbiol. 44:171-179. [DOI] [PubMed] [Google Scholar]
- 102.Olsen, J. A., R. Severinsen, T. B. Rasmussen, M. Hentzer, M. Givskov, and J. Nielsen. 2002. Synthesis of new 3- and 4-substituted analogues of acyl homoserine lactone quorum sensing autoinducers. Bioorg. Med. Chem. Lett. 12:325-328. [DOI] [PubMed] [Google Scholar]
- 103.Palmer, J. L., and R. H. Abeles. 1979. The mechanism of action of S-adenosylhomocysteinase. J. Biol. Chem. 254:1217-1226. [PubMed] [Google Scholar]
- 104.Pearson, J. P., C. Van Delden, and B. H. Iglewski. 1999. Active efflux and diffusion are involved in transport of Pseudomonas aeruginosa cell-to-cell signals. J. Bacteriol. 181:1203-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Perret, X., C. Staehelin, and W. J. Broughton. 2000. Molecular basis of symbiotic promiscuity. Microbiol. Mol. Biol. Rev. 64:180-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Pessi, G., F. Williams, Z. Hindle, K. Heurlier, M. T. Holden, M. Camara, D. Haas, and P. Williams. 2001. The global posttranscriptional regulator RsmA modulates production of virulence determinants and N-acylhomoserine lactones in Pseudomonas aeruginosa. J. Bacteriol. 183:6676-6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Piper, K. R., S. Beck Von Bodman, I. Hwang, and S. K. Farrand. 1999. Hierarchical gene regulatory systems arising from fortuitous gene associations: controlling quorum sensing by the opine regulon in Agrobacterium. Mol. Microbiol. 32:1077-1089. [DOI] [PubMed] [Google Scholar]
- 108.Piper, K. R., and S. K. Farrand. 2000. Quorum sensing but not autoinduction of Ti plasmid conjugal transfer requires control by the opine regulon and the antiactivator TraM. J. Bacteriol. 182:1080-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Podbielski, A., and B. Kreikemeyer. 2004. Cell density-dependent regulation: basic principles and effects on the virulence of Gram-positive cocci. Int. J. Infect. Dis. 8:81-95. [DOI] [PubMed] [Google Scholar]
- 110.Qin, Y., A. J. Smyth, S. Su, and S. K. Farrand. 2004. Dimerization properties of TraM, the antiactivator that modulates TraR-mediated quorum-dependent expression of the Ti plasmid tra genes. Mol. Microbiol. 53:1471-1485. [DOI] [PubMed] [Google Scholar]
- 111.Quinones, B., G. Dulla, and S. E. Lindow. 2005. Quorum sensing regulates exopolysaccharide production, motility, and virulence in Pseudomonas syringae. Mol. Plant-Microbe Interact. 18:682-693. [DOI] [PubMed] [Google Scholar]
- 112.Rasmussen, T. B., M. E. Skindersoe, T. Bjarnsholt, R. K. Phipps, K. B. Christensen, P. O. Jensen, J. B. Andersen, B. Koch, T. O. Larsen, M. Hentzer, L. Eberl, N. Hoiby, and M. Givskov. 2005. Identity and effects of quorum-sensing inhibitors produced by Penicillium species. Microbiology 151:1325-1340. [DOI] [PubMed] [Google Scholar]
- 113.Rasmussen, T. B., T. Bjarnsholt, M. E. Skindersoe, M. Hentzer, P. Kristoffersen, M. Kote, J. Nielsen, L. Eberl, and M. Givskov. 2005. Screening for quorum-sensing inhibitors (QSI) by use of a novel genetic system, the QSI selector. J. Bacteriol. 187:1799-1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Redfield, R. J. 2002. Is quorum sensing a side effect of diffusion sensing? Trends Microbiol. 10:365-370. [DOI] [PubMed] [Google Scholar]
- 115.Ren, D., J. J. Sims, and T. K. Wood. 2001. Inhibition of biofilm formation and swarming of Escherichia coli by (5Z)-4-bromo-5-(bromomethylene)-3-butyl-2(5H)-furanone. Environ. Microbiol. 3:731-736. [DOI] [PubMed] [Google Scholar]
- 116.Ren, D., R. Zuo, A. F. González Barrios, L. A. Bedzyk, G. R. Eldridge, M. E. Pasmore, and T. K. Wood. 2005. Differential gene expression for investigation of Escherichia coli biofilm inhibition by plant extract ursolic acid. Appl. Environ. Microbiol. 71:4022-4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Reverchon, S., B. Chantegrel, C. Deshayes, A. Doutheau, and N. Cotte-Pattat. 2002. New synthetic analogues of N-acyl homoserine lactones as agonists or antagonists of transcriptional regulators involved in bacterial quorum sensing. Bioorg. Med. Chem. Lett. 12:1153-1157. [DOI] [PubMed] [Google Scholar]
- 118.Rice, S. A., K. S. Koh, S. Y. Queck, M. Labbate, K. W. Lam, and S. Kjelleberg. 2005. Biofilm formation and sloughing in Serratia marcescens are controlled by quorum sensing and nutrient cues. J. Bacteriol. 187:3477-3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Rosenthal, G. A. 2001. l-Canavanine: a higher plant insecticidal allelochemical. Amino Acids 21:319-330. [DOI] [PubMed] [Google Scholar]
- 120.Ruzheinikov, S. N., S. K. Das, S. E. Sedelnikova, A. Hartley, S. J. Foster, M. J. Horsburgh, A. G. Cox, C. W. McCleod, A. Mekhalfia, G. M. Blackburn, D. W. Rice, and P. J. Baker. 2001. The 1.2 Å structure of a novel quorum-sensing protein, Bacillus subtilis LuxS. J. Mol. Biol. 313:111-122. [DOI] [PubMed] [Google Scholar]
- 121.Scarlato, V., B. Arico, A. Prugnola, and R. Rappuoli. 1991. Sequential activation and environmental regulation of virulence genes in Bordetella pertussis. EMBO J. 10:3971-3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Schaefer, A. L., D. L. Val, B. L. Hanzelka, J. E. Cronan, Jr., and E. P. Greenberg. 1996. Generation of cell-to-cell signals in quorum sensing: acyl homoserine lactone synthase activity of a purified Vibrio fischeri LuxI protein. Proc. Natl. Acad. Sci. USA 93:9505-9509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Schripsema, J., K. E. de Rudder, T. B. van Vliet, P. P. Lankhorst, E. de Vroom, J. W. Kijne, and A. A. van Brussel. 1996. Bacteriocin small of Rhizobium leguminosarum belongs to the class of N-acyl-l-homoserine lactone molecules, known as autoinducers and as quorum sensing cotranscription factors. J. Bacteriol. 178:366-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Shadel, G. S., R. Young, and T. O. Baldwin. 1990. Use of regulated cell lysis in a lethal genetic selection in Escherichia coli: identification of the autoinducer-binding region of the LuxR protein from Vibrio fischeri ATCC 7744. J. Bacteriol. 172:3980-3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Shaw, P. D., G. Ping, S. L. Daly, C. Cha, J. E. Cronan, Jr., K. L. Rinehart, and S. K. Farrand. 1997. Detecting and characterizing N-acyl-homoserine lactone signal molecules by thin-layer chromatography. Proc. Natl. Acad. Sci. USA 94:6036-6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sircili, M. P., M. Walters, L. R. Trabulsi, and V. Sperandio. 2004. Modulation of enteropathogenic Escherichia coli virulence by quorum sensing. Infect. Immun. 72:2329-2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Skwierczynski, R. D., and K. A. Connors. 1993. Demethylation kinetics of aspartame and l-phenylalanine methyl ester in aqueous solution. Pharm. Res. 10:1174-1180. [DOI] [PubMed] [Google Scholar]
- 128.Slock, J., D. VanRiet, D. Kolibachuk, and E. P. Greenberg. 1990. Critical regions of the Vibrio fischeri LuxR protein defined by mutational analysis. J. Bacteriol. 172:3974-3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Spaink, H. P. 2000. Root nodulation and infection factors produced by rhizobial bacteria. Annu. Rev. Microbiol. 54:257-288. [DOI] [PubMed] [Google Scholar]
- 130.Sperandio, V., J. L. Mellies, W. Nguyen, S. Shin, and J. B. Kaper. 1999. Quorum sensing controls expression of the type III secretion gene transcription and protein secretion in enterohemorrhagic and enteropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 96:15196-15201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Sperandio, V., A. G. Torres, J. A. Giron, and J. B. Kaper. 2001. Quorum sensing is a global regulatory mechanism in enterohemorrhagic Escherichia coli O157:H7. J. Bacteriol. 183:5187-5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Sperandio, V., A. G. Torres, B. Jarvis, J. P. Nataro, and J. B. Kaper. 2003. Bacteria-host communication: the language of hormones. Proc. Natl. Acad. Sci. USA 100:8951-8956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Stevens, A. M., K. M. Dolan, and E. P. Greenberg. 1994. Synergistic binding of the Vibrio fischeri LuxR transcriptional activator domain and RNA polymerase to the lux promoter region. Proc. Natl. Acad. Sci. USA 91:12619-12623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Suntharalingam, P., and D. G. Cvitkovitch. 2005. Quorum sensing in streptococcal biofilm formation. Trends Microbiol. 13:3-6. [DOI] [PubMed] [Google Scholar]
- 135.Surette, M. G., M. B. Miller, and B. L. Bassler. 1999. Quorum sensing in Escherichia coli, Salmonella typhimurium, and Vibrio harveyi: a new family of genes responsible for autoinducer production. Proc. Natl. Acad. Sci. USA 96:1639-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Swiderska, A., A. K. Berndtson, M. R. Cha, L. Li, G. M. Beaudoin III, J. Zhu, and C. Fuqua. 2001. Inhibition of the Agrobacterium tumefaciens TraR quorum-sensing regulator. Interactions with the TraM anti-activator. J. Biol. Chem. 276:49449-49458. [DOI] [PubMed] [Google Scholar]
- 137.Swift, S., M. K. Winson, P. F. Chan, N. J. Bainton, M. Birdsall, P. J. Reeves, C. E. Rees, S. R. Chhabra, P. J. Hill, J. P. Throup, et al. 1993. A novel strategy for the isolation of luxI homologues: evidence for the widespread distribution of a LuxR:LuxI superfamily in enteric bacteria. Mol. Microbiol. 10:511-520. [DOI] [PubMed] [Google Scholar]
- 138.Taga, M. E., and B. L. Bassler. 2003. Chemical communication among bacteria. Proc. Natl. Acad. Sci. USA 100:14549-14554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Taminiau, B., M. Daykin, S. Swift, M. L. Boschiroli, A. Tibor, P. Lestrate, X. De Bolle, D. O'Callaghan, P. Williams, and J. J. Letesson. 2002. Identification of a quorum-sensing signal molecule in the facultative intracellular pathogen Brucella melitensis. Infect. Immun. 70:3004-3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Teplitski, M., H. Chen, S. Rajamani, M. Gao, M. Merighi, R. T. Sayre, J. B. Robinson, B. G. Rolfe, and W. D. Bauer. 2004. Chlamydomonas reinhardtii secretes compounds that mimic bacterial signals and interfere with quorum sensing regulation in bacteria. Plant Physiol. 134:137-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Teplitski, M., J. B. Robinson, and W. D. Bauer. 2000. Plants secrete substances that mimic bacterial N-acyl homoserine lactone signal activities and affect population density-dependent behaviors in associated bacteria. Mol. Plant-Microbe Interact. 13:637-648. [DOI] [PubMed] [Google Scholar]
- 142.Toth, I. K., J. A. Newton, L. J. Hyman, A. K. Lees, M. Daykin, C. Ortori, P. Williams, and R. G. Fray. 2004. Potato plants genetically modified to produce N-acylhomoserine lactones increase susceptibility to soft rot erwiniae. Mol. Plant-Microbe Interact. 17:880-887. [DOI] [PubMed] [Google Scholar]
- 143.Tzfira, T., and V. Citovsky. 2002. Partners-in-infection: host proteins involved in the transformation of plant cells by Agrobacterium. Trends Cell Biol. 12:121-129. [DOI] [PubMed] [Google Scholar]
- 144.Uroz, S., C. D'Angelo-Picard, A. Carlier, M. Elasri, C. Sicot, A. Petit, P. Oger, D. Faure, and Y. Dessaux. 2003. Novel bacteria degrading N-acylhomoserine lactones and their use as quenchers of quorum-sensing-regulated functions of plant-pathogenic bacteria. Microbiology 149:1981-1989. [DOI] [PubMed] [Google Scholar]
- 145.Vannini, A., C. Volpari, and S. Di Marco. 2004. Crystal structure of the quorum-sensing protein TraM and its interaction with the transcriptional regulator TraR. J. Biol. Chem. 279:24291-24296. [DOI] [PubMed] [Google Scholar]
- 146.Vannini, A., C. Volpari, C. Gargioli, E. Muraglia, R. Cortese, R. De Francesco, P. Neddermann, and S. D. Marco. 2002. The crystal structure of the quorum sensing protein TraR bound to its autoinducer and target DNA. EMBO J. 21:4393-4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Wang, L. H., Y. He, Y. Gao, J. E. Wu, Y. H. Dong, C. He, S. X. Wang, L. X. Weng, J. L. Xu, L. Tay, R. X. Fang, and L. H. Zhang. 2004. A bacterial cell-cell communication signal with cross-kingdom structural analogues. Mol. Microbiol. 51:903-912. [DOI] [PubMed] [Google Scholar]
- 148.Watson, W. T., T. D. Minogue, D. L. Val, S. B. von Bodman, and M. E. Churchill. 2002. Structural basis and specificity of acyl-homoserine lactone signal production in bacterial quorum sensing. Mol. Cell 9:685-694. [DOI] [PubMed] [Google Scholar]
- 149.Weaks, T. E. 1977. Differences between strains of Rhizobium in sensitivity to canavanine. Plant Soil 48:387-395. [Google Scholar]
- 150.Wisniewski-Dye, F., and J. A. Downie. 2002. Quorum-sensing in Rhizobium. Antonie Leeuwenhoek 81:397-407. [DOI] [PubMed] [Google Scholar]
- 151.Wu, H., Z. Song, M. Hentzer, J. B. Andersen, S. Molin, M. Givskov, and N. Hoiby. 2004. Synthetic furanones inhibit quorum-sensing and enhance bacterial clearance in Pseudomonas aeruginosa lung infection in mice. J. Antimicrob. Chemother. 53:1054-1061. [DOI] [PubMed] [Google Scholar]
- 152.Zavil'gel'skii, G. B., and I. V. Manukhov. 2001. “Quorum sensing,” or how bacteria “talk” to each other. Mol. Biol. (Moscow) 35:268-277. [PubMed] [Google Scholar]
- 153.Zhang, H. B., C. Wang, and L. H. Zhang. 2004. The quormone degradation system of Agrobacterium tumefaciens is regulated by starvation signal and stress alarmone (p)ppGpp. Mol. Microbiol. 52:1389-1401. [DOI] [PubMed] [Google Scholar]
- 154.Zhang, H. B., L. H. Wang, and L. H. Zhang. 2002. Genetic control of quorum-sensing signal turnover in Agrobacterium tumefaciens. Proc. Natl. Acad. Sci. USA 99:4638-46343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Zhang, L. H. 2003. Quorum quenching and proactive host defense. Trends Plant Sci. 8:238-244. [DOI] [PubMed] [Google Scholar]
- 156.Zhang, R. G., T. Pappas, J. L. Brace, P. C. Miller, T. Oulmassov, J. M. Molyneaux, J. C. Anderson, J. K. Bashkin, S. C. Winans, and A. Joachimiak. 2002. Structure of a bacterial quorum-sensing transcription factor complexed with pheromone and DNA. Nature 417:971-974. [DOI] [PubMed] [Google Scholar]
- 157.Zhu, J., J. W. Beaber, M. I. More, C. Fuqua, A. Eberhard, and S. C. Winans. 1998. Analogs of the autoinducer 3-oxooctanoyl-homoserine lactone strongly inhibit activity of the TraR protein of Agrobacterium tumefaciens. J. Bacteriol. 180:5398-5405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Zhu, J., Y. Chai, Z. Zhong, S. Li, and S. C. Winans. 2003. Agrobacterium bioassay strain for ultrasensitive detection of N-acylhomoserine lactone-type quorum-sensing molecules: detection of autoinducers in Mesorhizobium huakuii. Appl. Environ. Microbiol. 69:6949-6953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Zhu, J., E. Dizin, X. Hu, A. S. Wavreille, J. Park, and D. Pei. 2003. S-Ribosylhomocysteinase (LuxS) is a mononuclear iron protein. Biochemistry 42:4717-4726. [DOI] [PubMed] [Google Scholar]
- 160.Zhu, J., P. M. Oger, B. Schrammeijer, P. J. Hooykaas, S. K. Farrand, and S. C. Winans. 2000. The bases of crown gall tumorigenesis. J. Bacteriol. 182:3885-3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Zhu, J., and S. C. Winans. 1999. Autoinducer binding by the quorum-sensing regulator TraR increases affinity for target promoters in vitro and decreases TraR turnover rates in whole cells. Proc. Natl. Acad. Sci. USA 96:4832-4837.10220379 [Google Scholar]
- 162.Zhu, J., and S. C. Winans. 2001. The quorum-sensing transcriptional regulator TraR requires its cognate signaling ligand for protein folding, protease resistance, and dimerization. Proc. Natl. Acad. Sci. USA 98:1507-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]