Abstract
Selenocysteine is incorporated into proteins via “recoding” of UGA from a stop codon to a sense codon, a process that requires specific secondary structures in the 3′ untranslated region, termed selenocysteine incorporation sequence (SECIS) elements, and the protein factors that they recruit. Whereas most selenoprotein mRNAs contain a single UGA codon and a single SECIS element, selenoprotein P genes encode multiple UGAs and two SECIS elements. We have identified evolutionary adaptations in selenoprotein P genes that contribute to the efficiency of incorporating multiple selenocysteine residues in this protein. The first is a conserved, inefficiently decoded UGA codon in the N-terminal region, which appears to serve both as a checkpoint for the presence of factors required for selenocysteine incorporation and as a “bottleneck,” slowing down the progress of elongating ribosomes. The second adaptation involves the presence of introns downstream of this inefficiently decoded UGA which confer the potential for nonsense-mediated decay when factors required for selenocysteine incorporation are limiting. Third, the two SECIS elements in selenoprotein P mRNA function with differing efficiencies, affecting both the rate and the efficiency of decoding different UGAs. The implications for how these factors contribute to the decoding of multiple selenocysteine residues are discussed.
Selenoprotein P is an enigma of genetic recoding. It is the only known protein in which multiple potential stop codons are recoded to function as sense codons. In selenoprotein mRNAs, UGA codons, which would normally signal the termination of protein synthesis, are decoded as the amino acid selenocysteine. This process involves a unique tRNA with an anticodon complementary to UGA and specialized secondary structures located in the 3′ untranslated regions (UTRs) of selenoprotein mRNAs, termed selenocysteine incorporation sequence (SECIS) elements (3). For selenocysteine incorporation to occur, the SECIS element must recruit a SECIS binding protein, SBP2 (6). SBP2, in turn, recruits a dedicated elongation factor, EFsec (7, 26), complexed with selenocysteyl-tRNA (2, 31). Most selenoprotein mRNAs contain a single UGA codon and a single SECIS element. Selenoprotein P mRNAs contain 10 to 18 UGA codons, depending on the species, and two SECIS elements differing in secondary structure. The majority of studies to date on the mechanism of selenoprotein synthesis have focused on selenoprotein mRNAs containing a single UGA codon and a single SECIS element. Thus, little is known about the mechanism for incorporating multiple selenocysteines.
The incorporation of selenium into selenoprotein P serves several important biological functions: it allows the accumulation of this essential but potentially toxic trace element in a biologically stable and nontoxic form, it plays a critical role in the transport of selenium from the liver via the circulation to target organs (13, 25), it is thought to function in sequestering and thereby detoxifying heavy metals in plasma (15, 16, 21, 24, 29, 30), and it functions as a stable repository and donor of selenium for selenoprotein biosynthesis in target tissues (23). Recent studies with selenoprotein P knockout mice showed that disrupting the delivery of selenium in the form of selenoprotein P to the brain resulted in impaired neurological development and subsequent lethality (13, 25), further illuminating the critical functions of this protein.
How are selenoprotein P mRNAs with their multiple selenocysteine codons, and thus multiple potential sites for termination, translated into full-length proteins? Selenoprotein P mRNAs utilize only two SECIS elements to direct incorporation at up to 18 UGA codons. Thus, selenoprotein P mRNAs may present a difficult challenge to the translation machinery. The efficiency of the selenocysteine incorporation process in vivo is not known. However, as evidence of the inherent difficulty in translating selenoprotein P mRNAs, immunoaffinity purification of selenoprotein P from rat plasma followed by mass spectrometry characterization revealed termination at the second, third, and seventh UGA codons (14, 18). Termination at these positions increased upon dietary selenium limitation. The requirement for de novo synthesis of selenoprotein P to incorporate selenium for delivery to target tissues, and the subsequent degradation of the protein for selenium reutilization, comes at a high energy cost to the cell. This would include not only the energy requirements for protein synthesis and degradation, but in addition the inefficient expenditure when selenium is limiting, resulting in the premature termination of protein synthesis. This high energy cost further underscores the paramount importance of the ability to efficiently incorporate multiple selenocysteines.
The selenoprotein P genes provide unique tools for addressing the mechanism and efficiency of translation for multiple UGA codons in naturally occurring selenoprotein genes. We report that selenoprotein P gene sequences have evolved several adaptations to increase the efficiency of selenocysteine incorporation. These adaptations include a conserved, inefficiently decoded UGA codon in the N-terminal region, the presence of introns in the pre-mRNA downstream of this UGA, and two SECIS elements differing in secondary structure. We show that the first UGA in selenoprotein P mRNAs is translated inefficiently in transiently transfected cells, resulting in high levels of termination. This codon appears to be served primarily by the 3′-proximal SECIS element (SECIS 2), which functions inefficiently compared to the upstream, coding region-proximal element (SECIS 1). Strikingly, the first UGA is the only position at which failure to decode as selenocysteine is capable of invoking nonsense-mediated decay; it is the only UGA followed by introns. Thus, the decision to translate or terminate at this position determines the ultimate fate of the mRNA: translation or mRNA degradation. We postulate that the first UGA codon serves both as an early checkpoint to determine whether the components required for selenocysteine incorporation are present in sufficient quantities and as a bottleneck to slow the progress of ribosomes to downstream UGA codons. We further speculate that the relatively slow or inefficient decoding by SECIS 2 is an evolutionary adaptation for ensuring this bottleneck function, resulting in more-efficient decoding of downstream UGAs.
MATERIALS AND METHODS
Constructs.
All constructs were derived from zebra fish selenoprotein P cDNA, cloned into pUHD10-3 vector (28). The glutathione S-transferase (GST)-coding sequence was introduced by PCR via a unique PstI site located downstream of the signal sequence and upstream of the first UGA codon (Fig. 1). Site-directed mutagenesis of SECIS elements (AUGA to AUCC) or the first UGA codon was performed by PCR, using two complementary mutagenic oligonucleotides and flanking 5′ and 3′ oligonucleotides, followed by subcloning of the mutated fragments via unique restriction sites. Deletion of the first SECIS element (D1) was carried out by SpeI digestion followed by self-ligation. Deletion of the second selenoprotein P SECIS element (D2) was achieved using two previously generated constructs with unique SphI sites introduced after the first or second SECIS element. The 1,831-bp HindIII-SphI fragment including both SECIS elements was replaced by a 1,354-bp fragment including only the first SECIS element. In this way, the region following the second SECIS element including the polyadenylation signal was retained in all constructs. Introduction of SECIS 1, SECIS 2, or both into wild-type (wt) selenoprotein P, D1, or D2 produced the constructs S1S1, S2S1, S2S2, and S1S2S1. For generation of the UGC no SECIS construct, a region corresponding to the last 25 amino acids of the coding region and the entire 3′ UTR was inverted by excision and religation of an EagI-NotI fragment, the EagI site being 75 nucleotides upstream of the termination codon and the NotI site in the vector multiple-cloning site. All regions amplified by PCR were sequenced to verify that no additional mutations had been introduced.
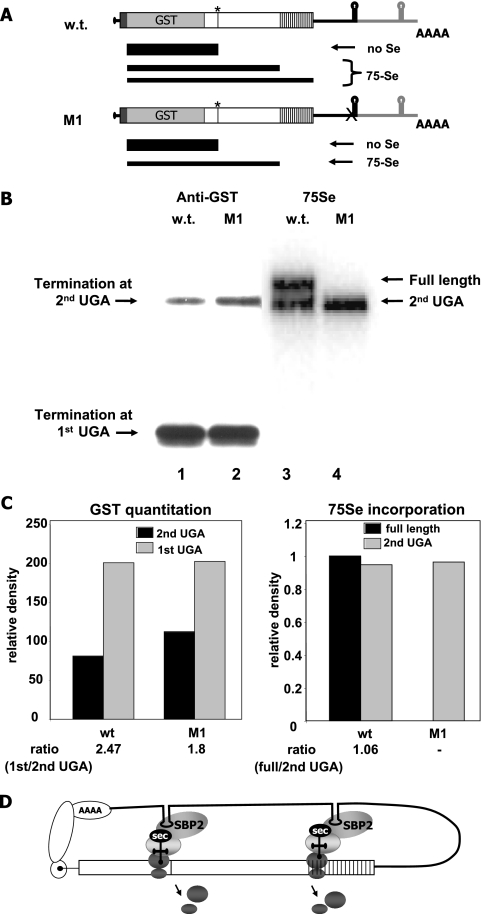
FIG. 1.
High frequency of termination and role of SECIS 1 in selenoprotein P translation. (A) Zebra fish selenoprotein P wt and SECIS 1 mutant (M1) mRNAs and predicted protein products. Open boxes indicate the coding regions, vertical lines represent UGA codons, the black boxes indicate the signal sequences, and the gray boxes indicate the GST sequences inserted just upstream of the first UGA. The 3′ untranslated regions are indicated by thin lines, with the first SECIS element and 5′ flanking regions in black and the second SECIS element and flanking regions in gray. A's indicate the polyadenylation sequence. Predicted termination and incorporation products are depicted below the mRNA schematics, with the thickness of the black bars representing the relative amounts of the products. The asterisks indicate the positions of the first UGA codon, and the X indicates point mutations in the SECIS elements. (B) Termination at the first UGA codon in selenoprotein P and effects of SECIS 1 mutation. Transient transfection of wt and M1 selenoprotein P constructs in HEK293 cells was carried out as described in Materials and Methods. Western blotting of GST-selenoprotein P secreted into media was performed with an anti-GST antibody, followed by chemiluminescence detection (left). Autoradiography for 75Se incorporation was performed on the same blot (right). (C) Quantitation of protein densities by GST Western blotting and 75Se incorporation. (D) Schematic showing putative decoding of the first UGA by SECIS 2 and downstream UGAs by SECIS 1. The open reading frame is depicted by the open box, with vertical lines representing UGA codons. Protein factors, ribosomes, and terminated ribosomal subunits are indicated by gray ovals. Circularization of the mRNA is depicted via interactions of proteins associated with the 5′ and 3′ ends.
Cell culture and transient transfections.
HEK293 cells were grown and maintained by standard tissue culture techniques in Dulbecco's modified Eagle's medium (GIBCO) supplemented with 10% fetal bovine serum (GIBCO). Cells were incubated in 5% CO2 at 37°C. Approximately 80 to 90% confluent monolayers of cells in 60-mm tissue culture dishes were transiently transfected using the calcium phosphate method, with 10 μg of the expression construct plasmid DNA in pUHD10-3 and 4 μg pUHD15-1 per dish. The pUHD10-3 expression vector (10) contains tetracycline operator sequences upstream of the polylinker region, whereas the pUHD15-1 plasmid encodes a tetracycline repressor-human papillomavirus VP16 activation domain fusion protein. Transfected cells were reseeded 24 h later onto four 100-mm plates (1:4 dilutions) to optimize the growth rate and yield of large polysomes. Cells were harvested 72 h after transfection.
75Se labeling.
75Se labeling was carried out as previously described (27). On day 2, media were exchanged for media without serum, with no added unlabeled selenium, at which time 75Se-sodium selenite (1,000 μCi/μg, 3 to 6 μCi/ml) was added. The media were harvested on day 3. Media were concentrated in VIVAspin columns (Vivascience Inc., Acton, MA) prior to analysis. Aliquots of media were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, followed by either autoradiography or Western blotting analysis.
GST affinity purification and Western blotting.
GST fusion proteins were purified and concentrated using Micro Spin GST purification module resin according to the manufacturer's protocol (Amersham Biosciences, Piscataway, NJ). Western blotting detection was performed using mouse monoclonal anti-GST antibody (UPSTATE Biotechnology, Lake Placid, NY) and anti-mouse peroxidase-labeled secondary antibody (Amersham Biosciences), followed by chemiluminescence detection of signal (ECL plus Western blotting detection system; Amersham Biosciences). Signal quantitation was carried out using a Molecular Dynamics scanning densitometer and ImageQuant software.
Cytoplasmic extracts.
For sucrose gradient analysis, cells were treated with 50 μg cycloheximide (Calbiochem) per dish for 25 min at 37°C prior to harvest. Media were decanted, and the cells were scraped into 5 ml ice-cold phosphate-buffered saline (GIBCO), washed once with phosphate-buffered saline, and resuspended in 750 μl low-salt buffer (200 mM Tris, pH 7.5, 100 mM NaCl, 30 mM MgCl2) supplemented with 200 U RNase inhibitor (Promega) and 5 μl 1 M dithiothreitol. The cells were then incubated on ice for 20 min, and 250 μl lysis buffer (200 mM Tris, pH 7.5, 100 mM NaCl, 30 mM MgCl2, 0.2 M sucrose, 1.2% Triton X-100) was added. Cells were transferred into a Kontes tissue homogenizer (2-ml capacity) and disrupted with 25 strokes. Cell debris was pelleted by centrifugation in a tabletop microcentrifuge for 5 min at 14,000 × g at +4°C. The supernatant was removed and stored on ice for further analysis.
Sucrose gradient preparation and centrifugation.
Sucrose density gradients were prepared as described previously (1), with 15 and 50% (wt/wt) sucrose (Sigma) in low-salt buffer. Gradients (4.8-ml volume) were prepared in Beckman 13- by 64-mm polyallomer tubes. Cytoplasmic extract (0.5 ml, equal to 700 μg RNA) was applied to the gradients and centrifuged at 45,000 rpm for 55 min in a Beckman SW-55 Ti rotor, using a Beckman L-8 M ultracentrifuge. For the gradient with the combined sample of SECIS 1 and SECIS 2 constructs, 350 μg RNA of each sample was applied in a final volume of 0.5 ml. Fractions of 0.25 ml were withdrawn from the top of the gradient and monitored for absorbance at 254 nm by using an ISCOsyringe pump with a UV-6 detector (Brandel). Fractions were frozen in liquid N2 and stored at −80°C until further analysis could be carried out.
RNA isolation and reverse transcription (RT)-PCR.
TRIzol reagent (Invitrogen) was used to extract total RNA from sucrose gradient fractions. Briefly, 250 μl of each fraction was added to 750 μl TRIzol reagent and shaken vigorously for 15 s. After 5 min of incubation at room temperature, 150 μl chloroform was added, followed by vigorous shaking and brief incubation at room temperature. Samples were then centrifuged at 13,000 × g for 10 min in a tabletop microcentrifuge. One microgram of nuclease-free glycogen (Roche) was added to 500 μl of the aqueous-phase concentration, and the nucleic acids were precipitated with the addition of equal volumes of 2-propanol. After centrifugation at 13,000 × g for 30 min at 4°C, the pellet was washed once with 75% ethanol and resuspended in 20 μl of nuclease-free, sterile water. One microgram of total RNA from each fraction was used as a substrate for oligonucleotide deoxyribosylthymine cDNA synthesis, using Superscript III reverse transcriptase (Invitrogen). The cDNA product was diluted with nuclease-free, sterile water to a final concentration of 50 ng/μl. For the PCR, the forward primer, 5′ CAGGGAGCATGCAATAATATCAG 3′, was used with either the reverse primer for S1, 5′ TGCTCTGTATGGCCCAAACA 3′, or the reverse primer for S2, 5′ ACATTTCCATACAGTTTCTCGGG 3′. PCRs utilized Go Green Taq PCR mixture (Promega) and 50 ng cDNA product derived from each fraction as a template. The number of cycles required for amplification in the linear range was determined for each primer pair and template. PCR products were electrophoresed through 1.5% agarose gels and visualized with ethidium bromide. Quantitation was carried out using Photoshop 8.0 software.
RESULTS
The goals of the present study were to assess selenocysteine incorporation and termination levels in a protein incorporating multiple selenocysteine residues and to investigate the roles of the two selenoprotein P SECIS elements in incorporation. We utilized the zebra fish selenoprotein P cDNA, which encodes 17 selenocysteine residues and two SECIS elements (Fig. 1A), for these studies. Previous studies have shown that premature termination of translation occurs at several of the UGA codons in selenoprotein P in the intact rat (see introduction) as well as in both rat and zebra fish selenoprotein P expressed by transient transfection of mammalian cells (27, 28). However, because 75Se labeling was used in these prior studies, the degree of termination at the first UGA codon, which would result in a product containing no selenium (Fig. 1A), could not be ascertained. Similarly, termination at subsequent UGA codons would result in products containing various numbers of selenocysteine residues. Thus, early-termination products would be underrepresented by 75Se labeling and may be difficult to detect, compared to late-terminating products containing more selenocysteine residues per protein. To circumvent these potential problems, we inserted GST coding sequences into the coding regions of wt and mutant zebra fish selenoprotein P expression constructs, downstream of the signal sequence but upstream of the first UGA codon. Selenoprotein P products in the media were then analyzed following transient transfection, both by Western blotting with anti-GST antibody and by assessment of 75Se incorporation.
High frequency of termination at the first UGA codon in selenoprotein P.
The expression of the wt zebra fish selenoprotein P construct produced high levels of a peptide terminating at the first UGA (Fig. 1B, lane 1), as determined by Western blotting with an anti-GST antibody. This product was not detected by 75Se labeling (Fig. 1B, lane 3). Two bands of slower mobilities were detected by 75Se labeling (Fig. 1B, lane 3, upper arrows). The slower of these migrated at the size corresponding to termination at the second UGA, as verified by its comigration with the product of a construct in which the first UGA was mutated to a UGC cysteine codon and the SECIS elements were inactivated by inversion (Fig. 2C). The uppermost band, migrating at the position predicted for full-length selenoprotein P, exhibited intense 75Se labeling but was detectable only by anti-GST staining upon overexposure (not shown). This is consistent with a low-abundance protein with high Se content (Fig. 1B, lane 3, upper arrow).
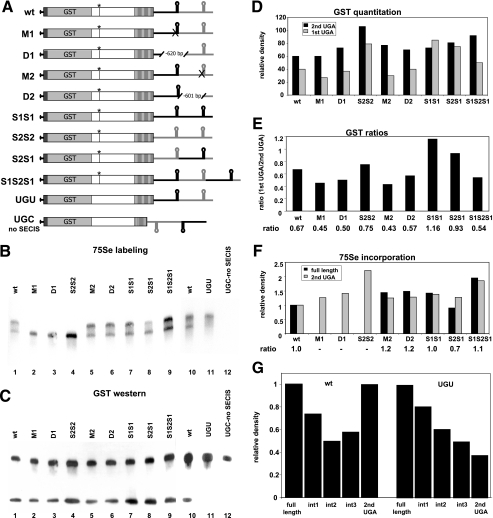
FIG. 2.
Effects of SECIS mutation, deletion, or position on termination at UGA codons in selenoprotein P. (A) Schematic representation of selenoprotein P expression constructs. Point mutations are indicated by Xs, deletions are indicated by hatch marks, and insertions or swapping of SECIS positions are indicated by black or gray SECIS elements. Mutation of the first UGA to UGU or UGC is indicated by the absence of the first vertical line. All other symbols are as defined in the legend to Fig. 1. (B) 75Se labeling of selenoprotein P transiently expressed from constructs in which SECIS elements were mutated, deleted, or duplicated or their positions swapped. Media from cells were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis on 7.5% acrylamide gels, followed by autoradiography. (C) Western blotting of GST-selenoprotein P on the same blot was performed with an anti-GST antibody, followed by chemiluminescence detection. (D) Densitometric quantitation of proteins detected by anti-GST Western blotting (GST western). (E) Ratios of products terminated at the first UGA to products terminated at the second UGA, as detected by anti-GST Western blotting. (F) Densitometric quantitation of proteins detected by 75Se labeling and ratios of full-length product to product terminating at the second UGA. (G) Densitometric quantitation of full-length, intermediate, and second-UGA-terminated products detected by 75Se labeling.
SECIS 1 is required for the production of full-length selenoprotein P.
We have previously shown that the conserved AUGA motif in SECIS elements is critical for its function, with point mutations in this sequence rendering the elements inactive (4, 20). To assess the functions of the two SECIS elements in selenoprotein P, we mutated the conserved AUGA motif in SECIS 1 to AUCC. The effect of this mutation, M1, as shown by 75Se labeling, was a complete loss of detectable full-length selenoprotein P (Fig. 1B, lane 4, upper arrow) and a corresponding slight increase in termination at the second UGA codon (Fig. 1B, lane 2 versus 1, and C). These results indicate that SECIS 1 is required for the production of full-length selenoprotein P and possibly for decoding all UGA codons beyond the first. SECIS 2, on the other hand, appears to function primarily to decode the first UGA codon, as indicated by the lack of incorporation at UGA codons beyond the first one when only SECIS 2 is present. Considering the two SECIS elements in their native context, circularization of the mRNA via interactions between the cap and poly(A) binding proteins at the 5′ and 3′ ends would bring SECIS 2 into the proximity of the first UGA codon in the N-terminal portion of the coding region, with SECIS 1 being brought into the proximity of the downstream UGA codons (Fig. 1D). The differential decoding efficiencies of the two SECIS elements are consistent with our previous demonstration that rat selenoprotein P SECIS 1 is at least threefold more efficient than SECIS 2 at directing selenocysteine incorporation into a heterologous reporter construct containing a single UGA codon (4).
Effects of SECIS mutations and deletions and SECIS position on selenocysteine incorporation.
To further assess the functions of the two SECIS elements, constructs containing point mutations, SECIS insertions or deletions, or manipulations of their positions were generated (Fig. 2A). Supporting the requirement for SECIS 1 in decoding all UGA codons beyond the first, loss of full-length protein was observed not only upon deletion of this element (Fig. 2B, lane 3), but even when a second copy of SECIS 2 was introduced in its place (Fig. 2B, lane 4). Thus, an additional copy of SECIS 2 does not compensate for the loss of SECIS 1, further indicating the different functions of these two elements. In contrast, mutation or deletion of SECIS 2 allowed production of full-length protein (Fig. 2B, lanes 5 and 6 versus 1), as did the insertion of a second copy of SECIS 1 in place of SECIS 2 (Fig. 2B, lane 7). However, when termination at the first and second UGA codons was quantitated by Western blotting for GST (Fig. 2C), a relative increase in termination at the first UGA codon was seen with both the S1S1 and the S2S1 constructs (Fig. 2D and E). Thus, SECIS 1 is required for production of the full-length protein, while SECIS 2 appears to be required for maximum efficiency in decoding the first UGA codon.
To ascertain the positional effects of the two elements, we swapped their positions relative to each other or introduced an additional copy of SECIS 1 downstream of SECIS 2. Swapping the positions of the two elements resulted in an increase in termination at the first UGA and a decrease in the ratio of full-length to second UGA termination product (Fig. 2B and C, lanes 8, and D, E, and F). This indicates that not only are both elements required for maximum efficiency, but also the positions of the elements relative to each other are crucial for optimal function. The introduction of an additional copy of SECIS 1 downstream of SECIS 2 had no apparent effect on incorporation (Fig. 2B and C, lanes 9, and D, E, and F), consistent with the finding that all selenoprotein P genes encode only two SECIS elements, regardless of the number of UGA codons.
An emerging pattern throughout is the presence of two distinct 75Se-labeled bands, corresponding to the full-length protein and termination at the second UGA codon, with minimal laddering between the two bands (Fig. 2B, lanes 1 and 5 to 10). This suggests that termination at intermediate UGA codons is minimal and that once incorporation occurs at the second UGA, translation is usually processive, proceeding through the remaining UGAs. High levels of termination at the first and second UGA codons were also consistently observed, and while the ratios of the two differed from one experiment to another (e.g., Fig. 1B versus 2C), they were consistent within a set of transfections for any given construct.
As alterations in RNA structure might affect the stability of the mRNAs, we carried out real-time RT-PCR to amplify zebra fish selenoprotein P mRNAs from all constructs as well as the housekeeping gene for hypoxanthine phosphoribosyltransferase and endogenous human selenoprotein P. All zebra fish constructs produced comparable levels of selenoprotein mRNAs, with the exception of the construct in which SECIS 2 was deleted (data not shown). This construct produced significantly lower levels of mRNA. The reason for this is not readily apparent, as an AU-rich element is removed in this construct, relative to those in the others.
The presence of the first UGA codon enhances the production of full-length selenoprotein P.
As termination occurs with a high frequency at the first UGA codon, mutation of this UGA to a UGU cysteine codon might be predicted to increase the production of the full-length protein. Surprisingly, the amount of full-length protein slightly decreased with this mutation, as assessed by 75Se labeling (Fig. 2B, lanes 10 and 11, in which the ratio of UGU to wt full-length proteins is 0.64:1). Quantitation of the intensities of 75Se-labeled intermediate products revealed slight increases in the relative levels of these products in the UGU mutant compared to those in the wild-type protein (Fig. 2G, note particularly the levels for the first and second intermediate bands, int1 and int2, respectively relative to those for each full-length band).
Polysome loading suggests slow decoding at the first UGA in selenoprotein P.
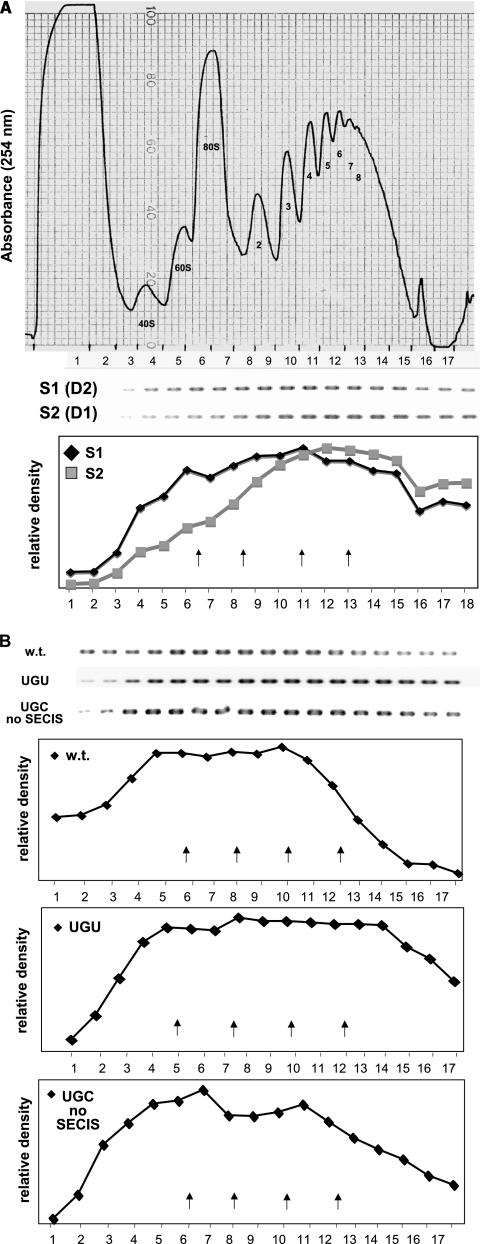
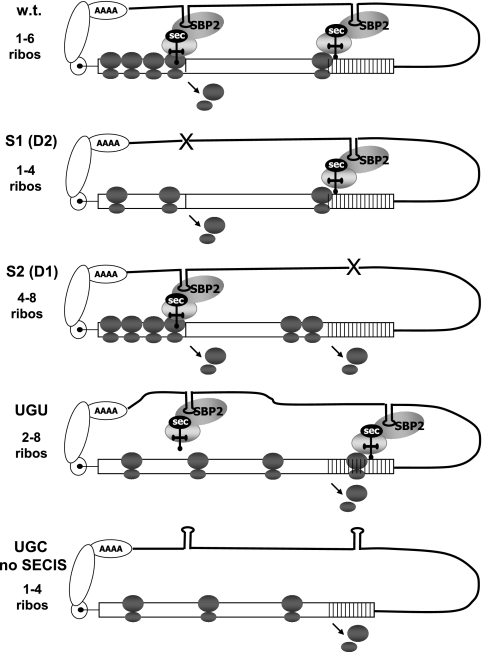
To further investigate the functions of the two SECIS elements and the first UGA codon in selenocysteine incorporation, we carried out sucrose gradient fractionation, followed by RT-PCR of gradient fractions, to determine polysome loadings on selenoprotein P mRNAs. Selenoprotein P mRNA containing SECIS 1 but lacking SECIS 2 was consistently associated with polysomes that were smaller (Fig. 3A, peak fractions 6 to 15, containing one to four ribosomes) than those associated with mRNA containing SECIS 2 but lacking SECIS 1 (Fig. 3A, peak fractions 9 to 16, containing four to eight or more ribosomes), despite the production of full-length protein by the former but not the latter. Since the latter mRNA does not permit detectable translation through the second UGA codon, this implies heavier ribosomal loading in the region upstream of this point (Fig. 4, S1 and S2). Peak fractions were identified as those in which the amounts of product were equal to 80% or more of that found in the fraction containing the highest level of product. How can we explain the low ribosomal occupancy of the mRNA containing SECIS 1 but lacking SECIS 2? In this case, SECIS 1 would be required for incorporation at all UGA codons. As depicted in Fig. 4, when the selenocysteine incorporation complex is translating downstream UGA codons, ribosomes arriving at the first UGA terminate and dissociate, such that only the ribosomes upstream of this point would remain associated with the mRNA.
FIG. 3.
Polysome profiles of wt and mutant selenoprotein P mRNAs. (A) Polysome profiles of mRNAs containing either SECIS 1 or SECIS 2 were assessed by sucrose gradient fractionation, followed by reverse transcription and PCR of gradient fractions, using primers specific for either SECIS element. The upper panel shows the gradient absorbance profile at 254 nm, with subunits, monosomes, and numbers of ribosomes indicated under each peak and the boundaries of collected fractions indicated across the bottom of the tracing. The middle panel shows the PCR products, and their quantitation by densitometry is shown in the lower panel. Arrows in the lower panel indicate the positions of the peaks of 80S monosomes and polysomes containing two, four, and eight ribosomes. (B) Polysome association for wt, UGU, and UGC no SECIS mRNAs. The upper panel shows the reverse transcription-PCR products of gradient fractions, and the lower panels show the quantitations of the products. Fractionation and quantitation were carried out as described above and in detail in Materials and Methods. Each experiment was carried out at least three times, with highly reproducible results.
FIG. 4.
Models for the functions of the two SECIS elements and the first UGA codon in selenoprotein P. Schematic representations of the decoding of UGA codons in wt selenoprotein mRNA, in mRNAs in which either SECIS is deleted, or in constructs in which the first UGA is mutated to a cysteine codon are shown. Ribosomes (ribos) stacking behind the first UGA codon in the wt and S2 constructs and behind the second UGA codon in the S2 construct are depicted. Dissociated subunits are shown at sites where termination may occur.
The decrease in full-length selenoprotein P production upon the mutation of the first UGA codon observed in Fig. 2B seems in contradiction with the inefficient decoding of this UGA, unless a selective advantage is conferred by its presence. For example, slow decoding of this UGA, coupled with alternating incorporation and termination events at this position, may serve to decrease ribosomal loading downstream. A decrease in the frequency of ribosomal loading might in turn increase incorporation efficiency at the downstream UGA codons by allowing increased time for reassembly of decoding complexes (Fig. 4, w.t.). Conversely, mutation of the first UGA codon to UGU might allow heavier ribosomal loading between this point and the downstream UGA codons, resulting in increased termination at these sites (Fig. 4, UGU). To assess this possibility, polysome analysis was carried out using the wt construct and constructs in which the first UGA was mutated to a cysteine codon, with both SECIS elements functional (UGU) or with the 3′ UTR inverted so that neither SECIS is functional (UGC no SECIS). The majority of wt selenoprotein P mRNA was found in polysomes containing one to six ribosomes (Fig. 3B, peak fractions 5 to 11), whereas the UGU mutant mRNA was present in fractions ranging from monosomes to the heaviest polysomes, with the peak in the two- to eight-ribosome range (Fig. 3B, peak fractions 5 to 14). The mRNA containing the UGC mutation and inverted 3′ UTR (UGC no SECIS) was found in fractions in the one- to four-ribosome range (Fig. 3B, peak fractions 4 to 12), similar to the pattern seen when only SECIS 1 is present. Both the UGC no SECIS construct and the construct containing only SECIS 2 result in termination at the same position, that occupied by the second UGA codon in the wt mRNA (Fig. 4, S2 and UGC no SECIS). However, as the first UGA is not present in the UGC no SECIS construct, the difference in ribosomal loadings on these two constructs, ∼4 ribosomes, must be due solely to the effects of the first UGA codon. Thus, the presence of the first UGA codon significantly increases ribosomal loading on the mRNA, consistent with this UGA undergoing slow decoding.
DISCUSSION
Selenoprotein P provides an ideal model for investigating how the translation machinery deals with multiple instances of dual codon functions within a single message. Through introduction of the GST epitope tag, manipulation of the two SECIS elements, and analysis of polysome loadings on the corresponding mRNAs, we have investigated the mechanism and efficiency of selenocysteine incorporation in selenoprotein P and the functions of the two SECIS elements in this process. Quantitation of the level of termination at the first UGA codon relative to that of incorporation products revealed that termination at this position could exceed incorporation by a factor of 2 to 2.5. This may be an underestimate if the termination product is rapidly degraded. The relative levels of termination and incorporation in transfected cells may not reflect the ratios for endogenous selenoprotein P expression, but they clearly indicate that the UGA codons in selenoprotein P are capable of significant levels of termination, in agreement with prior studies of intact animals (14, 18).
The conservation of the two SECIS elements in all selenoprotein P genes, regardless of the number of UGA codons, suggested that the two elements might serve different functions, perhaps different UGA codons. We previously reported that rat selenoprotein P SECIS 1 functions about threefold more efficiently than SECIS 2 in directing the translation of a single UGA codon in a reporter construct (4). This was further borne out by studies showing that selenoprotein P SECIS 1 exhibited greater efficiency than SECIS elements from four other selenoprotein genes and that cotransfection of SBP2 resulted in a more dramatic increase in selenocysteine incorporation by selenoprotein P SECIS 1 than in that by the other SECIS elements (17). Herein, we demonstrate that the two selenoprotein P SECIS elements are functionally distinct. SECIS 1 is required for the production of any detectable full-length selenoprotein P in transfected cells, and this occurs regardless of its position in the 3′ UTR. Deletion or mutation of this element resulted in the loss of full-length and near-full-length selenoprotein P, resulting primarily in the termination of products at the first or second UGA. Thus, it is clear that SECIS 1 serves the multiple downstream UGA codons of selenoprotein P. SECIS 2, on the other hand, appears to function primarily to decode the first UGA and does so inefficiently.
How might the translation machinery evoke differential efficiencies from the two elements? Studies investigating the affinity of SBP2 for different SECIS elements in vitro revealed only a twofold range in apparent Kd values among six SECIS elements, including selenoprotein P SECIS 1 (9). However, other factors present in vivo, including ribosomal protein L30 (5), may affect the interactions between SBP2-SECIS complexes and other components of the selenocysteine incorporation machinery, in agreement with our results showing that SECIS 1 exhibits significantly higher efficiency than other elements in transfected cells. As discussed in the introduction, SECIS 1 and 2 differ by the presence or absence of a short region of a secondary structure in the apical loop. We previously showed that manipulation or deletion of this region reduces or abolishes the SECIS function; thus, it is a prime candidate for interaction with factors other than SBP2 for enhancing the efficiency of selenocysteine incorporation (11).
The studies reported herein show that decoding of the first UGA codon by SECIS 2 is inefficient, inviting the question “why evolve inefficient decoding at this position?” Recognition of a stop codon as premature, invoking the nonsense-mediated decay pathway, requires that the stop codon be positioned upstream of an intron in the pre-mRNA (12, 22). Intriguingly, only the first UGA codon in all selenoprotein P genes sequenced to date is in a position to invoke nonsense-mediated decay if inefficiently decoded, as it is the only UGA codon followed by introns. All subsequent UGAs are present in the last exon. Further, the position of the first UGA codon is conserved in all selenoprotein P sequences reported to date and is found within the first 15% of the coding sequence. The fate of translation at the first UGA would thus serve as an early checkpoint, determining an outcome of either nonsense-mediated decay or survival of the mRNA, the former eliminating excess mRNAs when their translation would be inefficient, e.g., under conditions of selenium deficiency. Thus, in addition to its function in eliminating deleterious mRNAs, nonsense-mediated decay would in this case serve to attain the appropriate stoichiometry between selenoprotein mRNAs, selenocysteyl-tRNA, and the protein factors required for selenocysteine incorporation.
For mRNAs that escape nonsense-mediated decay and undergo translation,inefficient decoding at the first UGA directed by SECIS 2 could produce at least two effects. First, alternating termination and incorporation events would decrease overall ribosomal loading downstream. Second, slow decoding at this position could result in a bottleneck effect, as depicted in Fig. 4, impeding the progress of ribosomes to downstream UGA codons. We and others have previously demonstrated low ribosomal occupancy for several selenoprotein mRNAs, using polysome profile analysis (8, 19). Termination at the first UGA would clearly contribute to low ribosomal occupancy, whereas stacking of ribosomes at the putative bottleneck may or may not result in heavier polysomes, depending on the selenoprotein mRNA and the proximity of the UGA codon to the beginning of the open reading frame. In the case of selenoprotein P, the putative bottleneck effect could allow for sufficient intervals between ribosomes, such that following decoding at the first UGA mediated by SECIS 2, a ribosome would proceed to the second UGA and from that point could track with SECIS 1 to decode the downstream UGAs, producing a full-length protein. Ribosomes arriving at the second UGA before SECIS 1 becomes available for a subsequent round would terminate at this position, consistent with the significant levels of termination at the second UGA codon observed with all constructs. Thus, the ability of the first UGA codon to increase the intervals between ribosomes would theoretically increase the efficiency of full-length-protein production. Conversely, elimination of this bottleneck by mutation to a cysteine codon would result in increased termination at the second and subsequent UGA codons, as depicted in Fig. 4 and observed experimentally in Fig. 2. Future experiments aimed at unraveling exactly how SECISs 1 and 2 are distinguished by the selenocysteine incorporation machinery will surely provide exciting new insights into this fascinating process.
Acknowledgments
We thank Claire Kendall for critical reading of the manuscript and Ilko Stoytchev and Gabriel Forrey for assistance with the constructs.
This work was supported by Public Health Service grants DK-52963 and 47320 to M.J.B. from the NIH.
Footnotes
Published ahead of print on 25 September 2006.
REFERENCES
- 1.Abe, S., and E. Davies. 1986. Quantitative analysis of polysomes using a baseline from uncentrifuged blank gradients. Mem. Coll. Agric. Ehime Univ. 31:187-199. [Google Scholar]
- 2.Berry, M. J. 2000. Recoding UGA as selenocysteine, p. 763-783. In N. Sonenberg, J. W. B. Hershey, and M. B. Mathews (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 3.Berry, M. J., L. Banu, Y. Y. Chen, S. J. Mandel, J. D. Kieffer, J. W. Harney, and P. R. Larsen. 1991. Recognition of UGA as a selenocysteine codon in type I deiodinase requires sequences in the 3′ untranslated region. Nature 353:273-276. [DOI] [PubMed] [Google Scholar]
- 4.Berry, M. J., L. Banu, J. W. Harney, and P. R. Larsen. 1993. Functional characterization of the eukaryotic SECIS elements which direct selenocysteine insertion at UGA codons. EMBO J. 12:3315-3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chavatte, L., B. A. Brown, and D. M. Driscoll. 2005. Ribosomal protein L30 is a component of the UGA-selenocysteine recoding machinery in eukaryotes. Nat. Struct. Mol. Biol. 12:408-416. [DOI] [PubMed] [Google Scholar]
- 6.Copeland, P. R., J. E. Fletcher, B. A. Carlson, D. L. Hatfield, and D. M. Driscoll. 2000. A novel RNA binding protein, SBP2, is required for the translation of mammalian selenoprotein mRNAs. EMBO J. 19:306-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fagegaltier, D., N. Hubert, K. Yamada, T. Mizutani, P. Carbon, and A. Krol. 2000. Characterization of mSelB, a novel mammalian elongation factor for selenoprotein translation. EMBO J. 19:4796-4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fletcher, J. E., P. R. Copeland, and D. M. Driscoll. 2000. Polysome distribution of phospholipid hydroperoxide glutathione peroxidase mRNA: evidence for a block in elongation at the UGA/selenocysteine codon. RNA 6:1573-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fletcher, J. E., P. R. Copeland, D. M. Driscoll, and A. Krol. 2001. The selenocysteine incorporation machinery: interactions between the SECIS RNA and the SECIS-binding protein SBP2. RNA 7:1442-1453. [PMC free article] [PubMed] [Google Scholar]
- 10.Gossen, M., and H. Bujard. 1992. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl. Acad. Sci. USA 89:5547-5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grundner-Culemann, E., G. W. Martin III, J. W. Harney, and M. J. Berry. 1999. Two distinct SECIS structures capable of directing selenocysteine incorporation in eukaryotes. RNA 5:625-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hentze, M. W., and A. E. Kulozik. 1999. A perfect message: RNA surveillance and nonsense-mediated decay. Cell 96:307-310. [DOI] [PubMed] [Google Scholar]
- 13.Hill, K. E., J. Zhou, W. J. McMahan, A. K. Motley, J. F. Atkins, R. F. Gesteland, and R. F. Burk. 2003. Deletion of selenoprotein P alters distribution of selenium in the mouse. J. Biol. Chem. 278:13640-13646. [DOI] [PubMed] [Google Scholar]
- 14.Himeno, S., H. S. Chittum, and R. F. Burk. 1996. Isoforms of selenoprotein P in rat plasma. Evidence for a full-length form and another form that terminates at the second UGA in the open reading frame. J. Biol. Chem. 271:15769-15775. [DOI] [PubMed] [Google Scholar]
- 15.Koelman, J. H., W. H. Peeters, C. H. Koudstaal-Hol, P. S. Tjioe, and J. J. de Goeij. 1973. Mercury-selenium correlations in marine mammals. Nature 245:385-386. [DOI] [PubMed] [Google Scholar]
- 16.Kosta, L., A. R. Byrne, and V. Zelenko. 1975. Correlation between selenium and mercury in man following exposure to inorganic mercury. Nature 254:238-239. [DOI] [PubMed] [Google Scholar]
- 17.Low, S. C., E. Grundner-Culemann, J. W. Harney, and M. J. Berry. 2000. SECIS-SBP2 interactions dictate selenocysteine incorporation efficiency and selenoprotein hierarchy. EMBO J. 19:6882-6890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma, S., K. E. Hill, R. M. Caprioli, and R. F. Burk. 2002. Mass spectrometric characterization of full-length rat selenoprotein P and three isoforms shortened at the C terminus. Evidence that three UGA codons in the mRNA open reading frame have alternative functions of specifying selenocysteine insertion or translation termination. J. Biol. Chem. 277:12749-12754. [DOI] [PubMed] [Google Scholar]
- 19.Martin, G. W., III, and M. J. Berry. 2001. Selenocysteine codons decrease polysome association on endogenous selenoprotein mRNAs. Genes Cells 6:121-129. [DOI] [PubMed] [Google Scholar]
- 20.Martin, G. W., III, J. W. Harney, and M. J. Berry. 1998. Functionality of mutations at conserved nucleotides in eukaryotic SECIS elements is determined by the identity of a single nonconserved nucleotide. RNA 4:65-73. [PMC free article] [PubMed] [Google Scholar]
- 21.Mostert, V., I. Lombeck, and J. Abel. 1998. A novel method for the purification of selenoprotein P from human plasma. Arch. Biochem. Biophys. 357:326-330. [DOI] [PubMed] [Google Scholar]
- 22.Nagy, E., and L. E. Maquat. 1998. A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem. Sci. 23:198-199. [DOI] [PubMed] [Google Scholar]
- 23.Saito, Y., and K. Takahashi. 2002. Characterization of selenoprotein P as a selenium supply protein. Eur. J. Biochem. 269:5746-5751. [DOI] [PubMed] [Google Scholar]
- 24.Sasakura, C., and K. T. Suzuki. 1998. Biological interaction between transition metals (Ag, Cd and Hg), selenide/sulfide and selenoprotein P. J. Inorg. Biochem. 71:159-162. [DOI] [PubMed] [Google Scholar]
- 25.Schomburg, L., U. Schweizer, B. Holtmann, L. Flohe, M. Sendtner, and J. Kohrle. 2003. Gene disruption discloses role of selenoprotein P in selenium delivery to target tissues. Biochem. J. 370:397-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tujebajeva, R. M., P. R. Copeland, X. M. Xu, B. A. Carlson, J. W. Harney, D. M. Driscoll, D. L. Hatfield, and M. J. Berry. 2000. Decoding apparatus for eukaryotic selenocysteine insertion. EMBO Rep. 1:158-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tujebajeva, R. M., J. W. Harney, and M. J. Berry. 2000. Selenoprotein P expression, purification, and immunochemical characterization. J. Biol. Chem. 275:6288-6294. [DOI] [PubMed] [Google Scholar]
- 28.Tujebajeva, R. M., D. G. Ransom, J. W. Harney, and M. J. Berry. 2000. Expression and characterization of nonmammalian selenoprotein P in the zebrafish, Danio rerio. Genes Cells 5:897-903. [DOI] [PubMed] [Google Scholar]
- 29.Yan, J., and J. N. Barrett. 1998. Purification from bovine serum of a survival-promoting factor for cultured central neurons and its identification as selenoprotein-P. J. Neurosci. 18:8682-8691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoneda, S., and K. T. Suzuki. 1997. Equimolar Hg-Se complex binds to selenoprotein P. Biochem. Biophys. Res. Commun. 231:7-11. [DOI] [PubMed] [Google Scholar]
- 31.Zavacki, A. M., J. B. Mansell, M. Chung, B. Klimovitsky, J. W. Harney, and M. J. Berry. 2003. Coupled tRNASec dependent assembly of the selenocysteine decoding apparatus. Mol. Cell 11:773-781. [DOI] [PubMed] [Google Scholar]