Abstract
To study the genetic control of plant responses to cold stress, Arabidopsis thaliana mutants were isolated by a screen for mutations that impair cold-induced transcription of the CBF3-LUC reporter gene. We report here the characterization and cloning of a mutated gene, atnup160-1, which causes reduced CBF3-LUC induction under cold stress. atnup160-1 mutant plants display altered cold-responsive gene expression and are sensitive to chilling stress and defective in acquired freezing tolerance. AtNUP160 was isolated through positional cloning and shown to encode a putative homolog of the animal nucleoporin Nup160. In addition to the impaired expression of CBF genes, microarray analysis revealed that a number of other genes important for plant cold tolerance were also affected in the mutants. The atnup160 mutants flower early and show retarded seedling growth, especially at low temperatures. AtNUP160 protein is localized at the nuclear rim, and poly(A)-mRNA in situ hybridization shows that mRNA export is defective in the atnup160-1 mutant plants. Our study suggests that Arabidopsis AtNUP160 is critical for the nucleocytoplasmic transport of mRNAs and that it plays important roles in plant growth and flowering time regulation and is required for cold stress tolerance.
In eukaryotic cells, the genome is enclosed within the nucleus. Nucleocytoplasmic transport of macromolecules across the nuclear membrane occurs through channels formed by nuclear pore complexes (NPCs). Embedded in the double-lipid bilayer nuclear envelope, NPCs form a ringlike structure surrounding a central pore that is believed to facilitate the bidirectional transport of RNAs, proteins, and ribonucleoprotein particles and, at the same time, to allow the diffusion of small molecules and ions across the double membrane (reviewed in reference 4). The overall three-dimensional architecture and transport mechanisms seem to be highly conserved from yeasts to mammals. In Saccharomyces cerevisiae, NPCs are constructed from ∼30 different nucleoporins with a combined mass of ∼50 MDa (26). The mammalian NPCs are much larger complexes (∼120 MDa) composed of ∼80 different proteins (10). Although the structural organization of NPCs, the transport mechanism across the channels, and the function of individual nucleoporins have been extensively studied in yeast and vertebrates, very little is known about NPCs in plants.
Recently, it was reported that the Arabidopsis thaliana proteins MOS3/SAR3 and SAR1 share high sequence similarities with human nucleoporins Nup96 and Nup160, respectively (25, 34). The putative nucleoporin MOS3/SAR3 was localized at the nuclear rim. The studies suggested that nucleocytoplasmic trafficking plays an important role in plant disease resistance, hormone signaling, and development (25, 34). In the present report, we provide evidence that Arabidopsis nucleoporin AtNUP160/SAR1 controls nucleocytoplasmic transport of RNAs and plays important roles in seedling growth, flowering time regulation, and cold stress tolerance.
Low temperature is one of the most important environmental factors that greatly influences the growth, development, survival, and geographic distribution of plants. Most plants from temperate regions can increase their freezing tolerance by being exposed to low, nonfreezing temperatures (0°C to 10°C), i.e., by cold acclimation (20). Plant cold acclimation involves complicated biochemical and physiological changes, such as alterations in lipid composition and the accumulation of sugars and other osmolytes. Low temperatures induce the expression of diverse and numerous plant genes, many of which are known as COR (cold regulated), KIN (cold induced), LTI (low-temperature induced), or RD (responsive-to-desiccation) genes (5, 24, 33). Transcription factors known as CBFs or DREB1s can bind to the DRE/CRT cis elements in their promoters and activate the transcription of the COR/KIN/LTI/RD genes (33, 27). The CBF genes are also induced by low temperature, and the induction is transient and precedes that of the COR/KIN/LTI/RD genes (6, 21, 22). Ectopic overexpression of CBF1 and CBF3 in Arabidopsis results in the constitutive expression of the downstream cold-inducible genes even at warm temperatures and confers increased freezing tolerance (7, 14, 16, 21). These studies demonstrate an important role for the CBF regulon in cold acclimation.
Recently, a number of studies have investigated the regulation of the CBF regulon. ICE1 is a positive regulator of CBF genes and encodes a MYC-like basic helix-loop-helix transcriptional activator (1). The ice1 mutation reduces the expression of CBF3 and many other cold-responsive transcription factor genes, decreases the expression of numerous genes downstream of the transcription factors, and significantly reduces chilling and freezing tolerances (1, 17). On the other hand, the Arabidopsis HOS1 protein is a negative regulator of CBF genes. CBF genes and their downstream COR genes show enhanced cold induction in hos1 mutant plants (13). HOS1 encodes a RING-finger ubiquitin E3 ligase that targets ICE1 and possibly other positive regulators of CBF genes for ubiquitination and degradation (3, 19). In addition to ice1 and hos1, mutations in the translation elongation factor 2 (11) and an RNA helicase (9) also alter the expression of CBF genes under cold treatment.
To identify additional regulatory components in plant responses to cold stress, we performed a genetic screen to search for mutations that impair cold-induced expression of the CBF3-LUC reporter gene (1). One of the mutated genes, atnup160-1, was characterized in the present study. The atnup160-1 mutation greatly reduces CBF3-LUC expression in response to cold treatment, and the mutant plants are highly sensitive to chilling and freezing stresses. In addition, the atnup160-1 mutation also affects plant growth and flowering time. We cloned AtNUP160 using a map-based approach and found that it encodes a putative homolog of the human nucleoporin Nup160. AtNUP160 protein is enriched at the nuclear rim, and atnup160 mutant plants are defective in mRNA export from the nucleus. Our results suggest the possibility of a critical role for RNA export in cold stress responses and in the regulation of plant growth and development.
MATERIALS AND METHODS
Plant materials and mutant isolation.
Construction of Arabidopsis (ecotype Columbia gl1) plants expressing CBF3-LUC (referred to as the wild type), ethyl methanesulfonate mutagenesis, and mutant screening by luminescence imaging were carried out as described previously (1). Mutants were backcrossed to the wild type three times to eliminate other mutations from the genetic background. Plants were grown in a growth chamber set at 22°C, with 16 h light (70 μmol · m−2 · s−1), 8 h dark, and 75% relative humidity.
RNA expression analysis.
Two-week-old wild-type and mutant plants grown on agar plates with Murashige and Skoog (MS) salts plus 3% sucrose were treated at 4°C in light (20 μmol · m−2 · s−1). Total RNA was extracted from whole seedlings. Gene-specific probes used in the RNA analysis were described previously (1).
For Affymetrix GeneChip array analysis, 20 μg of total RNA from wild-type and atnup160-1 mutant seedlings grown on two separate halves of the sample agar plates treated with cold (0°C, 24 h) was extracted using the RNeasy plant minikit (QIAGEN) and used to make biotin-labeled cRNA targets. An Affymetrix Arabidopsis ATH1 genome array, the GeneChip array, which contains >22,500 probe sets representing ∼24,000 genes, was used, and hybridization, washing, and staining were carried out as instructed in the manufacturer's manual. Microarray data were obtained from scanned GeneChip images and analyzed using Microarray Suite version 5.0.1 software (Affymetrix) (1). The array experiment was repeated with independent plant materials.
Electrolyte leakage and cold tolerance assays.
The electrolyte leakage test was performed to compare membrane integrity and chilling sensitivity of wild-type and atnup160-1 plants. Two-week-old plants grown in soil were transferred to a cold room and treated at 4°C in the light. Several rosette leaves from either treated or untreated plants were detached and transferred to tubes with 10 ml of deionized water. The conductivity of the solution was measured after shaking overnight at room temperature. After measurement, the samples were autoclaved. After shaking at room temperature for 2 h, the conductivity of the solution was measured again. The percentage of electrolyte leakage was calculated as the percentage of the conductivity before autoclaving divided by that after autoclaving (18).
For observation of chilling tolerance, 2-week-old plants grown in soil under normal growth conditions were transferred to a cold room at 4°C in light. The freezing test was carried out as described previously (1).
Genetic mapping and cloning of AtNUP160.
For genetic mapping of the atnup160-1 mutation, the mutant plants from the Columbia ecotype were crossed with wild-type plants of the Ler ecotype. A total of 1,011 homozygous atnup160-1 mutants were chosen from the segregated F2 population. Genomic DNA extracted from these seedlings was used for PCR-based mapping with simple sequence polymorphism markers.
A cDNA containing a full-length open reading frame (ORF) of AtNUP160 was obtained by reverse transcription (RT)-PCR and cloned into a vector (pEGAD) in frame with a gene encoding green fluorescent protein (GFP). The transient expression of GFP-AtNUP160 was conducted as previously described (2). Photographs were taken using a confocal laser scanning microscope (Leica TCS SP2).
Poly(A)-mRNA in situ localization.
Poly(A)-mRNA in situ hybridization was conducted as described previously (8). The samples were observed under a confocal laser scanning microscope (Leica TCS SP2).
RESULTS
Identification of the atnup160-1 mutant plants.
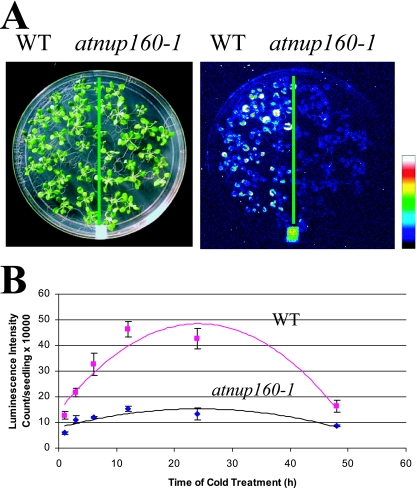
To study the genetic control of plant responses to cold stress, we previously generated transgenic Arabidopsis plants expressing the firefly luciferase reporter gene under control of the cold-responsive CBF3 promoter (1). The transgenic plants emit bioluminescence in response to low temperatures. Using these plants as the starter line (herein referred to as the wild type), mutants with an altered luminescence response to cold treatment were selected from ethyl methanesulfonate-mutagenized plants by luciferase imaging. Preliminary screening identified a number of mutant genes that showed abnormal responses to the cold regulation of CBF3-LUC expression. One of these, subsequently identified as an Arabidopsis homolog of human nucleoporin, Nup160, was designated as AtNUP160. Figure 1A shows a comparison of luminescence images of atnup160-1 mutant seedlings and wild-type seedlings under cold treatment (4°C) in the light. The luminescence images of atnup160-1 mutant seedlings appear less bright than those of the wild type. After 6 h of cold treatment, atnup160-1 mutant plants emitted only ∼30% of the luminescence of the wild-type plants (Fig. 1B).
FIG. 1.
The atnup160-1 mutation impairs cold-induced luminescence expression from CBF3-LUC. (A) Luminescence images of wild-type (WT, left) and atnup160-1 (right) mutant plants taken after cold treatment at 4°C for 6 h. The color scale at the right shows the luminescence intensity, from dark blue (lowest) to white (highest). (B) Time course of luminescence induction in wild-type and atnup160-1 mutant plants by cold treatment. Shown are the means ± standard deviations from 20 individual seedlings.
The atnup160-1 mutant plants were backcrossed with the wild type, and the resulting F1 seedlings were examined for CBF3-LUC luminescence after 6 h of cold treatment at 4°C. All of the F1 plants showed a wild-type level of luminescence. The F2 population from the self-fertilized F1 plant segregated in ∼3:1 ratio of wild-type to mutant plants, respectively (data not shown), indicating that the mutant is caused by a recessive mutation in a single nuclear gene. atnup160-1 mutant plants were backcrossed to the wild type three times to remove possible unlinked mutations. All subsequent characterizations were performed using the mutants that had been backcrossed.
Effect of the atnup160 mutation on endogenous cold-regulated gene expression.
We conducted Northern blot hybridization to determine the transcript levels of cold-induced CBF genes and their downstream genes. Total RNA was extracted from atnup160-1 mutant seedlings and wild-type seedlings that were treated with low temperatures. The transcript level of CBF3 as well as that of CBF1 and CBF2 was substantially reduced in the mutant plants. Wild-type plants showed induction of the CBF genes after 1 h of cold treatment, and the expression peaked at 3 h. For atnup160-1 plants, the transcript levels of CBF genes also peaked at 3 h after cold treatment, but the levels were greatly reduced compared to those of the wild type (see Fig. S1 in the supplemental material). This observation of reduced endogenous CBF gene expression in the atnup160-1 mutant plants is consistent with that of the reduced luminescence of the CBF3-LUC transgene in the mutant plants (Fig. 1). We also examined the transcript levels of two CBF target genes and found that the transcript levels of COR47 and RD29A in the atnup160 mutants were not lower than those in wild-type plants. Rather, COR47 and RD29A transcript levels were higher in the mutant plants at the 12- and 48-h time points during cold treatment (see Fig. S1 in the supplemental material).
To examine additional alterations of the gene expression of atnup160-1, we used Affymetrix GeneChip arrays containing about 24,000 genes per chip. Transcript levels of atnup160-1 were analyzed after plants were transferred from 22°C to 0°C at the 24-h time point. Comparisons between transcript levels of atnup160-1 and those of wild-type plants indicated that the transcript levels of 39 genes decreased and those of 17 genes increased by at least twofold for atnup160-1 mutant plants relative to those for the wild-type (see Table S1 in the supplemental material).
The GeneChip analysis revealed that other genes important for plant cold tolerance were affected in the atnup160-1 mutant plants. These included a cold-induced LEA gene (At1g52690) and genes involved in sugar metabolism or responsive to oxidative stress. During cold acclimation, sugars, including sucrose and raffinose, accumulate in Arabidopsis (7, 32), and the genes with roles in sugar synthesis, sugar metabolism, and sugar transport are up-regulated in response to low temperatures. The transcript levels of sucrose synthase (At3g43190), polygalacturonase (At1g48100), and arabinogalactan proteins (At5g53250 and At4g26320) in atnup160-1 were at least twofold lower than those in the wild type (see Table S1 in the supplemental material). In addition, the expression of several genes known to be responsive to oxidative stress was lower in atnup160-1 mutant plants. These include a glutathione S-transferase (At1g49860), peroxidases (At5g64110 and At5g67400), and a stromal ascorbate peroxidase (At4g08390). In contrast, the transcript of an oxidoreductase (At3g49620) was higher in the mutant (see Table S1 in the supplemental material). In summary, the atnup160-1 mutation affects the expression of a number of stress-related genes, although its impact on global gene expression appears to be rather limited.
The atnup160-1 mutation renders the plant sensitive to chilling and freezing stresses.
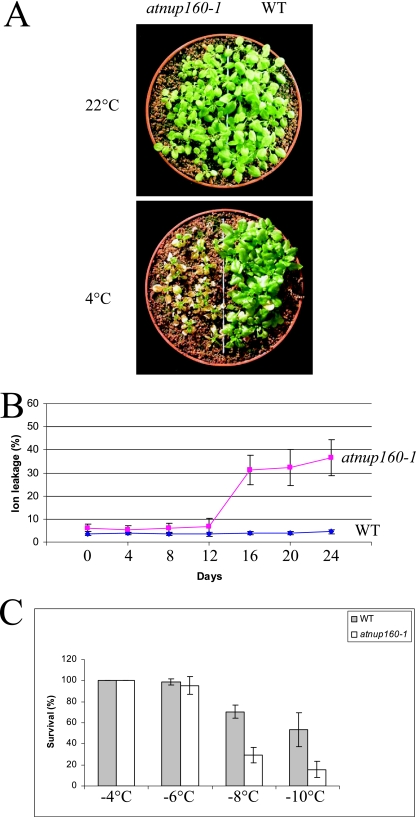
Impaired cold-regulated gene expression of atnup160-1 prompted us to investigate the cold stress tolerance of the mutant plants. Under normal growth conditions, the atnup160-1 mutant plant was like the wild type, although the mutant seedlings were slightly smaller (Fig. 2A). When plants were treated at 4°C in the light, the growth of the atnup160-1 mutant plants was dramatically retarded, and dead leaves appeared after 3 weeks of cold treatment (Fig. 2A). In contrast, the chilling treatment caused no apparent damage to wild-type plants (Fig. 2A).
FIG. 2.
Cold sensitivity of atnup160-1 mutant plants. (A) Fourteen-day-old wild-type (WT) and atnup160-1 mutant plants grown at 22°C were shifted to 4°C in the light for 21 days. (B) Ion leakage was assayed using wild-type and atnup160-1 mutant leaves from 14-day-old plants grown in soil at 22°C and then shifted to 4°C in the light for the indicated number of days. Data shown are means with standard errors (n = 10). (C) Survival rates for wild-type and atnup160-1 mutant plants were quantified 2 days after freezing treatment following a 4-day cold acclimation. Data shown are means with standard errors (n = 10).
To examine the extent of chilling injury to atnup160-1 mutant plants, leaves excised from soil-grown plants treated at 4°C in the light for different time periods were used for electrolyte leakage assays (18). As shown in Fig. 2B, wild-type leaves showed little increase in electrolyte leakage during a 24-day cold treatment, while atnup160-1 mutant leaves showed a drastic increase in electrolyte leakage after only 12 days of cold treatment.
We analyzed the effect of cold acclimation on the freezing tolerance of atnup160-1 mutant plants. Without cold acclimation, wild-type and atnup160-1 mutant plants had the same level of freezing tolerance (data not shown). Both the wild-type and the atnup160-1 mutant plants survived −4°C freezing, but neither survived −8°C freezing. Ten-day-old atnup160-1 mutant and wild-type seedlings grown on separate halves of the same agar plates were cold acclimated at 4°C for 4 days and were then subjected to a freezing tolerance assay. The atnup160-1 mutants were less tolerant to freezing than the wild types at all freezing temperatures (Fig. 2C). For example, most of the atnup160-1 mutant plants did not survive freezing at −8°C. On the contrary, ∼80% of the wild-type plants survived the −8°C freezing. Even after freezing at −10°C, about half of the wild-type plants survived. These results demonstrate that the atnup160-1 mutant is defective in cold acclimation, i.e., the mutant is impaired in acquired freezing tolerance.
Early flowering and growth phenotypes in atnup160-1 mutant plants.
As shown in Fig. 3, atnup160-1 mutant plants flowered earlier and produced fewer rosette leaves than wild-type plants (average times to flowering after germination were 32.8 ± 1.83 days for wild-type plants and 21.9 ± 1.10 days for atnup160-1 mutant plants; average numbers of rosette leaves were 11.2 ± 0.79 for wild-type plants and 5.7 ± 0.48 for atnup160-1 mutant plants). Under our standard growth conditions, wild-type Arabidopsis plants produced a mean of 11 rosette leaves before they formed inflorescences, and the first flowers opened on average 33 days after germination. In contrast, atnup160-1 mutant plants began flowering 11 days earlier and formed about 6 rosette leaves (see above) (Fig. 3).
FIG. 3.
Early flowering phenotype of atnup160-1 mutant plants. Wild-type (WT) and atnup160-1 mutant plants gown under normal conditions were photographed at indicated time points after imbibition.
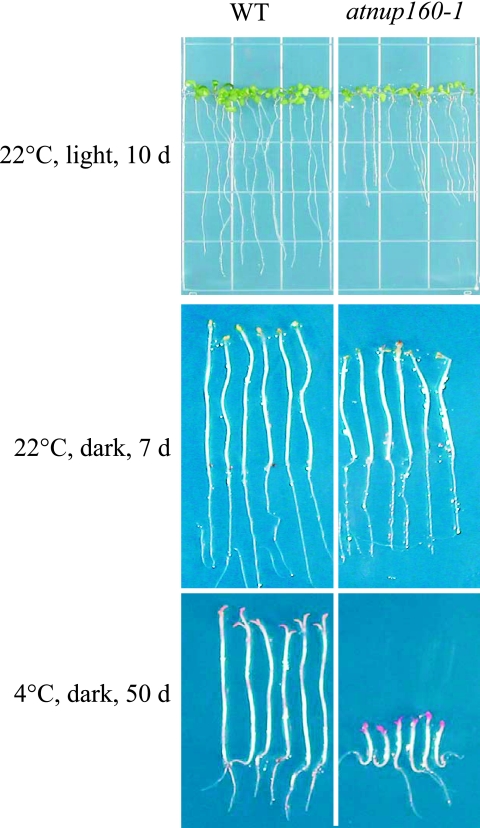
Mature atnup160-1 mutant plants were slightly smaller than the wild-type plants. The lengths of adult inflorescences and seedling roots and hypocotyls were decreased in the mutant plants (Table 1). The hypocotyl of etiolated (dark-grown) seedlings is an extremely rapidly expanding organ. To analyze the effect of the atnup160-1 mutation on hypocotyl elongation, we measured the lengths of hypocotyls of 7-day-old, dark-grown seedlings. As shown in Fig. 4, the atnup160-1 mutant hypocotyls reached only ∼72% of the length of those of the wild type. Interestingly, when the atnup160-1 mutant seedlings were treated at 4°C in the dark for 50 days, the lengths of the hypocotyls in the atnup160-1 mutant seedlings was only ∼25% of that of the wild type, indicating that hypocotyl elongation in atnup160-1 mutants is hypersensitive to inhibition by chilling stress.
TABLE 1.
Average length of seedling hypocotyls and main roots and adult inflorescences of wild-type and atnup160-1 mutant plantsa
| Genotype | Avg length (mm) of
|
||
|---|---|---|---|
| Hypocotyls | Roots | Inflorescences | |
| Wild type | 15.2 ± 0.77 | 25.6 ± 1.55 | 259.5 ± 39.54 |
| atnup160-1 | 11.0 ± 0.53 | 13.7 ± 1.58 | 110.0 ± 21.60 |
Values are means ± SD.
FIG. 4.
Retarded seedling growth of atnup160-1 mutant seedlings. Wild-type (WT) and atnup160-1 mutant seedlings grown under different conditions were compared. Growth conditions are indicated at left.
Cloning and characterization of AtNUP160.
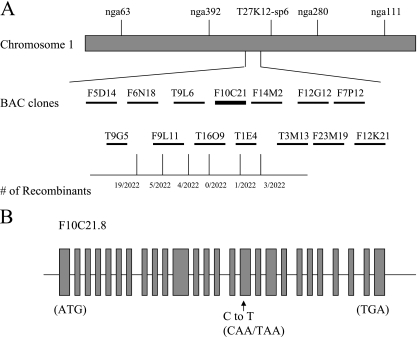
To isolate the AtNUP160 gene, a map-based cloning strategy was used. atnup160-1 mutant plants of the Columbia ecotype were crossed with wild-type plants of the Ler ecotype. The F1 plants from the cross were self-fertilized, and the resulting F2 seeds were obtained. From the segregating F2 population, a total of 1,011 atnup160-1 mutant plants were identified based on their morphology and chilling sensitivity. An initial mapping with polymorphic markers placed AtNUP160 at the middle of chromosome 1, between simple sequence length polymorphism markers nga392 and T27K12-Sp6. Subsequent mapping by using markers K6N18, F9L11, T9L6, T16O9, T1E4, and F14M2 narrowed the position of AtNUP160 to a contig of two bacterial artificial chromosome (BAC) clones (T16O9 and F10C21) (Fig. 5). The ORFs of candidate genes in this region were sequenced from the atnup160-1 mutant plants. Sequence analysis revealed a single nucleotide substitution in the atnup160-1 mutation in the F10C21.8 gene, i.e., a C-to-T change at position 37549 of the BAC clone. The mutation creates a premature stop codon (CAA to TAA) at exon 16 and thus truncates the encoded protein (Fig. 5). No mutation was found in the mutant plants in any other sequenced genes. The predicted protein shares sequence homology in its entire length with human nucleoporin Nup160 (25, 30) (see Fig. S2 in the supplemental material), indicating that Arabidopsis AtNUP160 might be a homolog of animal Nup160. Recently, Parry et al. (2006) found that the Arabidopsis suppressor of auxin resistance1 (SAR1) gene also encodes AtNUP160 (25).
FIG. 5.
Positional cloning of AtNUP160. (A) Genetic mapping with PCR-based markers shows the position of atnup160 on the contig of BAC clones F10C21 and T16O9. The numbers of recombination events out of the total numbers of chromosomes examined are indicated. (B) Sequence analysis identified a single nucleotide change (C37549 to T37549) that creates a stop codon in At1g33410 (F10C21.8).
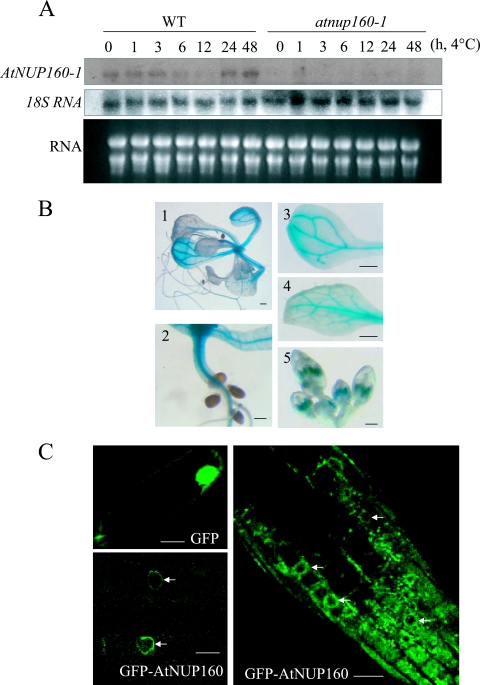
RNA hybridization was carried out to examine AtNUP160 transcript levels in Arabidopsis seedlings. In wild-type plants, AtNUP160 mRNA was detected in both the control and the cold-treated plants, and AtNUP160 expression was not cold regulated. The AtNUP160 transcript was not detected in the mutant plants, indicating that the mutation renders the AtNUP160 mRNA unstable (Fig. 6A). To examine AtNUP160 expression in various plant tissues, a 2.0-kb genomic DNA fragment upstream of the AtNUP160 start codon was fused with the β-glucuronidase reporter gene (GUS), and the resulting construct was introduced into wild-type Arabidopsis plants. All transgenic lines (a total of nine lines examined) showed similar GUS staining patterns. GUS expression was observed in all plant tissues examined (Fig. 6B). The expression was particularly strong in the vascular tissues of cotyledons, leaves, and hypocotyls. Strong GUS activity was also detected in the anthers of the flowers (Fig. 6B).
FIG. 6.
Gene expression and protein localization of AtNUP160. (A) AtNUP160 expression in wild-type (WT) and atnup160-1 mutant plants. Fourteen-day-old plants were treated at 4°C for the indicated times. A cDNA fragment of AtNUP160 3′-UTR was used as a probe. Included are 18S RNA and ethidium bromide-stained rRNA as loading controls. (B) AtNUP160 gene expression pattern as indicated by promoter-GUS analysis. Twenty-day-old seedlings grown on MS plates and flower buds from soil-grown mature plants were stained for GUS activity. Panels 1 to 5 show a seedling, a hypocotyl, a cotyledon, a leaf, and flowers, respectively. Scale bar, 0.5 mm. (C) Subcellular localization of AtNUP160. The GFP (upper left panel) and the GFP-AtNUP160 fusion gene (lower left panel) were transiently expressed in onion epidermal cells by particle bombardment. (Right panel) The root of a stable transgenic Arabidopsis plant expressing GFP-AtNUP160. Samples were observed under a confocal microscope. Arrows point to the nuclear rim. Scale bar, 50 μm.
In order to determine the subcellular localization of the AtNUP160 protein, a cDNA containing the AtNUP160 full-length ORF was amplified by RT-PCR and fused in-frame to the C terminus of GFP under the control of the 35S promoter of the cauliflower mosaic virus. The construct was transiently expressed in plant cells via particle bombardment, and GFP fluorescence was found predominantly at the nuclear rim (Fig. 6C). As a control, GFP alone from the 35S-GFP-expressing cells was found in both the cytoplasm and the nucleus (Fig. 6C). Confocal laser scanning analysis revealed a similar pattern of AtNUP160 protein localization at the nuclear rim in root tip cells of stable transgenic Arabidopsis plants expressing the GFP-AtNUP160 fusion protein (Fig. 6C).
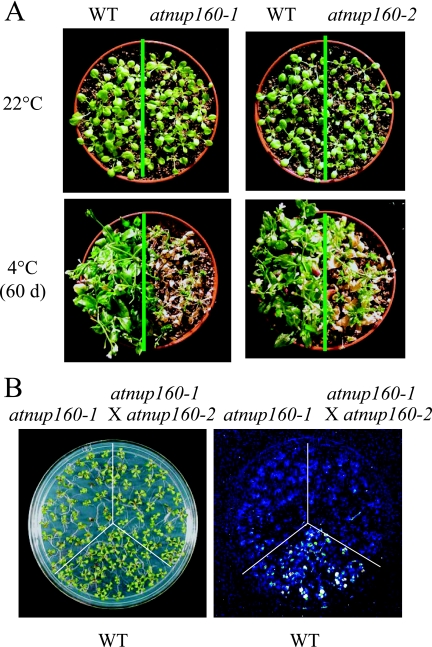
A T-DNA insertion mutant in AtNUP160 (Salk_016091) (see Fig. S3 in the supplemental material) was obtained from the Salk collection and designated atnup160-2. To examine the cold stress sensitivity of atnup160-2, the mutant plants were treated at 4°C in the light together with the wild-type and atnup160-1 mutant plants. A chilling sensitivity phenotype for atnup160-2 mutant plants was similar to that observed for atnup160-1 mutant plants (Fig. 7A). In addition, the atnup160-2 mutant plants also flowered earlier than the wild type. When the atnup160-2 mutant plants were crossed with atnup160-1 mutant plants, the resulting F1 plants showed a reduced CBF3-LUC expression in the cold, compared to that of the wild type (Fig. 7B). These results confirm that the AtNUP160 gene is responsible for CBF-LUC expression, chilling sensitivity, and early flowering phenotypes.
FIG. 7.
Chilling sensitivity of atnup160-2 mutant plants. (A) Comparison of the chilling sensitivity phenotypes of the atnup160-1 and the atnup160-2 mutants. Fourteen-day-old wild-type plants (WT), atnup160-1 mutant plants, and atnup160-2 mutant plants grown at 22°C were shifted to 4°C in the light for 2 months. (B) Luminescence images of the WT, the atnup160-1 mutant plants, and the F1 progeny of the atnup160-1 mutant crossed with the atnup160-2 mutant. Photographs and luminescence images were taken after cold treatment at 4°C for 6 h.
Defective nucleocytoplasmic transport of mRNAs in the atnup160-1 mutant.
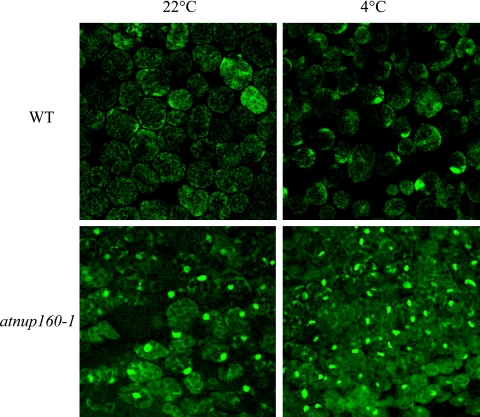
The sequence similarity between AtNUP160 and human nucleoporin Nup160 and its nuclear rim localization suggested that the Arabidopsis AtNUP160 could be a functional homolog of the human nucleoporin. To determine the potential role of AtNUP160 in mRNA export, we carried out in situ hybridization to localize poly(A)-mRNA in wild-type and atnup160-1 mutant plants. Leaves of atnup160-1 mutant and wild-type plants grown under normal conditions for 3 weeks in soil were hybridized with a 45-mer oligo-(dT) probe end-labeled with fluorescein. After hybridization and washing, the leaf cells were observed under a confocal microscope. As shown in Fig. 8, strong nuclear poly(A) signals were observed in atnup160-1 mutant leaf cells but not in the wild type, indicating that the export of mRNA is defective in atnup160-1 mutant plants. We also compared the nuclear poly(A) signals of atnup160-1 mutant plants to those of wild-type plants after cold treatment. Cold-treated atnup160-1 mutant plants accumulated strong poly(A)-mRNA in the nuclei, whereas the cold-treated wild-type leaf cells showed little mRNA accumulation in the nuclei (Fig. 8). These results show that Arabidopsis AtNUP160 is required for mRNA export under both warm and cold temperatures. The observation of mRNA export in the mutant at warm temperatures is support that the atnup160-1 mutation is not a cold-sensitive allele, consistent with the lack of AtNUP160 transcript in the mutant.
FIG. 8.
Defect of atnup160-1 mutant plants in mRNA export. Poly(A)-mRNA export is blocked in atnup160-1 mutant cells. Wild-type (WT) and atnup160-1 mutant plants were grown at 22°C for 3 weeks. In situ hybridization with fluorescein-labeled oligo(dT) probe was performed using leaves with or without cold treatment (22°C or 4°C, respectively).
To examine the effect of the atnup160-1 mutation on the nucleocytoplasmic transport of proteins, the mutant was crossed with a transgenic line tagged with a nucleus-localized GFP-ICE1. ICE1 was known to be an upstream regulator of the cold-induced CBF3 gene (1). A homozygous atnup160-1 mutant line expressing GFP-ICE1 was identified from the resulting F2 population and was examined under a confocal microscope. The result indicated that the nuclear localization of GFP-ICE1 was not significantly affected by the atnup160-1 mutation (not shown).
DISCUSSION
The nucleoporin Nup160 is conserved in various eukaryotic organisms. These include the human Nup160 (KIAA0197), a putative 160-kDa mouse protein (AAD17922), a 160-kDa Drosophila protein (AAF53075.1), and a more distantly related 176-kDa Caenorhabditis elegans protein (AAB37803.1) (30). Studies of vertebrates showed that Nup160 exists together with other nucleoporins in assembled nuclear pores as a complex and that the Nup160 complex plays important roles in nuclear pore assembly, intranuclear architecture, and export of RNA (12, 30, 31). Studies in yeast and animal cell cultures were limited to single cells, and the physiological roles of Nup160 in multicellular organisms are not known. In this study, we took advantage of the power of forward genetics to screen Arabidopsis and explored the in vivo function of AtNUP160 in the growth, development, and environmental responses of a multicellular organism. Our results show that AtNUP160 is important for flowering time regulation and for plant tolerance to cold stress. Recently, Parry et al. (2006) reported that AtNUP160/SAR1 also plays an important role in auxin signaling (25).
AtNUP160 appears to be the only Nup160 gene in Arabidopsis. The atnup160-1 mutation truncates most of the C-terminal half of the protein, and the nonsense mutation renders the AtNUP160 transcript undetectable in the mutant, probably due to nonsense-mediated decay. Therefore, atnup160-1 appears to be a null allele. Consistent with a critical role for AtNUP160 in RNA export, the mutation causes mRNA accumulation in the nuclei of the mutant plant. Nevertheless, the mutant plants can complete their life cycles and do not exhibit severe defects in growth and development. Considering the wide range of essential functions of the nuclear pore complex, it is likely that other nucleoporins can partially compensate for the loss of function of atnup160-1. This notion is supported by the additive effects observed with the sar1 sar3 double mutant, in which the development of the plants was more severely affected than in either of the single mutants (25). Considering their similarities in the nuclear rim localization of AtNUP160, AtNUP96 (34, 25), and the RNA helicase-like protein LOS4 (8) and their similar roles in RNA export and in the early flowering phenotypes of their loss-of-function mutants, it is likely that these proteins function together or in the same pathway which is important for plant growth, development, disease resistance, and response to cold stress.
In contrast to its largely unessential role in plant growth and development, AtNUP160 is necessary for plants to survive cold stress. The atnup160-1 mutant plants are not only defective in acquired freezing tolerance but are also deficient in chilling tolerance. The molecular mechanisms of chilling tolerance in plants are poorly understood (23, 28). Membrane lipid fatty acid desaturation has been shown to be important for chilling tolerance (23). In addition, mutational analysis showed that an Arabidopsis 16S rRNA methylase is required for the development of chloroplasts at low temperatures, indicating that there is a chilling sensitivity step in ribosome biogenesis, which is compensated by the 16S rRNA methylase (29). Our results suggest that nucleocytoplasmic transport is another cellular process critical for chilling tolerance.
The defect in chilling and freezing tolerance is probably related to the altered cold regulation of gene expression observed in the atnup160 mutant. The CBF transcription factor genes have been shown to be important for acquired freezing tolerance (7, 16, 17, 21). All three cold-induced CBF genes show substantially reduced expression in atnup160-1 mutant plants. Intriguingly, two downstream genes of the CBF transcription factors, COR47 and RD29A, do not show reduced expression in the mutant. Recent studies indicate that these downstream genes are controlled by both CBF-dependent and CBF-independent pathways (35, 36). It seems that the atnup160-1 mutation influences both pathways that may have antagonistic effects on downstream target genes like COR47 and RD29A. Microarray analysis showed that the transcript levels of other genes potentially important for plant cold tolerance are decreased in cold-treated atnup160-1 mutant plants versus similarly treated wild-type plants. These include a cold-responsive LEA-like gene (At1g52690), genes involved in sugar metabolism and oxidative stress detoxification, and genes encoding possible signal transduction pathway components (see Table S1 in the supplemental material). The chilling and freezing sensitivity phenotypes of the atnup160-1 mutant under cold conditions may be attributed to the altered expression levels of these genes. For example, the reduced expression of several oxidative stress-related genes such as glutathione S-transferases, peroxidase, and stromal ascorbate peroxidase in the atnup160-1 mutant likely contributes to the mutant phenotypes. Lipid peroxidation produces phospholipid hydroperoxides that form clusters and results in an increase in membrane permeability to electrolytes (15). Increased membrane permeability resulting from the lipid peroxidation by reactive oxygen species may contribute to the chilling sensitivity phenotype of atnup160-1 mutant plants.
The effect of the atnup160-1 mutation on cold-regulated gene expression, cold tolerance, and flowering time may be due to the defect in RNA export seen in the mutant plants. Compared to other genes in general, certain regulators of cold responses and flowering time may have a more critical RNA export requirement. In addition, the structure or transport properties of the nuclear pores may be influenced by cold temperatures such that AtNUP160 may become more important in cold-remodeled nuclear pores.
Supplementary Material
Acknowledgments
We thank Rebecca Stevenson and Xuhui Hong for excellent technical assistance.
This work was supported by grants MCB-0241450 and USDA NRI 2003-00751 to J.-K. Zhu.
Footnotes
Published ahead of print on 9 October 2006.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Chinnusamy, V., M. Ohta, S. Kanrar, B. H. Lee, X. Hong, M. Agarwal, and J. K. Zhu. 2003. ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 17:1043-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dong, C. H., B. Kost, G. Xia, and N. H. Chua. 2001. Molecular identification and characterization of the Arabidopsis AtADF1, AtADF5 and AtADF6 genes. Plant Mol. Biol. 45:517-527. [DOI] [PubMed] [Google Scholar]
- 3.Dong, C. H., M. Agarwal, Y. Zhang, Q. Xie, and J. K. Zhu. 2006. The negative regulator of plant cold responses, HOS1, is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1. Proc. Natl. Acad. Sci. USA 103:8281-8286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fahrenkrog, B., J. Koser, and U. Aebi. 2004. The nuclear pore complex: a jack of all trades? Trends Biochem. Sci. 29:175-182. [DOI] [PubMed] [Google Scholar]
- 5.Gilmour, S. J., N. N. Artus, and M. F. Thomashow. 1992. cDNA sequence analysis and expression of two cold-regulated genes of Arabidopsis thaliana. Plant Mol. Biol. 18:13-32. [DOI] [PubMed] [Google Scholar]
- 6.Gilmour, S. J., D. G. Zarka, E. J. Stockinger, M. P. Salazar, J. M. Houghton, and M. F. Thomashow. 1998. Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J. 16:433-442. [DOI] [PubMed] [Google Scholar]
- 7.Gilmour, S. J., A. M. Sebolt, M. P. Salazar, J. D. Everard, and M. F. Thomashow. 2000. Overexpression of the Arabidopsis CBF3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation. Plant Physiol. 124:1854-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gong, Z., C. H. Dong, H. Lee, J. Zhu, L. Xiong, D. Gong, B. Stevenson, and J. K. Zhu. 2005. A DEAD box RNA helicase is essential for mRNA export and important for development and stress responses in Arabidopsis. Plant Cell 17:256-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gong, Z., H. Lee, L. Xiong, A. Jagendorf, B. Stevenson, and J. K. Zhu. 2002. RNA helicase-like protein as an early regulator of transcription factors for plant chilling and freezing tolerance. Proc. Natl. Acad. Sci. USA 99:11507-11512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gorlich, D., and U. Kutay. 1999. Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 15:607-660. [DOI] [PubMed] [Google Scholar]
- 11.Guo, Y., L. Xiong, M. Ishitani, and J. K. Zhu. 2002. An Arabidopsis mutation in translation elongation factor 2 causes superinduction of CBF/DREB1 transcription factor genes but blocks the induction of their downstream targets under low temperature. Proc. Natl. Acad. Sci. USA 99:7786-7791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harel, A., A. V. Orjalo, T. Vincent, A. Lachish-Zalait, S. Vasu, S. Shah, E. Zimmerman, M. Elbaum, and D. J. Forbes. 2003. Removal of a single pore subcomplex results in vertebrate nuclei devoid of nuclear pores. Mol. Cell 11:853-864. [DOI] [PubMed] [Google Scholar]
- 13.Ishitani, M., L. Xiong, H. J. Lee, B. Stevenson, and J. K. Zhu. 1998. HOS1, a genetic locus involved in cold-responsive gene expression in Arabidopsis. Plant Cell 10:1151-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jaglo-Ottosen, K. R., S. J. Gilmour, D. G. Zarka, O. Schabenberger, and M. F. Thomashow. 1998. Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 280:104-106. [DOI] [PubMed] [Google Scholar]
- 15.Kagan, V. E. 1988. Lipid peroxidation in biomembranes. CRC Press, Boca Raton, Fla.
- 16.Kasuga, M., Q. Liu, S. Miura, K. Yamaguchi-Shinozaki, and K. Shinozaki. 1999. Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat. Biotechnol. 17:287-291. [DOI] [PubMed] [Google Scholar]
- 17.Lee, B. H., D. A. Henderson, and J. K. Zhu. 2005. The Arabidopsis cold-responsive transcriptome and its regulation by ICE1. Plant Cell 17:3155-3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee, H., Y. Guo, M. Ohta, L. Xiong, B. Stevenson, and J. K. Zhu. 2002. LOS2, a genetic locus required for cold-responsive gene transcription encodes a bifunctional enolase. EMBO J. 21:2692-2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee, H., L. Xiong, Z. Gong, M. Ishitani, B. Stevenson, and J. K. Zhu. 2001. The Arabidopsis HOS1 gene negatively regulates cold signal transduction and encodes a RING finger protein that displays cold-regulated nucleo-cytoplasmic partitioning. Genes Dev. 15:912-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Levitt, J. 1980. Responses of plants to environmental stress, vol. 1: chilling, freezing, and high temperature stress. Academic Press, New York, N.Y.
- 21.Liu, Q., M. Kasuga, Y. Sakuma, H. Abe, S. Miura, K. Yamaguchi-Shinozaki, and K. Shinozaki. 1998. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10:1391-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Medina, J., M. Bargues, J. Terol, M. Perez-Alonso, and J. Salinas. 1999. The Arabidopsis CBF gene family is composed of three genes encoding AP2 domain-containing proteins whose expression is regulated by low temperature but not by abscisic acid or dehydration. Plant Physiol. 119:463-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nishida, I., and N. Murata. 1996. Chilling sensitivity in plants and cyanobacteria: the crucial contribution of membrane lipids. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47:541-568. [DOI] [PubMed] [Google Scholar]
- 24.Nordin, K., T. Vahala, and E. T. Palva. 1993. Differential expression of two related, low-temperature-induced genes in Arabidopsis thaliana (L.) Heynh. Plant Mol. Biol. 21:641-653. [DOI] [PubMed] [Google Scholar]
- 25.Parry, G., S. Ward, A. Cernac, S. Dharmasiri, and M. Estelle. 2006. The Arabidopsis suppressor of auxin resistance proteins are nucleoporins with an important role in hormone signaling and development. Plant Cell 18:1590-1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rout, M. P., J. D. Aitchison, A. Suprapto, K. Hjertaas, Y. Zhao, and B. T. Chait. 2000. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J. Cell Biol. 148:635-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stockinger, E. J., S. J. Gilmour, and M. F. Thomashow. 1997. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl. Acad. Sci. USA 94:1035-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tokuhisa, J., and J. Browse. 1999. Genetic engineering of plant chilling tolerance. Genet. Eng. (New York) 21:79-93. [DOI] [PubMed] [Google Scholar]
- 29.Tokuhisa, J. G., P. Vijayan, K A. Feldmann, and J. A. Browse. 1998. Chloroplast development at low temperatures requires a homolog of DIM1, a yeast gene encoding the 18S rRNA dimethylase. Plant Cell 10:699-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vasu, S., S. Shah, A. Orjalo, M. Park, W. H. Fischer, and D. J. Forbes. 2001. Novel vertebrate nucleoporins Nup133 and Nup160 play a role in mRNA export. J. Cell Biol. 155:339-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walther, T. C., A. Alves, H. Pickersgill, I. Loiodice, M. Hetzer, V. Galy, B. B. Hulsmann, T. Kocher, M. Wilm, T. Allen, I. W. Mattaj, and V. Doye. 2003. The conserved Nup107-160 complex is critical for nuclear pore complex assembly. Cell 113:195-206. [DOI] [PubMed] [Google Scholar]
- 32.Wanner, L. A., and O. Junttila. 1999. Cold-induced freezing tolerance in Arabidopsis. Plant Physiol. 120:391-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamaguchi-Shinozaki, K., and K. Shinozaki. 1994. A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 6:251-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang, Y., and X. Li. 2005. A putative nucleoporin 96 is required for both basal defense and constitutive resistance responses mediated by suppressor of npr1-1, constitutive 1. Plant Cell 17:1306-1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu, J., H. Shi, B. H. Lee, B. Damsz, S. Cheng, V. Stirm, J. K. Zhu, P. M. Hasegawa, and R. A. Bressan. 2004. An Arabidopsis homeodomain transcription factor gene, HOS9, mediates cold tolerance through a CBF-independent pathway. Proc. Natl. Acad. Sci. USA 101:9873-9878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu, J., P. E. Verslues, X. Zheng, B. H. Lee, X. Zhan, Y. Manabe, Y. Zhu, C. H. Dong, J. K. Zhu, P. M. Hasegawa, and R. A. Bressan. 2005. HOS10 encodes an R2R3-type MYB transcription factor essential for cold acclimation in plants. Proc. Natl. Acad. Sci. USA 102:9966-9971. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.