Abstract
The nucleoprotein filament formed by Rad51 polymerization on single-stranded DNA is essential for homologous pairing and strand exchange. ATP binding is required for Rad51 nucleoprotein filament formation and strand exchange, but ATP hydrolysis is not required for these functions in vitro. Previous studies have shown that a yeast strain expressing the rad51-K191R allele is sensitive to ionizing radiation, suggesting an important role for ATP hydrolysis in vivo. The recruitment of Rad51-K191R to double-strand breaks is defective in vivo, and this phenotype can be suppressed by elimination of the Srs2 helicase, an antagonist of Rad51 filament formation. The phenotype of the rad51-K191R strain is also suppressed by overexpression of Rad54. In vitro, the Rad51-K191R protein exhibits a slight decrease in binding to DNA, consistent with the defect in presynaptic filament formation. However, the rad51-K191R mutation is dominant in heterozygous diploids, indicating that the defect is not due simply to reduced affinity for DNA. We suggest the Rad51-K191R protein either forms an altered filament or is defective in turnover, resulting in a reduced pool of free protein available for DNA binding.
The recognition and repair of DNA damage are vital to maintaining genome integrity. Homologous recombination is required to repair double-strand breaks (DSBs) and single-strand gaps that arise spontaneously during normal cellular processes or by treatment of cells with DNA damaging agents. The RAD52 group of genes was originally identified for Saccharomyces cerevisiae by the sensitivity of mutants to ionizing radiation (IR), and subsequent studies have shown that these genes are required for homologous recombination (57). Homologues of most of the Rad52 group of proteins have been found in other eukaryotes, indicating functional conservation of the homologous recombination pathway. RAD51 encodes a structural and functional homologue of the RecA protein, which is essential for homologous recombination in Escherichia coli (1, 2, 45). Like RecA, Rad51 polymerizes on single- and double-stranded DNA (ssDNA and dsDNA) to form nucleoprotein filaments in which the DNA is in an extended conformation (34, 55). The Rad51-ssDNA filament is active in homologous pairing and strand exchange to form heteroduplex DNA, a central intermediate in homologous recombination. The binding of Rad51 to DNA requires ATP but is independent of ATP hydrolysis (8, 31, 40, 53, 56). Complexes between human Rad51 and ssDNA or dsDNA are stabilized by the nonhydrolyzable ATP analog, AMP-PNP, and strand exchange with oligonucleotide substrates is more efficient in the presence of AMP-PNP (8, 40). Substitution of Mg2+ with Ca2+ also stabilizes Rad51-DNA complexes and stimulates strand exchange by slowing the rate of ATP hydrolysis (4). Because inhibition of ATP hydrolysis stabilizes Rad51-DNA filaments, it has been proposed that the function of ATP hydrolysis by Rad51 is in disassembly of filaments, as suggested previously for the RecA protein (8, 26, 40).
To address the role of ATP binding and hydrolysis in Rad51 function, the invariant lysine residue within the Walker A motif of the yeast Rad51 and human Rad51 proteins has been substituted with alanine or arginine. The yeast Rad51-K191A protein is unable to hydrolyze ATP or bind to ssDNA in vitro, and this allele confers a rad51 null phenotype in vivo and is dominant negative in the presence of wild-type RAD51 (10, 29, 45, 56). Although the human Rad51-K133A protein binds to DNA, the filaments formed are inactive, and as in yeast, the rad51-K133A allele is phenotypically null and confers a dominant-negative phenotype in RAD51+/+ mouse embryonic stem (ES) cells (8, 30, 48, 49). By contrast, the yeast Rad51-K191R and human Rad51-K133R proteins are active for DNA binding and strand exchange (8, 30, 56). The phenotype of yeast cells expressing the rad51-K191R allele depends on the expression level of the mutant allele. When expressed in single copy from the native RAD51 promoter, a rad51-K191R strain shows high sensitivity to IR and is defective for mating-type switching, a DSB-induced gene conversion event (29). However, when expressed at high levels, the rad51-K191R allele fully complements the IR and methyl methanesulfonate sensitivity of a rad51 null strain (29, 56). Expression of the human Rad51-K133R protein in a chicken DT40 cell line was able to support proliferation after depletion of the native protein in some cell lines, suggesting that ATP hydrolysis is not required for the essential function of Rad51 in vertebrates; however, the mutant protein was expressed at higher levels than wild-type Rad51 in the viable cell lines (30). A dominant-negative effect on homologous recombination was observed when the human RAD51-K133R allele was expressed in mouse ES cells, consistent with an important role for ATP hydrolysis in RAD51 function (48, 49).
In the case of RecA, the RecA-K72R protein shows highly attenuated ATPase activity but is still able to bind nucleotide cofactor (dATP), forms extended filaments on ssDNA, and promotes limited strand exchange (38, 42). However, the recA-K72R allele is unable to complement the UV sensitivity of a recA null strain (24). While these results suggest ATP hydrolysis is essential for RecA function in vivo, there are other ATPase-defective alleles of recA that confer less-profound DNA repair and recombination deficiency in vivo (5).
In a previous study we reported suppression of the IR sensitivity of haploid rad51-K191R cells by expression of both mating-type alleles (MATa and MATα) or by high-copy-number expression of RAD54 (29). Although the suppression by mating-type heterozygosity is not understood, it is likely to function by increasing the activity of Rad51, because some other DNA repair mutants that are suppressed by mating-type heterozygosity (rad55Δ, rad57Δ, rad52-20, and rad52-327) are also suppressed by overexpression of RAD51 (3, 13, 16, 25, 41). Rad54 stabilizes the Rad51 nucleoprotein filament in vitro, but the major function of Rad54 in recombination is thought to be postsynaptic, because Rad51 is still able to associate with DSBs in vivo in rad54 mutants (20, 27, 28, 52). Furthermore, Rad54 is able to displace Rad51 from duplex DNA, leading to the suggestion that one function of Rad54 is in turnover of Rad51 by removing it from heteroduplex joints following strand exchange (47). Thus, one hypothesis for the rad51-K191R defect in vivo is an inability to turn over at the end of recombination. Such a defect would be consistent with the suppression by high levels of Rad54 and also with overexpression of the Rad51-K191R protein (29, 56). Studies with the human Rad51-K133R protein have shown formation of more stable D loops by the mutant protein, which could be explained by more stable binding to the heteroduplex DNA joint molecule following strand exchange (46). The biochemical studies predict that strand exchange is initiated but not completed in the rad51-K191R mutant, resulting in the mitotic recombination and repair defects. To test this hypothesis, we analyzed the recruitment of Rad51-K191R to DSBs in vivo, as well as genetic interactions with Srs2, a helicase that displaces Rad51 from ssDNA (19, 60). In contrast to our expectations, the defect caused by the rad51-K191R allele is at an early step, preventing normal association of Rad51 with ssDNA. We suggest that the Rad51-K191R protein is unable to form a competent nucleoprotein filament and this can be overcome by high levels of Rad54 or by deletion of SRS2.
MATERIALS AND METHODS
Media, growth conditions, and genetic methods.
Rich medium (yeast extract-peptone-dextrose [YPD]), synthetic complete medium (SC) lacking the appropriate amino acids or nucleic acid bases, sporulation medium, and genetic methods were as described previously (43). Raffinose (2%) was substituted for glucose as a nonrepressing carbon source in SC medium minus tryptophan (Trp) that was used for the galactose induction of HO in the physical analysis of mating-type switching. Yeast extract-peptone (YP) medium containing 2% lactate (pH 5.5) was used for the galactose induction of HO in the chromatin immunoprecipitation assays. Transformation of yeast cells was performed by the lithium acetate method (15).
Yeast strains and plasmids.
Saccharomyces cerevisiae strains used in this study are listed in Table 1. All strains are in the RAD5-corrected W303 background (his3-11,15 leu2-3,112 trp1-1 ura3-1 ade2-1 can1-100 RAD5) except JKM179 and its derivatives, LSY1750-1 and LSY1751. The yellow fluorescent protein (YFP) fusion strains were made by crossing the appropriate haploid parents, sporulating the resulting diploids, and screening the haploid progeny for the correct phenotype; the expression of YFP was confirmed by epifluorescence microscopy. To construct LSY1576 and LSY1752, pRS406-rad51-K191R or pRS406-rad51-K191A, respectively, was cut with Bsu36I and transformed into the YFP-RAD51 strain, W4121-20D (20). The resulting Ura+ transformants were patched to synthetic medium containing 5-fluoroorotic acid to select for pop-out events and then screened for ionizing radiation sensitivity conferred by the rad51-K191 or rad51-K191A allele. The RAD51 locus of these strains was PCR amplified and sequenced to confirm the substitution of the RAD51 allele with either rad51-K191R or rad51-K191A. LSY1750-1 was made by the same method, using pRS406-rad51-K191R and transforming into strain JKM179 (52) (kindly provided by J. Haber). LSY1751 was made by a one-step gene replacement of the RAD51 locus in the JKM179 strain with a linear PCR fragment containing homologous 5′ and 3′ flanking sequences from the rad51::KANMX deletion strain, resulting in the integration of the KANMX marker and the loss of the wild-type RAD51 allele. LSY1753-1C was made by mating LSY1102-1B and LSY977 and screening the haploid segregants from sporulated diploids for the correct phenotype. The presence of the 3× hemagglutinin (HA) epitope tag at the carboxy terminus encoded by RAD52 was confirmed by PCR using primers 5′AAGAACTGGGGCCTCATATG and 5′GATCCCCGGGAATTGCCATGAGCGTAATCTGGAACGTC. LSY1755-1 was made by transforming the HIS3-containing pRS413 or the LEU2-containing pRS415 vector into LSY1309-1 or LSY1205-2A, respectively, and mating the resulting transformants of each strain to each other. Diploids were selected on SC medium minus histidine and leucine.
TABLE 1.
Yeast strains used in this work
| Strain | Genotypea | Source or reference |
|---|---|---|
| W1588-4C | MATa | R. Rothstein |
| W1588-4A | MATα | R. Rothstein |
| W4121-20D | MATaADE2 bar1::LEU2 YFP-RAD51 | 20 |
| W3778-2B | MATaADE2 bar1::LEU2 RAD55-YFP | 20 |
| W4644-4A | MATaADE2 bar1::LEU2 RAD54-YFP | 20 |
| W5857-15A | MATaADE2 bar1::LEU2 YFP-RAD51 RAD54-CFP rad55 | R. Rothstein |
| J894 | MATaRAD52-HA | R. Rothstein |
| HKY590-1D | MATasrs2::HIS3 | H. Klein |
| HKY598-2C | MATarad57::LEU2 | H. Klein |
| HKY1434-4A | MATaADE2 CAN1 srs2-K41A | 18 |
| JKM179 | MATα Δho Δhml::ADE1 Δhmr::ADE1 ade1-100 leu2-3,112 lys5 trp1::hisG ura3-52 ade3::GAL10-HO | J. Haber |
| LSY411 | MATα rad51::URA3 | 37 |
| LSY977 | MATarad51-K191R-URA3-rad51-K191R | 29 |
| LSY979 | MATarad51-K191R | 29 |
| LSY1102-1B | MATα RAD59-V5-URA3-rad59::LEU2 RAD52-HA rad51::HIS3 met17-s | 9 |
| LSY1197 | MATarad51-K191R-URA3-rad51-K191R rad57::LEU2 | This study |
| LSY1205-2A | MATα ade2-n-URA3-ade2-a rad51-K191R | This study |
| LSY1309-1 | matΔ::URA3 | This study |
| LSY1388-1C | MATα rad51-K191R-URA3-rad51-K191R srs2::HIS3 | This study |
| LSY1391 | MATarad57::LEU2 srs2::HIS3 | |
| LSY1405-1A | MATarad51::LEU2 srs2::TRP1 | This study |
| LSY1576 | MATaADE2 bar1::LEU2 YFP-rad51-K191R | This study |
| LSY1596-4A | MATarad51-K191R-URA3-rad51-K191R rad57::LEU2 srs2::HIS3 | This study |
| LSY1752 | MATaADE2 bar1::LEU2 YFP-rad51-K191A | This study |
| LSY1743-5C | MATaADE2 RAD54-YFP rad51-K191R-URA3-rad51-K191R | This study |
| LSY1745 | MATα RAD54-YFP rad51-K191A-URA3-rad51-K191A | This study |
| LSY1754 | MATα ADE2 RAD54-YFP rad51-K191R-URA3-rad51-K191R srs2::HIS3 | This study |
| LSY1748 | MATabar1::LEU2 RAD54-YFP rad51-K191A-URA3-rad51-K191A srs2::HIS3 | This study |
| LSY1744-1A | MATaADE2 bar1::LEU2 RAD55-YFP rad51-K191R-URA3-rad51-K191R | This study |
| LSY1746-1A | MATaADE2 bar1::LEU2 RAD55-YFP rad51-K191A-URA3-rad51-K191A | This study |
| LSY1750-1 | MATα ΔhoΔhml::ADE1 Δhmr::ADE1 ade1-100 | This study |
| leu2-3,112 lys5 trp1::hisG ura3-52 ade3::GAL10-HO rad51-K191R | ||
| LSY1751 | MATα ΔhoΔhml::ADE1 Δhmr::ADE1 ade1-100 | This study |
| leu2-3,112 lys5 trp1::hisG ura3-52 ade3::GAL10-HO rad51::kanMX6 | ||
| LSY1753-1C | MATaRAD52-HA rad51-K191R-URA3-rad51-K191R | This study |
| LSY1755-1 | matΔ::URA3/MATα RAD51/rad51-K191R | This study |
| LSY1759 | matΔ::URA3/MATα RAD51/RAD51 | This study |
| LSY1781 | matΔ::URA3/MATα RAD51/rad51::HIS3 | This study |
All strains are in the RAD5-corrected W303 background (his3-11,15 leu2-3,112 trp1-1 ura3-1 ade2-1 can1-100 RAD5) except JKM179, LSY1750-1, and LSY1751; only differences from this genotype are noted.
The plasmid for expression and purification of Rad51 from Escherichia coli (pEZ5139) was a gift from S. Kowalczykowski (64). Plasmids pR51.4 (rad51-K191A) and pR51.5 (rad51-K191R) were gifts from P. Sung (56). To construct the expression plasmid for Rad51-K191R, the Bsu36I-BstEII fragment of pR51.5 was cloned into pEZ5139. To construct pRS406-rad51-K191R, a SpeI-SacI-digested PCR fragment containing rad51-K191R was made using pR51.5 as the PCR template. The PCR fragment was cloned into the pGEM-T Easy vector (Promega, Madison, WI) and subsequently cloned into the pRS406 vector using the SacI and NotI restriction sites. The plasmid pRS406-rad51-K191A was made by the same method using pR51.4 as the PCR template. Plasmid pFH800 (TRP1 ARS1 CEN4 GAL1p-HO) was provided by J. Nickoloff (33). Plasmids pR54.4 (2μm ADC1-RAD54), pR54.5 (2μm ADC1-rad54-K341A), and pR54.6 (2μm ADC1-rad54-K341R) were kindly provided by P. Sung (36).
Gamma irradiation survival assays.
Cells were grown in liquid medium to mid-log phase. The cultures were serially diluted, and aliquots of each dilution were plated on solid medium. The plates were irradiated in a Gammacell-220 irradiator containing 60Co for the designated dose. The plates were incubated for 3 days at 30°C before survivors were counted. For spot assays, cells were grown as described above, serially diluted, and spotted onto YPD plates. The plates were irradiated and incubated at 30°C for 3 days.
Microscopy.
Cells were grown in SC medium or SC lacking the appropriate amino acid until an optical density at 600 nm (OD600) of 0.2, at which time the liquid cultures were exposed to the defined doses of γ-rays in a Gammacell-220 60Co irradiator or left unirradiated, and aliquots of the cultures were processed immediately for imaging or for the time indicated postirradiation as described previously (21). Live cell images were captured as described previously (20). YFP fluorescence was acquired using Openlab software (Improvision) and quantified using Volocity software (Improvision).
Physical analysis of mating-type switching.
Strains W1588-4A, LSY1205-2A, and LSY1388-1C were transformed to Trp+ with plasmid pFH800 (33). Cells were grown in raffinose-containing SC medium minus Trp to an OD600 of 0.5. Induction of the HO endonuclease and hourly sample collections were carried out as described previously (23). Genomic DNA was extracted and digested with StyI, and DNA fragments were separated on 0.8% agarose gels and transferred to nylon membranes. Membranes were hybridized with a PCR fragment generated by amplification of a 405-bp sequence distal to the HO cut site (23).
Rad51 antibody affinity purification.
Anti-Rad51 polyclonal antibodies from rabbit were purified from crude serum by affinity chromatography as described previously (12). The chromatography column was prepared by concentrating approximately 3 mg of Rad51 protein in 600 μl of 0.1 M HEPES (pH 7.0) buffer and covalently coupled to a 200-μl bed volume of Affi-gel 15 beads (Bio-Rad).
ChIP.
Cells were grown in YP lactate at 30°C to an OD600 between 0.33 and 0.50, at which time galactose was added to a final concentration of 2% for induction of the HO endonuclease. Chromatin immunoprecipitation (ChIP) was carried out as previously described (50) with the following modifications. The lysis buffer contained 50 mM HEPES (pH 7.5), 150 mM NaCl, 1 mM EDTA, 0.1% sodium dodecyl sulfate, 1% Triton X-100, and 0.1% sodium deoxycholate. Proteins were cross-linked with 1% formaldehyde (final concentration) in 45 ml of cell culture for 10 min, followed by quenching with 125 mM glycine (final concentration) for 5 min. Cells were lysed with glass beads in 400 μl of lysis buffer with 1 mM phenylmethylsulfonyl fluoride, 1 mM dithiothreitol, 0.5 μl/ml octanol, and 4 μg/ml leupeptin-pepstatin A-aprotinin. The volume of the extracts was adjusted to 1.2 ml with lysis buffer plus protease inhibitors and sonicated for a total of 100 s, with 0.9 s pulse-on and 7 s pulse-off (model 250; Branson). For immunoprecipitation (IP), 10 μl of affinity-purified anti-Rad51 antibody was incubated with 1.2 mg of extract in 1 ml of lysis buffer overnight at 4°C and then bound to 60 μl of 50% (vol/vol) protein A-agarose beads for 2 h at 4°C. The protein-bound beads were separated by centrifugation, and 300 μl of the unbound extracts was set aside as input samples and stored at −20°C. The beads were taken through a series of washes, followed by elution of the proteins. The IP and input samples were then incubated at 65°C for 6 h to reverse the cross-links. The samples were subjected to proteinase-K treatment, phenol extraction, and ethanol precipitation.
PCR amplification.
The immunoprecipitated DNAs (undiluted) were quantified by real-time PCR amplification as described previously (6). MAT Z-specific primers, located 239 to 486 bp CEN distal to the HO cut site, were MATZ-5′ (ATGTGAACCGCATGGGCAGT)and MATZ-3′ (ACCCTTATCTACTTGCCTCT) (6). Primers specific for the ACT1 promoter were ACT1-5′ (TTTGAAACCAAACTCGCCTCTCTC) and ACT1-3′ (CTTGGTTTGAGTAGAAAGGGGAAGG) (6). The relative IP is the signal from immunoprecipitated DNA divided by the signal from the corresponding input DNA (1:1,000 dilutions). Each resulting IP/input ratio from the MAT locus was divided by the IP/input ratio obtained at the control ACT1 locus. The final number represents the n-fold enrichment over the value for the zero time point.
Coimmunoprecipitation.
Cells were grown to mid-log phase in YPD at 30°C, at which time 50 ml of cells were collected, washed twice with 20 mM Tris (pH 7.5)-400 mM NaCl, and stored at −80°C. Cells were resuspended in 400 μl lysis buffer (50 mM HEPES-KOH [pH 7.5], 400 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP-40, 1:1,000 dilution of Protease Cocktail IV [Calbiochem], 1 mM phenylmethylsulfonyl fluoride, 1 mM benzamidine, and 1 μg/ml leupeptin). Extracts were prepared and immunoprecipitated as previously described (50). For immunoprecipitations of native Rad51 or Rad51-K191R proteins, extracts were incubated with 12.5 μl anti-Rad51 crude serum antibody at 4°C for 3 h and then incubated with 60 μl 50% (vol/vol) protein A-agarose beads at 4°C for 1 h. Immunoprecipitated complexes were analyzed by Western blotting using a 1:500 dilution of anti-HA monoclonal antibody (12CA5; Roche) to detect Rad52-HA.
Protein purification.
Rad51 was expressed in E. coli strain BL21(DE3)/pLysS (Novagen, Inc., Madison, WI) using plasmid pEZ5139 and purified as previously described (64). The Rad51-K191R protein was purified by the same procedure.
DNA binding assay.
DNA binding was measured by retention of protein-DNA complexes on nitrocellulose filters as previously described (63). pUC19 DNA was linearized with EcoRI and end labeled with Klenow fragment in the presence of [α-32P]dATP. Single-stranded DNA was made by heat denaturing the labeled DNA and then quenching on ice. Increasing amounts of protein were incubated with 100 nM of ssDNA in binding buffer (40 mM morpolineethanesulfonic acid [pH 6.4], 4 mM magnesium chloride, 1 mM dithiothreitol, 1 mM ATP, 5% glycerol). Reactions were incubated at 37°C for 15 min and then passed through the nitrocellulose and DEAE membranes in a vacuum manifold. DNA binding was measured as the amount of DNA retained on the nitrocellulose membrane over the total amount retained on both membranes. Data were quantified with a Molecular Dynamics Storm 445 SI PhosphorImager and IMAGE-QUANT software. The data shown represent averages for three trials.
RESULTS
Hydrolysis of ATP by Rad54 is required for suppression of ionizing radiation sensitivity of the rad51-K191R strain.
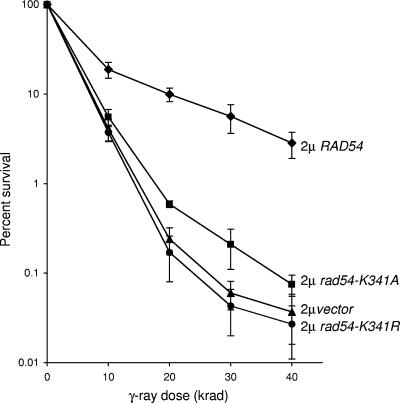
Previous studies demonstrated suppression of the IR sensitivity of the rad51-K191R mutant by RAD54 present on a high-copy-number plasmid (29). The suppression by Rad54 overexpression could occur by stabilization of the Rad51-K191R-ssDNA complex prior to synapsis, by enhancement of the pairing and strand exchange activity of Rad51-K191R, or by displacement of Rad51-K191R from duplex DNA postsynaptically. If the first hypothesis is correct, we predicted that overexpression of a Rad54 mutant protein that is unable to hydrolyze ATP should also suppress the IR sensitivity of the rad51-K191R strain, because the Rad54-K341R mutant protein is still proficient for stabilization of Rad51-ssDNA complexes in vitro (27, 61). However, overexpression of either the rad54-K314R or rad54-K341A allele was unable to suppress the IR sensitivity of the rad51-K191R strain to the same level as RAD54, indicating that the translocase activity of Rad54 is required for the observed suppression (Fig. 1). High expression of the rad51-K341R allele resulted in a slight dominant-negative effect, as reported previously (17).
FIG. 1.
ATP hydrolysis by Rad54 is required for suppression of the γ-ray sensitivity conferred by the rad51-K191R allele. Strain LSY979 (rad51-K191R) containing plasmids expressing the RAD54, rad54-K341R, or rad54-K341A allele was irradiated as described in Materials and Methods.
Defective recruitment of Rad51-K191R to DSBs in vivo.
The translocase activity of Rad54 could stimulate strand exchange by the Rad51-K191R protein or could act postsynaptically to increase turnover of the mutant protein (47, 59). To determine whether Rad51-K191R is proficient for initiation of recombination but defective for a later step, we monitored recruitment of the mutant protein to break sites in vivo. The RAD52 group proteins form DNA damage-induced foci that colocalize with DSBs and regions of single-stranded DNA in vivo (20, 21, 52). To assess the association of Rad51 and Rad51-K191R with DSBs in live cells, the proteins were fused to YFP and foci formation monitored by microscopy after exposing the cells to γ irradiation. Although the YFP-Rad51 fusion protein is not fully functional, it is recruited to DSBs with kinetics similar to that of untagged Rad51 as monitored by ChIP (20, 52). Both Rad51 and Rad51-K191R form foci in response to DNA damage, but Rad51-K191R foci are about six times less bright than Rad51 foci (Fig. 2). By Western blot analysis, the steady-state level of the YFP-Rad51-K191R protein was similar to that of YFP-Rad51 (data not shown). In coimmunoprecipitation experiments, Rad51-K191R retains normal association with Rad52 (data not shown); therefore, the dimmer Rad51-K191R foci are not a result of less-efficient recruitment of Rad51 by Rad52 at resected DSBs and most likely result from a reduced pool of protomers or reduced retention of Rad51-K191R at break sites. The Rad51-K191A protein, which fails to bind DNA in vitro (56, 58), was fused to YFP as a control for foci formation. Whereas every budded RAD51 and rad51-K191R cell contains at least 1 focus after exposure to irradiation, only 1 focus was observed in about 50 irradiated rad51-K191A cells. Furthermore, the rare Rad51-K191A foci are very dim compared to those formed by Rad51 or Rad51-K191R (Fig. 2A).
FIG. 2.
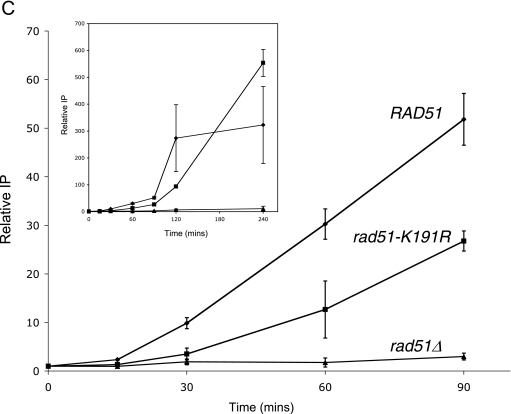
Rad51-K191R is recruited to DSBs less efficiently than Rad51. (A) Ionizing radiation-induced focus formation. YFP fusions were made with Rad51 and Rad51 mutant proteins (strains W4121-20D, LSY1576, and LSY1752, respectively). The strains were exposed to 20 krads of gamma radiation, followed by microscopy to monitor focus formation. (B) Distribution of focus brightness. The brightness of each focus was quantified and plotted; at least 30 foci were analyzed for each strain. The mean focus brightness for each strain is represented by a black bar. (C) Defective recruitment of Rad51-K191R to an HO-induced DSB. HO endonuclease was induced in a donorless strain that cannot repair the DSB at the MAT locus, and recruitment of native Rad51 (JKM179; diamonds) or Rad51-K191R (LSY1750-1; squares) was monitored by chromatin immunoprecipitation. A rad51 null strain (LSY1751; triangles) was used as a negative control. HO was induced at time zero and samples analyzed at 30-min time intervals after HO induction. The relative IP at each time point represents the amount of immunoprecipitated DNA divided by the amount of input DNA, normalized to the control ACT signal at time zero.The data represent averages for three independent trials.
Because YFP-Rad51 fusions are not fully functional, the recruitment of untagged Rad51 and Rad51-K191R proteins to DSBs was also monitored by ChIP. Strains lacking both HML and HMR homologous donor sequences were used to enhance the detection of protein recruitment to an unrepaired DSB at the MAT locus. Strains were grown in YP lactate media until mid-log phase, and galactose was then added to induce expression of the HO endonuclease. Native proteins were immunoprecipitated with affinity-purified Rad51 antibodies. In a wild-type strain, within 30 min of induction, Rad51 is detected at the MAT region, using a set of primers specific to the MAT Z region, from 239 bp to 486 bp CEN distal of the HO cut site. Consistent with results obtained using the YFP fusions, the rate of Rad51-K191R recruitment to the DSB was significantly less than that of Rad51. At 90 min after induction, three times less Rad51-K191R was recruited to the DSB than Rad51 (Fig. 2C); however, the level of Rad51-K191R recruited 4 h after HO induction was comparable to the wild-type level. A rad51 null strain was used as a control for antibody specificity (Fig. 2C).
Deletion of SRS2 suppresses IR sensitivity of the rad51-K191R strain.
The localization studies suggest Rad51-K191R forms a partial or unproductive filament on ssDNA, preventing initiation of recombination. Because the Srs2 helicase has been shown to displace Rad51 from ssDNA, we reasoned that Srs2 would be more effective in removing Rad51-K191R if the mutant protein exhibits less-stable binding to ssDNA in vivo. Consistent with this hypothesis, deletion of SRS2 resulted in an almost complete suppression of the IR sensitivity of the rad51-K191R strain (Fig. 3A). The srs2 rad51Δ double mutant showed a sensitivity to IR equivalent to that of the rad51Δ strain, indicating suppression requires Rad51 and is not due to activation of an alternate pathway (data not shown). The ATPase-defective srs2-K41A strain (18) also suppressed the IR sensitivity of the rad51-K191R strain to the same level as that of the srs2 null strain (data not shown), suggesting that the removal of the Srs2 ssDNA translocase activity and the resulting stabilization of the Rad51-K191R-ssDNA filament is responsible for the rescue of the rad51-K191R DNA repair defect. We have shown previously that the rad51-K191R strain is defective in mating-type switching, a gene conversion event that is initiated by a DSB made by the HO endonuclease (29). In addition to suppressing the IR sensitivity of rad51-K191R, the deletion of SRS2 also rescued the mating-type switching defect of the rad51-K191R strain (Fig. 3B), suggesting that the absence of Srs2 stabilizes the Rad51-K191R filament at DSB sites.
FIG. 3.
The DNA repair defect of the rad51-K191R strain is suppressed by deletion of SRS2. (A) Deletion of SRS2 suppresses the γ-ray sensitivity of the rad51-K191R strain. The wild-type strain is W1588-4C; the srs2::HIS3 strain is HKY590-1D; the rad51-K191R strain is LSY977, and the rad51-K191R srs2 strain is LSY1388-1C. (B) The mating-type switching defect of the rad51-K191R strain is suppressed by the deletion of SRS2. A cartoon of the MATα locus showing the location of the HO cut site and probe for Southern blots is shown in the upper panel. Switching to MATa produces a novel 0.9-kb StyI fragment. Kinetic analysis of mating-type switching in RAD51 (W1588-4A), rad51-K191R (LSY1205-2A), and rad51-K191R srs2 (LSY1388-1C) strains is shown in the lower panel. Galactose was added to the cultures for 1 h at time zero to induce expression of HO, and samples were removed every hour for DNA analysis.
The rad51-K191R strain is defective in assembly of IR-induced Rad54 and Rad55 foci.
As a further test of the hypothesis that the rad51-K191R allele confers a defect in the initiation of recombination, the assembly of two factors inferred to function downstream of Rad51 was monitored by microscopy using fusions of YFP to either Rad54 or Rad55. These fusions retain full biological activity as assessed by radiation resistance (20). Lisby et al. (20) showed that Rad51 is required for the formation of IR-induced Rad54 and Rad55 foci, indicating that recruitment of these proteins to DSBs requires Rad51 interaction with ssDNA. No IR-induced or spontaneous Rad54 or Rad55 foci were detected in the rad51-K191R strain at 30 min or up to 4 h postirradiation (Fig. 4; also data not shown). Thus, even though Rad51-K191R is eventually recruited to DSBs (Fig. 2C), the filament formed is not competent for recruitment of Rad54. The rad51-K191A strain, which is phenotypically null, was used as a control. Based on the srs2 suppression results described above, we expected srs2 to restore bright foci to the strain expressing YFP-Rad51-K191R. Although bright spontaneous foci were observed in the srs2 YFP-rad51-K191R strain, even brighter foci were found in the srs2 YFP-RAD51 strain, and in both cases large unusual structures were seen, especially following irradiation (see Fig. S1 in the supplemental material). To avoid potential artifacts caused by the YFP-Rad51 fusions, the formation of IR-induced Rad54-YFP foci in the srs2 rad51-K191R mutant was monitored instead. The srs2 mutation restored Rad54 foci in the rad51-K191R strain but not in the rad51-K191A strain (Fig. 4). We also observed the rescue of Rad55 foci in the rad51-K191R strain by srs2 (data not shown). These results suggest that Rad51-K191R is defective in forming a competent nucleoprotein filament and therefore cannot effectively recruit Rad54 and Rad55 to DSBs.
FIG. 4.
The rad51-K191R strain is defective in assembly of IR-induced Rad54 foci. YFP fusions with the Rad54 protein were made in various genetic backgrounds: RAD51 (W4644-4A), rad51-K191R (LSY1743-5C), rad51-K191A (LSY1745), rad51-K191R srs2 (LSY1754), and rad51-K191A srs2 (LSY1748). Rad54 focus formation was monitored by microscopy after irradiation at 20 krads. Each gray bars represents the total number of nuclei analyzed; each black bar represents the number of nuclei that contained at least one Rad54 focus. At least 60 cells were analyzed for each strain.
The phenotype of the rad51-K191R mutant is similar to those of the rad55 and rad57 mutants.
Because the Rad51-K191R mutant protein appears to be defective for presynaptic filament formation in vivo, we compared the rad51-K191R strain with other mutants that have been shown to exhibit a similar defect. The Rad55 and Rad57 proteins form an obligate heterodimer and have been implicated as mediators in the assembly and/or stabilization of the Rad51 nucleoprotein filament (11, 13, 16, 54). Sugawara et al. found a significant delay in the recruitment of Rad51 to a DSB in a rad55Δ strain (52), similar to the delay seen in the recruitment of the Rad51-K191R protein by chromatin immunoprecipitation (Fig. 2C). We determined the fluorescence intensity for IR-induced Rad51-YFP foci in a rad55Δ strain and found the foci to exhibit the same brightness as Rad51-K191R foci in a RAD55 background (Fig. 2B).
Consistent with these results, the rad55 and rad57 mutant strains showed sensitivities to IR similar to that of the rad51-K191R strain (Fig. 5; also data not shown). As in the case of rad51-K191R, the IR sensitivity of rad57 can be partially suppressed by deletion of SRS2 (Fig. 5) or by RAD54 present in high copy number (29), suggesting that the Rad51 filament formed in the rad57 strain has characteristics similar to those of the Rad51-K191R filament even in the presence of the Rad55-Rad57 mediator. The suppression of the IR sensitivity of the rad57 mutant by srs2 is less than that observed for the rad51-K191R strain. It is possible that the Rad55-Rad57 heterodimer has other functions in addition to the mediator role that cannot be rescued by the deletion of SRS2 (22). While the deletion of SRS2 can suppress the IR sensitivity of the rad51-K191R or rad57 single mutants, srs2 cannot suppress the IR sensitivity of a rad51-K191R rad57 double mutant (Fig. 5), indicating that Rad51 filament formation is severely impaired when both the Rad55-Rad57 heterodimer and Rad51 ATP hydrolysis are absent. In addition to suppression by srs2, the IR sensitivity of rad51-K191R, rad55, or rad57 single mutants can be suppressed by mating-type heterozygosity (16, 25, 29). However, like srs2, mating-type heterozygosity is unable to suppress the IR sensitivity of the rad51-K191R rad57 double mutant (data not shown).
FIG. 5.
The γ-ray sensitivity of the rad57 rad51-K191R srs2 triple mutant is more severe than that of either the rad57 srs2 or rad51-K191R srs2 double mutant. Serial dilutions of the strains were spotted onto YPD plates and left unirradiated or irradiated at 40 krads. Survival was assessed following growth for 3 days at 30°C. Strains used were W1588-1C (wild type), LSY411 (rad51::URA3), LSY977 (rad51-K191R), HKY598-2C (rad57::LEU2), LSY1197 (rad51-K191R rad57::LEU2), LSY1391 (rad57::LEU2 srs2::HIS3), LSY1388-1C (rad51-K191R srs2), and LSY1596-4A (rad51-K191R rad57::LEU2 srs2::HIS3).
The rad51-K191R allele exhibits semidominance.
Studies with mouse cells have shown a dominant-negative phenotype for mitomycin C sensitivity and DSB-induced recombination by expression of the human RAD51-K133R allele in a RAD51+/+ mouse ES cell line (48, 49). To determine whether the rad51-K191R allele exerts dominance in yeast, a haploid MATα rad51-K191R strain was mated to a RAD51 haploid strain with a deletion of the MAT locus rendering it a default a-mater (51). The use of a matΔ strain was necessary because heterozygosity at the MAT locus (the normal situation in diploids) suppresses the phenotype conferred by the rad51-K191R allele (29). The resulting diploid strain (MATα/matΔ rad51-K191R/RAD51) shows greater sensitivity to IR than a MATα/matΔ rad51Δ/RAD51 diploid, indicating semidominance of the rad51-K191R allele (Fig. 6).
FIG. 6.
The rad51-K191R allele is semidominant. Serial dilutions of the strains were spotted onto YPD plates and left unirradiated or irradiated at 20 krads. Survival was assessed following growth for 3 days at 30°C. Strains used were W1588-1C (wild type), LSY977 (rad51-K191R), LSY1759 (RAD51/RAD51), LSY1755-1 (RAD51/rad51-K191R), and LSY1781 (RAD51/rad51::HIS3).
The Rad51-K191R protein shows reduced DNA binding activity.
We interpreted the reduced localization, srs2 suppression, and synergism with rad57 as indicative of either less-efficient binding of the Rad51-K191R mutant protein to ssDNA at DSB sites in vivo or a reduced pool of protomers available for binding. To determine whether the Rad51-K191R protein has reduced DNA binding activity in vitro, the protein was purified and DNA binding assayed by binding to alkali-treated nitrocellulose filters (45). The Rad51-K191R protein shows less binding to ssDNA than wild-type Rad51 (Fig. 7), and in the presence of physiological salt concentration, binding by Rad51-K191R is greatly reduced.
FIG. 7.
Rad51-K191R shows reduced DNA binding compared with that of Rad51. (A) Binding of wild-type Rad51 (squares) or Rad51-K191R (triangles) to single-stranded DNA. Increasing concentrations of either wild-type Rad51 or Rad51-K191R were incubated with a constant amount of ssDNA and passed through alkali-treated nitrocellulose and DEAE filters. The percent bound represents the amount of DNA retained on nitrocellulose compared with the total DNA. (B) The same as in panel A except in the presence of 150 mM NaCl.
DISCUSSION
The Rad51 protein, like all members of the RecA family of recombinases, is a DNA-dependent ATPase that catalyzes strand exchange. RecA ATP hydrolysis is important for filament disassembly, extensive long-tract DNA strand exchange, four-strand DNA exchange, and the bypass of heterologous regions or structural barriers (26). In contrast, Rad51 does not require ATP hydrolysis for extensive strand exchange, and it is deficient in the other functions that are promoted by RecA ATP hydrolysis (32, 39, 56). In addition, the rate of Rad51 ATP hydrolysis is 30- to 40-fold less than that of RecA (53). While there is evidence that the Rad51 protein can perform strand exchange reactions in vitro without ATP hydrolysis, the ATPase-defective rad51-K191R mutant strain is sensitive to ionizing radiation and defective in mating-type switching and shows reduced rates of spontaneous recombination (data not shown) (29, 56). The goal of this study was to determine at which step(s) in the Rad51-catalyzed recombination reaction ATP hydrolysis is required. We found less recruitment of the Rad51-K191R protein at sites of DSBs than of Rad51 (Fig. 2). The Rad51-K191R filament formed at DSBs cannot support the assembly of Rad54 and Rad55, two factors inferred to function downstream of filament formation in homologous recombination (Fig. 4) (20). Furthermore, the IR sensitivity and Rad54 recruitment defect of the rad51-K191R mutant can be suppressed by stabilizing the filament via the removal of the Srs2 helicase (Fig. 3A and 4) (19, 60). These results implicate ATP hydrolysis by Rad51 as being important in presynaptic filament formation.
By chromatin immunoprecipitation and live imaging of YFP-tagged Rad51-K191R, we detected reduced recruitment of Rad51-K191R to DSBs generated by HO endonuclease or IR (Fig. 2). A twofold reduction in the number of IR-induced Rad51 foci was previously reported for chicken DT40 cells expressing the human Rad51-K133R protein, but these cell lines have elevated expression of the mutant protein (30). We did not detect reduced numbers of IR-induced foci in the YFP-rad51-K191R strain, but the foci were six- to sevenfold less bright than in the YFP-RAD51 strain. In live cells, several DSBs are recruited to recombination centers represented by foci; therefore, by this method we are unable to distinguish between reduced numbers of DSBs with Rad51-K191R recruited or less Rad51-K191R at all DSBs (21). The defect in mating-type switching (Fig. 3B) is more consistent with formation of an incomplete or inactive filament at most lesions. Furthermore, the inability to detect IR-induced Rad55 or Rad54 foci in the rad51-K191R mutant suggests a defect in Rad51-K191R filament formation at the majority of break sites (Fig. 4). By Western blot analysis, the steady-state level of Rad51-K191R is similar to that of wild-type Rad51; thus, the reduced recruitment in vivo is not due simply to lower expression of the mutant protein (29). Consistent with a defect in an early step during homologous recombination, we found that the rad51-K191R mutation suppresses the synthetic lethality of srs2 rad54 double mutants (data not shown). Because the srs2 rad54 synthetic lethality is suppressed by deletion of RAD51, RAD52, or RAD55, all early recombination functions, this suggests the rad51-K191R defect is also at an early step.
The defect in recruitment of Rad51-K191R to break sites is similar to the defect reported for Rad51 recruitment in rad55 mutants (20, 52, 62). The rad51-K191R and rad57 (phenotypically the same as rad55) mutants show similar sensitivity to IR and are suppressed by srs2, by RAD54 expressed from a high-copy-number plasmid, or by mating-type heterozygosity (Fig. 5) (29). The rad55 and rad57 mutants are also suppressed by overexpression of Rad51 or by rad51 alleles that encode gain-of-function proteins with a higher affinity for DNA (11, 13, 16). These data suggest the Rad51 filament formed in rad55 and rad57 mutants is incomplete or less stable, and this impediment can be overcome by increasing the pool of free Rad51 or creating a more stable presynaptic filament. Because of the similarity in phenotype between rad51-K191R and rad55 (or rad57), we suspect the Rad51-K191R filament to be incomplete or less stable than Rad51. In vitro, we observed slightly reduced DNA binding to ssDNA by the Rad51-K191R protein compared with results for Rad51, and formation of Rad51-K191R-ssDNA complexes was sensitive to high salt levels (Fig. 7). The defect in ssDNA binding could be the result of a lower affinity of the Rad51-K191R protein for ATP (W. D. Heyer, personal communication). Although formation of Rad51-K191R DNA complexes is sensitive to high salt levels, once formed the complexes show greater stability than those formed by wild-type Rad51 (W. D. Heyer, personal communication). In one study, the human Rad51-K133R protein was reported to show slightly reduced binding to DNA (30), but in a more recent article, the human Rad51-K133R protein demonstrated binding affinity similar to that of wild-type Rad51 and was shown to form more stable complexes with DNA than was observed for wild-type Rad51 (8). Furthermore, the human Rad51 protein forms very stable DNA complexes when ATP hydrolysis is inhibited by Ca2+ ions or the nonhydrolyzable ATP analog, AMP-PNP (8, 40). Binding of yeast Rad51 to ssDNA is not supported by AMP-PNP or stimulated by Ca2+ indicating distinct differences between the yeast and human proteins (4, 31). The in vitro ssDNA-binding defect observed for yeast Rad51-K191R could result in reduced recruitment of the mutant protein to break sites in vivo and is consistent with the suppression by srs2 or by overexpression of the rad51-K191R allele. However, the observation of a semidominant phenotype for the rad51-K191R allele when expressed in diploids suggests that the DNA recombination defect is not due simply to less-efficient recruitment of Rad51-K191R at break sites, and there is an additional impediment that results from a lack of ATP hydrolysis. Because the human Rad51-K133R and yeast Rad51-K191R proteins form more stable complexes with DNA (8) (W. D. Heyer, personal communication), it is possible that the dominant-negative phenotype observed for yeast and mouse cells results from poor turnover of the mixed filament after strand invasion. Removal of Srs2 results in an almost complete suppression of the rad51-K191R phenotype (Fig. 3A and 5), and this could result from a reduced pool of free Rad51-K191R due to a defect in turnover of the mutant protein. Srs2 only displaces Rad51 from ssDNA (19, 60); therefore, we interpret the suppression by srs2 as stabilization of the Rad51-K191R-ssDNA filament rather than increasing the pool of free Rad51-K191R by mobilization from dsDNA.
Another possible explanation for the similarity in phenotype of the rad51-K191R and rad55 or rad57 mutants is that Rad55 and Rad57 promote ATP hydrolysis by Rad51. The human Xrcc2 protein enhances ADP/ATP processing by human Rad51, resulting in more-efficient strand exchange (44). Although no stimulation of Rad51 ssDNA binding by Xrcc2 was observed, the Rad51 filament might be more dynamic in vivo, requiring multiple cycles of binding and release in order to form an active presynaptic filament. By this scenario, the dominant-negative effect might be due to trapping of the wild-type protein with Rad51-K191R in unproductive partial filaments that are unable to undergo the necessary cycles of binding and release to create the active filament.
Previously, we found the IR sensitivity of the rad51-K191R mutant to be suppressed by RAD54 in high copy number (29), and in this study we determined that the suppression requires Rad54 ATPase activity (Fig. 1). Because high-copy-number expression of the rad54-K341R allele failed to suppress the IR sensitivity of the rad51-K191R strain, it seems unlikely that the Rad54 suppressive effect is in stabilization of the Rad51-K191R filament. Indeed, we found that Rad54 overexpression cannot rescue the reduction in brightness of Rad51-K191R-YFP foci (data not shown). The failure to restore normal YFP-Rad51-K191R foci argues against a stabilization role by Rad54 overexpression and also is inconsistent with mobilizing Rad51-K191R bound to dsDNA. We had expected the Rad51-K191R protein to show a greater association with non-DSB-site DNA by ChIP if it was trapped on chromatin due to decreased turnover, but this was not observed. Although we cannot rule out the possibility that overexpression of Rad54 mobilizes Rad51-K191R from dsDNA to increase the protomer pool available for ssDNA binding, we currently have no evidence in support of this hypothesis. A recent study showed accumulation of the meiosis-specific RecA homolog, Dmc1, on chromatin in rdh54/tid1 mutants consistent with a role for Rdh54/Tid1 (a homolog of Rad54) in maintaining a pool of Dmc1 protomers by disruption of unproductive associations of Dmc1 with dsDNA (14). Like Rad54, Rdh54/Tid1 disrupts Rad51-dsDNA complexes in vitro and stimulates the strand exchange activity of Rad51 (7, 35). High-copy-number expression of RDH54 resulted in a weak suppression of the IR sensitivity of the rad51-K191R strain, but the suppression was much less than observed for high-copy-number RAD54 (data not shown). Rad54 has been shown to enhance Rad51-mediated strand exchange under suboptimal conditions, and it is possible that additional Rad54 supports strand exchange for the suboptimal Rad51-K191R filament, resulting in the observed suppression (59).
In summary, the recombination defect conferred by the rad51-K191R allele appears to be due to inefficient presynaptic filament formation. This defect can be suppressed by removal of the Srs2 helicase, overexpression of Rad54, or by overexpression of the mutant protein. Increased stability of Rad51-K191R-DNA complexes could prevent turnover of the protein, resulting in a reduced pool of free Rad51-K191R for binding to ssDNA generated at DSBs. Alternatively, the filament formed by Rad51-K191R could be qualitatively different from the filament formed by wild-type Rad51 and unable to undergo the necessary structural transitions to form a competent presynaptic filament.
Acknowledgments
We thank R. Rothstein (Columbia University) and members of the Rothstein lab for assistance with microscopy and B. Laurent (Mount Sinai Medical Center) for assistance with the chromatin immunoprecipitation experiments. We thank J. Haber, M. Lisby, R. Rothstein, and P. Sung for yeast strains and plasmids and W. D. Heyer for communication of unpublished results.
This research was supported by Public Health Service grants GM054099 (L.S.S.), T32 GM08224 (C.W.F.), and T32 CA09503 (G.S.F. and C.W.F.) from the National Institutes of Health.
Footnotes
Published ahead of print on 9 October 2006.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Aboussekhra, A., R. Chanet, A. Adjiri, and F. Fabre. 1992. Semidominant suppressors of Srs2 helicase mutations of Saccharomyces cerevisiae map in the RAD51 gene, whose sequence predicts a protein with similarities to procaryotic RecA proteins. Mol. Cell. Biol. 12:3224-3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Basile, G., M. Aker, and R. K. Mortimer. 1992. Nucleotide sequence and transcriptional regulation of the yeast recombinational repair gene RAD51. Mol. Cell. Biol. 12:3235-3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boundy-Mills, K. L., and D. M. Livingston. 1993. A Saccharomyces cerevisiae RAD52 allele expressing a C-terminal truncation protein: activities and intragenic complementation of missense mutations. Genetics 133:39-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bugreev, D. V., and A. V. Mazin. 2004. Ca2+ activates human homologous recombination protein Rad51 by modulating its ATPase activity. Proc. Natl. Acad. Sci. USA 101:9988-9993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campbell, M. J., and R. W. Davis. 1999. On the in vivo function of the RecA ATPase. J. Mol. Biol. 286:437-445. [DOI] [PubMed] [Google Scholar]
- 6.Chai, B., J. Huang, B. R. Cairns, and B. C. Laurent. 2005. Distinct roles for the RSC and Swi/Snf ATP-dependent chromatin remodelers in DNA double-strand break repair. Genes Dev. 19:1656-1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chi, P., Y. Kwon, C. Seong, A. Epshtein, I. Lam, P. Sung, and H. L. Klein. 2006. Yeast recombination factor Rdh54 functionally interacts with the Rad51 recombinase and catalyzes Rad51 removal from DNA. J. Biol. Chem. 281:26268-26279. [DOI] [PubMed] [Google Scholar]
- 8.Chi, P., S. Van Komen, M. G. Sehorn, S. Sigurdsson, and P. Sung. 2006. Roles of ATP binding and ATP hydrolysis in human Rad51 recombinase function. DNA Repair (Amsterdam) 5:381-391. [DOI] [PubMed] [Google Scholar]
- 9.Davis, A. P., and L. S. Symington. 2003. The Rad52-Rad59 complex interacts with Rad51 and replication protein A. DNA Repair (Amsterdam) 2:1127-1134. [DOI] [PubMed] [Google Scholar]
- 10.Donovan, J. W., G. T. Milne, and D. T. Weaver. 1994. Homotypic and heterotypic protein associations control Rad51 function in double-strand break repair. Genes Dev. 8:2552-2562. [DOI] [PubMed] [Google Scholar]
- 11.Fortin, G. S., and L. S. Symington. 2002. Mutations in yeast Rad51 that partially bypass the requirement for Rad55 and Rad57 in DNA repair by increasing the stability of Rad51-DNA complexes. EMBO J. 21:3160-3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harlow, E., and D. Lane. 1999. Using antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 13.Hays, S. L., A. A. Firmenich, and P. Berg. 1995. Complex formation in yeast double-strand break repair: participation of Rad51, Rad52, Rad55, and Rad57 proteins. Proc. Natl. Acad. Sci. USA 92:6925-6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holzen, T. M., P. P. Shah, H. A. Olivares, and D. K. Bishop. 2006. Tid1/Rdh54 promotes dissociation of Dmc1 from nonrecombinogenic sites on meiotic chromatin. Genes Dev. 20:2593-2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ito, H., Y. Fukuda, K. Murata, and A. Kimura. 1983. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153:163-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson, R. D., and L. S. Symington. 1995. Functional differences and interactions among the putative RecA homologs Rad51, Rad55, and Rad57. Mol. Cell. Biol. 15:4843-4850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim, P. M., K. S. Paffett, J. A. Solinger, W. D. Heyer, and J. A. Nickoloff. 2002. Spontaneous and double-strand break-induced recombination, and gene conversion tract lengths, are differentially affected by overexpression of wild-type or ATPase-defective yeast Rad54. Nucleic Acids Res. 30:2727-2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krejci, L., M. Macris, Y. Li, S. Van Komen, J. Villemain, T. Ellenberger, H. Klein, and P. Sung. 2004. Role of ATP hydrolysis in the antirecombinase function of Saccharomyces cerevisiae Srs2 protein. J. Biol. Chem. 279:23193-23199. [DOI] [PubMed] [Google Scholar]
- 19.Krejci, L., S. Van Komen, Y. Li, J. Villemain, M. S. Reddy, H. Klein, T. Ellenberger, and P. Sung. 2003. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature 423:305-309. [DOI] [PubMed] [Google Scholar]
- 20.Lisby, M., J. H. Barlow, R. C. Burgess, and R. Rothstein. 2004. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell 118:699-713. [DOI] [PubMed] [Google Scholar]
- 21.Lisby, M., R. Rothstein, and U. H. Mortensen. 2001. Rad52 forms DNA repair and recombination centers during S phase. Proc. Natl. Acad. Sci. USA 98:8276-8282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu, Y., J. Y. Masson, R. Shah, P. O'Regan, and S. C. West. 2004. RAD51C is required for Holliday junction processing in mammalian cells. Science 303:243-246. [DOI] [PubMed] [Google Scholar]
- 23.Llorente, B., and L. S. Symington. 2004. The Mre11 nuclease is not required for 5′ to 3′ resection at multiple HO-induced double-strand breaks. Mol. Cell. Biol. 24:9682-9694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Logan, K. M., and K. L. Knight. 1993. Mutagenesis of the P-loop motif in the ATP binding site of the RecA protein from Escherichia coli. J. Mol. Biol. 232:1048-1059. [DOI] [PubMed] [Google Scholar]
- 25.Lovett, S. T., and R. K. Mortimer. 1987. Characterization of null mutants of the RAD55 gene of Saccharomyces cerevisiae: effects of temperature, osmotic strength and mating type. Genetics 116:547-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lusetti, S. L., and M. M. Cox. 2002. The bacterial RecA protein and the recombinational DNA repair of stalled replication forks. Annu. Rev. Biochem. 71:71-100. [DOI] [PubMed] [Google Scholar]
- 27.Mazin, A. V., A. A. Alexeev, and S. C. Kowalczykowski. 2003. A novel function of Rad54 protein. Stabilization of the Rad51 nucleoprotein filament. J. Biol. Chem. 278:14029-14036. [DOI] [PubMed] [Google Scholar]
- 28.Miyazaki, T., D. A. Bressan, M. Shinohara, J. E. Haber, and A. Shinohara. 2004. In vivo assembly and disassembly of Rad51 and Rad52 complexes during double-strand break repair. EMBO J. 23:939-949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morgan, E. A., N. Shah, and L. S. Symington. 2002. The requirement for ATP hydrolysis by Saccharomyces cerevisiae Rad51 is bypassed by mating-type heterozygosity or RAD54 in high copy. Mol. Cell. Biol. 22:6336-6343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morrison, C., A. Shinohara, E. Sonoda, Y. Yamaguchi-Iwai, M. Takata, R. R. Weichselbaum, and S. Takeda. 1999. The essential functions of human Rad51 are independent of ATP hydrolysis. Mol. Cell. Biol. 19:6891-6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Namsaraev, E. A., and P. Berg. 1998. Binding of Rad51p to DNA. Interaction of Rad51p with single- and double-stranded DNA. J. Biol. Chem. 273:6177-6182. [DOI] [PubMed] [Google Scholar]
- 32.Namsaraev, E. A., and P. Berg. 2000. Rad51 uses one mechanism to drive DNA strand exchange in both directions. J. Biol. Chem. 275:3970-3976. [DOI] [PubMed] [Google Scholar]
- 33.Nickoloff, J. A., E. Y. Chen, and F. Heffron. 1986. A 24-base-pair DNA sequence from the MAT locus stimulates intergenic recombination in yeast. Proc. Natl. Acad. Sci. USA 83:7831-7835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ogawa, T., X. Yu, A. Shinohara, and E. H. Egelman. 1993. Similarity of the yeast RAD51 filament to the bacterial RecA filament. Science 259:1896-1899. [DOI] [PubMed] [Google Scholar]
- 35.Petukhova, G., P. Sung, and H. Klein. 2000. Promotion of Rad51-dependent D-loop formation by yeast recombination factor Rdh54/Tid1. Genes Dev. 14:2206-2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Petukhova, G., S. Van Komen, S. Vergano, H. Klein, and P. Sung. 1999. Yeast Rad54 promotes Rad51-dependent homologous DNA pairing via ATP hydrolysis-driven change in DNA double helix conformation. J. Biol. Chem. 274:29453-29462. [DOI] [PubMed] [Google Scholar]
- 37.Rattray, A. J., and L. S. Symington. 1994. Use of a chromosomal inverted repeat to demonstrate that the RAD51 and RAD52 genes of Saccharomyces cerevisiae have different roles in mitotic recombination. Genetics 138:587-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rehrauer, W. M., and S. C. Kowalczykowski. 1993. Alteration of the nucleoside triphosphate (NTP) catalytic domain within Escherichia coli recA protein attenuates NTP hydrolysis but not joint molecule formation. J. Biol. Chem. 268:1292-1297. [PubMed] [Google Scholar]
- 39.Rice, K. P., A. L. Eggler, P. Sung, and M. M. Cox. 2001. DNA pairing and strand exchange by the Escherichia coli RecA and yeast Rad51 proteins without ATP hydrolysis: on the importance of not getting stuck. J. Biol. Chem. 276:38570-38581. [DOI] [PubMed] [Google Scholar]
- 40.Ristic, D., M. Modesti, T. van der Heijden, J. van Noort, C. Dekker, R. Kanaar, and C. Wyman. 2005. Human Rad51 filaments on double- and single-stranded DNA: correlating regular and irregular forms with recombination function. Nucleic Acids Res. 33:3292-3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schild, D. 1995. Suppression of a new allele of the yeast RAD52 gene by overexpression of RAD51, mutations in srs2 and ccr4, or mating-type heterozygosity. Genetics 140:115-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shan, Q., M. M. Cox, and R. B. Inman. 1996. DNA strand exchange promoted by RecA K72R. Two reaction phases with different Mg2+ requirements. J. Biol. Chem. 271:5712-5724. [DOI] [PubMed] [Google Scholar]
- 43.Sherman, F., G. Fink, and J. Hicks. 1986. Methods in yeast genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 44.Shim, K. S., C. Schmutte, G. Tombline, C. D. Heinen, and R. Fishel. 2004. hXRCC2 enhances ADP/ATP processing and strand exchange by hRAD51. J. Biol. Chem. 279:30385-30394. [DOI] [PubMed] [Google Scholar]
- 45.Shinohara, A., H. Ogawa, and T. Ogawa. 1992. Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell 69:457-470. [DOI] [PubMed] [Google Scholar]
- 46.Sigurdsson, S., S. Van Komen, G. Petukhova, and P. Sung. 2002. Homologous DNA pairing by human recombination factors Rad51 and Rad54. J. Biol. Chem. 277:42790-42794. [DOI] [PubMed] [Google Scholar]
- 47.Solinger, J. A., K. Kiianitsa, and W. D. Heyer. 2002. Rad54, a Swi2/Snf2-like recombinational repair protein, disassembles Rad51:dsDNA filaments. Mol. Cell 10:1175-1188. [DOI] [PubMed] [Google Scholar]
- 48.Stark, J. M., P. Hu, A. J. Pierce, M. E. Moynahan, N. Ellis, and M. Jasin. 2002. ATP hydrolysis by mammalian RAD51 has a key role during homology-directed DNA repair. J. Biol. Chem. 277:20185-20194. [DOI] [PubMed] [Google Scholar]
- 49.Stark, J. M., A. J. Pierce, J. Oh, A. Pastink, and M. Jasin. 2004. Genetic steps of mammalian homologous repair with distinct mutagenic consequences. Mol. Cell. Biol. 24:9305-9316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Strahl-Bolsinger, S., A. Hecht, K. Luo, and M. Grunstein. 1997. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 11:83-93. [DOI] [PubMed] [Google Scholar]
- 51.Strathern, J., J. Hicks, and I. Herskowitz. 1981. Control of cell type in yeast by the mating type locus. The alpha 1-alpha 2 hypothesis. J. Mol. Biol. 147:357-372. [DOI] [PubMed] [Google Scholar]
- 52.Sugawara, N., X. Wang, and J. E. Haber. 2003. In vivo roles of Rad52, Rad54, and Rad55 proteins in Rad51-mediated recombination. Mol. Cell 12:209-219. [DOI] [PubMed] [Google Scholar]
- 53.Sung, P. 1994. Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science 265:1241-1243. [DOI] [PubMed] [Google Scholar]
- 54.Sung, P. 1997. Yeast Rad55 and Rad57 proteins form a heterodimer that functions with replication protein A to promote DNA strand exchange by Rad51 recombinase. Genes Dev. 11:1111-1121. [DOI] [PubMed] [Google Scholar]
- 55.Sung, P., and D. L. Robberson. 1995. DNA strand exchange mediated by a RAD51-ssDNA nucleoprotein filament with polarity opposite to that of RecA. Cell 82:453-461. [DOI] [PubMed] [Google Scholar]
- 56.Sung, P., and S. A. Stratton. 1996. Yeast Rad51 recombinase mediates polar DNA strand exchange in the absence of ATP hydrolysis. J. Biol. Chem. 271:27983-27986. [DOI] [PubMed] [Google Scholar]
- 57.Symington, L. S. 2002. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 66:630-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Van Komen, S., G. Petukhova, S. Sigurdsson, S. Stratton, and P. Sung. 2000. Superhelicity-driven homologous DNA pairing by yeast recombination factors Rad51 and Rad54. Mol. Cell 6:563-572. [DOI] [PubMed] [Google Scholar]
- 59.Van Komen, S., G. Petukhova, S. Sigurdsson, and P. Sung. 2002. Functional cross-talk among Rad51, Rad54, and replication protein A in heteroduplex DNA joint formation. J. Biol. Chem. 277:43578-43587. [DOI] [PubMed] [Google Scholar]
- 60.Veaute, X., J. Jeusset, C. Soustelle, S. C. Kowalczykowski, E. Le Cam, and F. Fabre. 2003. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature 423:309-312. [DOI] [PubMed] [Google Scholar]
- 61.Wolner, B., and C. L. Peterson. 2005. ATP-dependent and ATP-independent roles for the Rad54 chromatin remodeling enzyme during recombinational repair of a DNA double strand break. J. Biol. Chem. 280:10855-10860. [DOI] [PubMed] [Google Scholar]
- 62.Wolner, B., S. van Komen, P. Sung, and C. L. Peterson. 2003. Recruitment of the recombinational repair machinery to a DNA double-strand break in yeast. Mol. Cell 12:221-232. [DOI] [PubMed] [Google Scholar]
- 63.Wong, I., and T. M. Lohman. 1993. A double-filter method for nitrocellulose-filter binding: application to protein-nucleic acid interactions. Proc. Natl. Acad. Sci. USA 90:5428-5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zaitseva, E. M., E. N. Zaitsev, and S. C. Kowalczykowski. 1999. The DNA binding properties of Saccharomyces cerevisiae Rad51 protein. J. Biol. Chem. 274:2907-2915. [DOI] [PubMed] [Google Scholar]