Abstract
Eukaryotic cells deal with accumulation of unfolded proteins in the endoplasmic reticulum (ER) by the unfolded protein response, involving the induction of molecular chaperones, translational attenuation, and ER-associated degradation, to prevent cell death. Here, we found that the autophagy system is activated as a novel signaling pathway in response to ER stress. Treatment of SK-N-SH neuroblastoma cells with ER stressors markedly induced the formation of autophagosomes, which were recognized at the ultrastructural level. The formation of green fluorescent protein (GFP)-LC3-labeled structures (GFP-LC3 “dots”), representing autophagosomes, was extensively induced in cells exposed to ER stress with conversion from LC3-I to LC3-II. In IRE1-deficient cells or cells treated with c-Jun N-terminal kinase (JNK) inhibitor, the autophagy induced by ER stress was inhibited, indicating that the IRE1-JNK pathway is required for autophagy activation after ER stress. In contrast, PERK-deficient cells and ATF6 knockdown cells showed that autophagy was induced after ER stress in a manner similar to the wild-type cells. Disturbance of autophagy rendered cells vulnerable to ER stress, suggesting that autophagy plays important roles in cell survival after ER stress.
The endoplasmic reticulum (ER) is the compartment in which protein folding occurs prior to transport to the extracellular surface or different intracellular organelles. A number of cellular stress conditions lead to the accumulation of unfolded or misfolded proteins in the ER lumen, constituting a fundamental threat to the cell. Such accumulation of incorrectly folded proteins in the ER triggers the unfolded protein response (UPR) to avoid cell damage. The UPR involves at least three distinct components, namely transcriptional induction of genes encoding ER-resident chaperones to facilitate protein folding, translational attenuation to decrease the demands made on the organelle, and ER-associated degradation (ERAD) to degrade the unfolded proteins accumulated in the ER (11, 21, 31). Recent evidence indicates that ER stresses are associated with generic or degenerative disorders including Alzheimer's disease (10, 23) and Parkinson's disease (7).
In mammalian cells, PERK, IRE1, and ATF6 sense the presence of unfolded proteins in the ER lumen and transduce signals to the cytoplasm and the nucleus (32). PERK activation leads to the phosphorylation of the α subunit of the translation initiation factor, eIF2α, which inhibits the assembly of the 80S ribosome and inhibits protein synthesis. Activation of IRE1 and ATF6 promotes transcription of UPR target genes. IRE1 processes XBP1 mRNA to generate mature XBP1 mRNA. Spliced XBP1 binds directly to the ER stress response element and the unfolded protein response elements and activates transcription of ER molecular chaperones, such as BiP/GRP78 (BiP), or ERAD-related genes, such as EDEM and ERdj4. ATF6 is cleaved by site 1 and site 2 proteases (S1P and S2P) in response to ER stress. The cleaved ATF6 N-terminal fragment migrates to the nucleus to activate the transcription of BiP through direct binding to the ER stress response element. Recently, it was reported that transmembrane transcription factors localized to the ER membrane (OASIS, CREB-H, and Tisp40/AIbZIP) function as tissue-specific ER stress transducers (1, 13, 42).
Despite extensive characterization of the regulatory signaling for the UPR, the morphological changes and determination of cell fate due to damage caused by ER stress are not well understood. Moreover, it also remains unknown whether other signaling pathways are activated in response to ER stress to deal with the unfolded proteins accumulated in the ER.
Autophagy plays an important physiological role in eukaryotic cells. A double-membrane structure, which is called the autophagosome or autophagic vacuole, is formed de novo to sequester cytoplasm. Then, the vacuole membrane fuses with the lysosomal membrane to deliver the contents into the autolysosome, where they are degraded and the resulting macromolecules recycled. Autophagy gene (ATG)-related proteins coordinate specific steps in autophagy induction and sequestration. The process is initiated when an isolation membrane is created under the direction of the class III PI3-kinase complex and ATG proteins. Two ubiquitin-like protein conjugation pathways cause the expansion of the isolation membrane. Microtubule-associated protein light chain 3 II (LC3-II), which is formed by phosphatidylethanolamine conjugation of LC3-I, translocates to the autophagosome membrane, the process of which is essential for the autophagosome formation. Digestion of the sequestered cytoplasmic contents is initiated when a lysosome fuses with the outer membrane of the autophagosome and lysosomal hydrolases are introduced (25). Autophagy occurs at basal levels in most tissues and contributes to the routine turnover of cytoplasmic components. However, autophagy can be induced by a change of environmental conditions, such as nutrient depletion. In addition to turnover of cellular components, autophagy is involved in development, differentiation, and tissue remodeling in various organisms (17). Autophagy is also implicated in certain human diseases (15, 22, 30, 36).
In this study, we examined morphological changes of the cells under ER stress using electron microscopy and found out that autophagosome formation is accelerated in the cells under ER stress. Disturbance of autophagy rendered cells vulnerable to ER stress, suggesting that autophagy plays important roles in cell survival after ER stress.
MATERIALS AND METHODS
Plasmids, transfection, reagents, and antibodies.
The GFP-LC3 expression vector was a kind gift from T. Yoshimori (National Institute of Genetics, Shizuoka, Japan). An IRE1α delta-RNase cDNA was created from the IRE1α full vector by PCR and inserted into pcDNA3.1+. The primers used for this experiment were 5′-TCACTATAGGGAGACCCAAGCTGG-3′(sense) and 5′-CTCGAGCCAGAAGAACGGGTGTTTGAGCACG-3′(antisense). Cells were transfected with each expression plasmid using the Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA). For back-transfection experiments, full-length IRE1α or IRE1α mutants (C-terminally truncated cytoplasmic region mutant, K599A mutant, and RNase L domain-truncated mutant) were individually transfected into IRE1α-deficient mouse embryonic fibroblasts (MEFs). Cells were treated with tunicamycin (Tm) (Sigma, St. Louis, MO) or thapsigargin (TG) (Alomone Laboratories Ltd., Jerusalem, Israel) to induce ER stress. To induce amino acid starvation, the medium was exchanged for Hanks' balanced salt solution (Invitrogen). 3-Methyladenine (3-MA) was purchased from Sigma. The c-Jun N-terminal kinase (JNK) inhibitor SP600125 and rapamycin were purchased from Calbiochem (San Diego, CA). For Western blotting analysis, anti-caspase-3, anti-JNK, anti-phosphorylated JNK (Cell Signaling Technology, Beverly, MA), anti-mouse immunoglobulin G (IgG) (Sigma), and anti-rabbit IgG (Sigma) antibodies were used. The anti-LC3 antibody was used as described previously (19).
Cell culture and assessment of cell death.
SK-N-SH cells were maintained in α-modified Eagle's medium (MP Biomedicals, Irvine, CA) containing 10% fetal calf serum at 37°C. ATG5−/− MEFs, PERK−/− MEFs, and ATF6αβ knockdown MEFs were kind gifts from N. Mizushima (The Tokyo Metropolitan Institute of Medical Science, Tokyo, Japan), D. Ron (New York University, New York, NY), and Laurie H. Glimcher (Harvard School of Public Health, Boston, MA), respectively. MEFs were maintained in Dulbecco's modified Eagle's medium (Invitrogen) containing 20% fetal calf serum at 37°C. For assessment of cell death, cells were treated with tunicamycin and thapsigargin for the indicated times and then stained with 100 μM Hoechst 33258 (Wako Pure Chemical Co., Tokyo, Japan) in phosphate-buffered saline (PBS) for 20 min. A total of 500 cells were counted randomly, and apoptotic cells were determined by fluorescence microscopy.
Quantification of the GFP-LC3 punctate area.
Green fluorescent protein (GFP)-LC3 transfectants were imaged using a fluorescence microscope (ECLIPSE TE2000-U; Nikon, Kanagawa, Japan) equipped with a charge-coupled-device camera (ORCA-ER-1394 system; Hamamatsu Photonics K.K., Shizuoka, Japan). To quantify the amount of GFP-LC3 dots, each dot signal was detected by eye and the area was measured using the Lumina Vision software (Mitani Corporation, Fukui, Japan). The dot structures were quantified by two of the authors in a blinded manner. The ratios of the total areas of GFP-LC3 dots to the overall cellular areas are presented as percentages.
Western blotting.
Cells were washed with PBS, harvested and lysed in Triton X-100 lysis buffer (0.5% Triton X-100, 10 mM HEPES, pH 7.9, 50 mM NaCl, 100 mM EDTA, 0.5 M sucrose, and 0.1% protease inhibitor cocktail [Sigma]). The lysates were then incubated on ice for 30 min and centrifuged at 8,000 × g for 10 min. Equal amounts of protein were subjected to 10 to 15% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to polyvinylidene difluoride membranes, and immunoblotted with each primary antibody. The membranes were washed with PBS-Tween 20 and then incubated with a peroxidase-conjugated secondary antibody. The corresponding bands were detected using an ECL Plus kit (Amersham Biosciences Corp., Piscataway, NJ).
Knockdown of IRE1α and ATG5.
Annealed double-stranded small interfering RNAs (siRNAs) for IRE1α and ATG5 were purchased from Proligo Japan K.K. (Kyoto, Japan). The sequences of the IRE1α and ATG5 siRNAs were described previously (see references 27 and 3, respectively). A lamin A/C siRNA (QIAGEN, Valencia, CA) was used as a control. HeLa cells at 60% confluence in 24-well plates were transfected with 75 ng of each of the above siRNAs using the HyperFect transfection reagent (QIAGEN) according to the manufacturer's protocol. The transfected cells were incubated at 37°C for 12 h and then stimulated by ER stressors.
Electron microscopy.
After the indicated treatments, SK-N-SH cells were fixed in phosphate buffer (pH 7.4) containing 2.5% glutaraldehyde and 2% paraformaldehyde at room temperature for 60 min. The cells were postfixed in 1% OsO4 at room temperature for 60 min, dehydrated through graded ethanol solutions, and embedded in Quetol 812 (Nissin EM Co., Tokyo, Japan). Areas containing cells were block mounted and cut into 70-nm sections. The sections were stained with uranyl acetate (saturated aqueous solution) and lead citrate and examined with a transmission electron microscope (H-7100; Hitachi, Ibaraki, Japan).
Immunoelectron microscopy.
GFP-LC3 was detected using a preembedding silver-enhanced immunogold method (4), with slight modifications as follows. SK-N-SH cells were cultured on coverslips and transfected with an expression vector for GFP-LC3. At 30 h after the transfection, the cells were treated with 2 μg/ml tunicamycin for 6 h and then fixed with 4% paraformaldehyde and 0.01% glutaraldehyde in PBS for 30 min. Samples were permeabilized with 0.1% Triton X-100 in PBS for 5 min. After incubation with blocking buffer containing 5% bovine serum albumin, 5% normal goat serum, and 0.02% NaN3 in PBS for 30 min, the cells were incubated with a rat anti-GFP antibody (Nacalai Tesque Inc., Kyoto, Japan) at a dilution of 1:100 in blocking buffer for 14 h. After four rinses with blocking buffer over 30 min, the cells were incubated with a goat anti-rat IgG labeled with 1.4-nm gold particles (Nanoprobes Inc., Yaphank, NY) at a dilution of 1:50 in blocking buffer for 18 h. After four rinses with PBS over 30 min, the cells were fixed with 1% glutaraldehyde in PBS for 15 min. After washing, the gold particles were silver enhanced using an HQ silver enhancement kit (Nanoprobes Inc.) for 8 min at 20°C in the dark. After washing with distilled water, the cells were postfixed in 0.1% OsO4 for 30 min, dehydrated through a graded ethanol series, and embedded in epoxy resin.
Amino acid uptake assay.
Amino acid transport was measured by a modification of the method of Kim et al. (12). MEF cells were treated with 2 μg/ml Tm or 1 μM TG for 2 h, followed by incubation with 1 μCi/ml of 3H-amino acids mixture (Amersham Biosciences Corp.) for 1 h. At the end of the incubation, the cells were washed three times with cold PBS and then lysed in 0.1% Triton X-100 for subsequent liquid scintillation counting.
RESULTS
Increased autophagosome formation during ER stress.
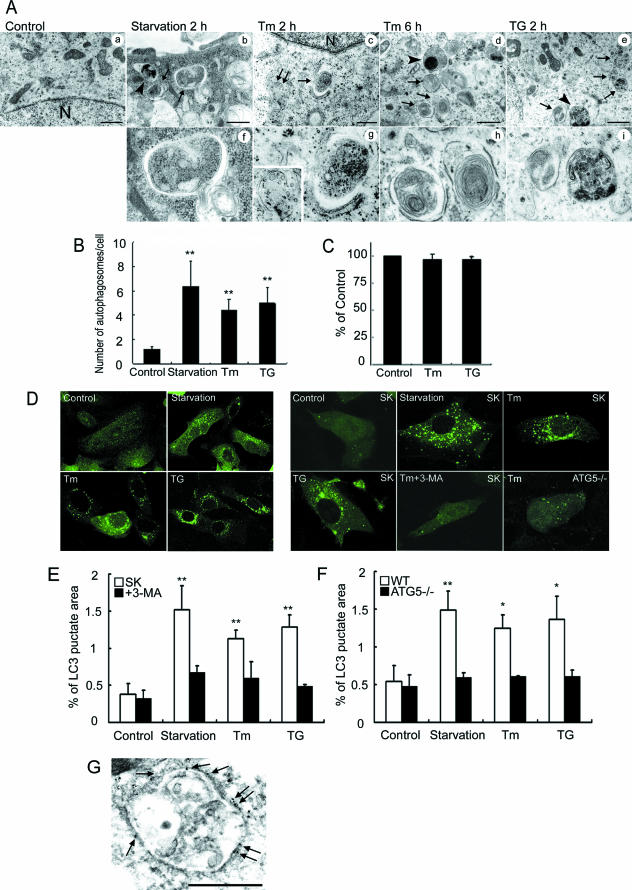
Initially, an electron microscopic analysis was performed using neuroblastoma SK-N-SH cells exposed to ER stressors, namely 2 μg/ml tunicamycin, an inhibitor of N glycosylation, and 1 μM thapsigargin, an inhibitor of ER Ca2+-ATPase. Treatments with these ER stressors for 2 and 6 h induced the formation of autophagosomes, which were recognized at the ultrastructural level as double-membrane vacuolar structures containing visible cytoplasmic contents (Fig. 1A, panels c to e and g to i). These ultrastructural findings were also observed in cells deprived of amino acids (Fig. 1A, panels b and f), a condition that is known to induce autophagy. In contrast, we rarely observed early autophagosomes and saw no evidence of later structures of the autophagy pathway in nontreated cells (Fig. 1A, panel a). The number of autophagosomes was significantly increased in cells treated with ER stressors compared with nontreated cells (Fig. 1B). Autolysosomes, which are recognized as single-membrane vacuolar structures containing high-density materials, also appeared in cells exposed to the ER stressors (Fig. 1A, panels d, e, and i). Some of the cells treated with the ER stressors revealed the formation of multivesicular body-like vesicles (Fig. 1A, panels c and g) and often contained multilamellar structures within the autophagosomes, findings that differ from those for cells deprived of amino acids. To confirm that treatment with tunicamycin or thapsigargin does not induce a starvation phenotype, we examined nutrient uptake in cells exposed to ER stress. These cells did not show changes in nutrient uptake compared with that for nontreated control cells, at least 2 h after treatment with ER stressors, when autophagy was induced (Fig. 1C). The results indicated that autophagosome formation is due to ER stress but not to a starvation phenotype.
FIG. 1.
Autophagosome formation is activated by ER stress. (A) Electron microscopic analysis of SK-N-SH cells exposed to 2 μg/ml Tm, 1 μM TG, or amino acid deprivation for the indicated times. Panels f to i are high magnification images of panels b to e, respectively. Typical autophagosomes (arrows), autolysosomes (arrowheads), and multivesicular bodies (indicated by double arrows in c and shown in the inset in g) are indicated. Scale bars, 1 μm. (B) The numbers of autophagosomes in SK-N-SH cells after exposure to the indicated stimuli for 2 h. A total of 30 electron microscopical sections were prepared, and autophagosomes were defined as double- or multiple-membrane structures in the cytosol. The values are the means ± standard deviations of three independent experiments. Asterisks indicate a significant difference from control cells (**, P < 0.01) (Student's t test). (C) Nutrient uptake after ER stress. MEF cells were treated with 2 μg/ml Tm or 1 μM TG for 2 h and then subjected to amino acid uptake assay as described in Materials and Methods. Results were expressed as the percentages of the counts compared with those in the absence of Tm or TG. The values are the means ± standard deviations of three independent experiments. (D) Autophagosome formation in SK-N-SH cells or ATG5-deficient (ATG5−/−) MEFs treated with ER stressors (left panels, whole fields; right panels, high magnification images). Cells were transfected with an expression vector for GFP-LC3 and then stimulated by each stressor for 2 h. The formation of GFP-LC3-labeled punctate structures is observed in cells exposed to Tm, TG, and amino acid starvation. Note that the GFP-LC3 dot formation is inhibited by treatment with 10 mM 3-MA and in ATG5−/− cells. (E and F) Quantification of the GFP-LC3-labeled punctate areas in the SK-N-SH cells (E) and MEFs (F) shown in panel D. The ratios of the total GFP-LC3 dot areas to the overall cellular areas are shown as percentages. The values are the means ± standard deviations of three independent experiments. Asterisks indicate a significant difference from control cells (**, P < 0.01; *, P < 0.05) (Student's t test). WT, wild type. (G) Immunoelectron microscopic analysis of SK-N-SH cells transiently transfected with an expression vector for GFP-LC3. Cells were exposed to 2 μg/ml Tm for 6 h. Immunogold particles of GFP-LC3 (arrows) are present on the membranes of autophagosomes. Scale bar, 1 μm.
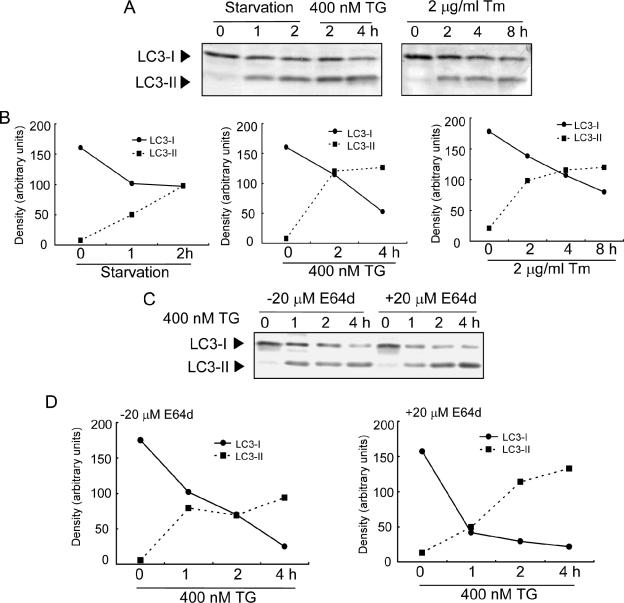
Next, GFP-LC3 fluorescence was used to monitor autophagy in cells transiently transfected with an expression vector for GFP-LC3 (8). The formation of GFP-LC3-labeled structures (GFP-LC3 “dots”), representing autophagosomes, was extensively induced in cells exposed to ER stress (Fig. 1D). Morphometric analysis of the GFP fluorescence images revealed that the GFP-LC3-positive areas occupied ∼0.38% and ∼1.29% of the total cytoplasmic area in the absence and presence of thapsigargin, respectively, indicating that the area increased by about 3.4-fold after treatment with thapsigargin (Fig. 1E). The same effects were also observed after treatment with tunicamycin. Immunoelectron microscopic analysis revealed the presence of immunogold particles, indicating GFP-LC3-positive signals, on the membranes of the autophagosomes (Fig. 1G). Furthermore, Western blotting analysis demonstrated that the amount of membrane-bound LC3-II (the phosphatidylethanolamine-conjugated form; 9) was increased in cells exposed to ER stress (Fig. 2A and B). The level of LC3-II in cells incubated in the presence of a lysosomal inhibitor, E64d, was significantly increased (Fig. 2C and D), suggesting that some LC3-II molecules are subject to lysosomal-dependent turnover. These experiments indicate that LC3 is activated after ER stress and that ER stress induces the formation of autophagosomes, consistent with the results obtained by microscopy.
FIG. 2.
Conversion of LC3 by ER stress. (A) Western blotting of endogenous LC3 in MEFs after exposure to various stimuli. Each stress results in conversion of LC3-I to LC3-II. (B) Quantitative analysis of intensities of Western blotting. Values are arbitrary intensities of each band of Western blotting. (C) Western blotting of endogenous LC3 in MEFs after exposure to TG in the presence or absence of E64d. Note that treatment with E64d promotes accumulation of LC3-II. (D) Quantitative analysis of intensities of Western blotting.
GFP-LC3 dot formation was inhibited by pretreatment with 3-MA (Fig. 1D and E), a nucleotide derivative that inhibits the earliest stages of autophagosome formation (33), similar to the effect of amino acid starvation, indicating that the target of 3-MA is required for the autophagic vacuole formation induced by ER stress, similar to the cases for amino acid starvation and treatment with rapamycin (data not shown).
Finally, to confirm whether the autophagy signaling pathway was also required for ER stress-dependent autophagy, we investigated autophagosome formation after ER stress in ATG5-deficient cells (16). ATG5 is an acceptor molecule for the ubiquitin-like molecule ATG12 and is required for elongation of the autophagic isolation membrane (20). The GFP-LC3 expression vector was transfected into ATG5-deficient MEFs, and GFP-LC3 dot formation was examined. The area of the GFP-LC3 dots increased in wild-type MEFs after ER stress, whereas no change was observed in ATG5-deficient MEFs (Fig. 1D and F). Moreover, no endogenous LC3 processing was observed in ATG5−/− cells during ER stress (data not shown). Taken together, we conclude that ER stress activates the formation of autophagosomes mediated by essential molecules for autophagy, including ATG5.
IRE1 signaling pathway is required for activation of autophagy induced by ER stress.
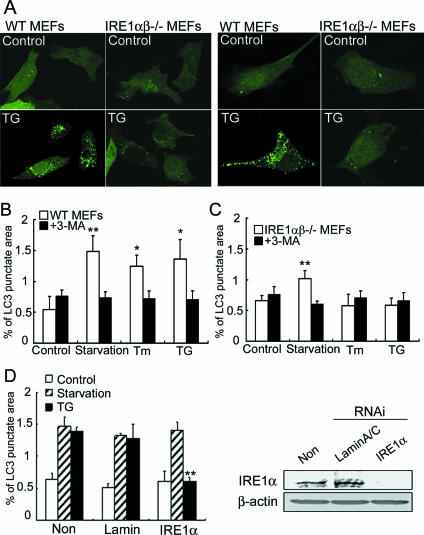
The three major transducers of the UPR are IRE1, PERK, and ATF6, which all sense the presence of unfolded proteins in the ER lumen and transduce signals to the nucleus or cytosol. Therefore, we postulated that one of these transducers must activate the signaling required for the formation of autophagosomes in response to ER stress. We tested this hypothesis using IREα-, IRE1αβ-, and PERK-deficient cells, as well as ATF6αβ knockdown cells. Among these cells, IRE1αβ-deficient MEFs (Fig. 3A to C) and IRE1α-deficient MEFs (data not shown) showed no elevation of GFP-LC3 dot formation during ER stress. In contrast, amino acid deprivation induced the formation of LC3-labeled structures in IRE1-deficient MEFs, and this formation was inhibited by pretreatment with 3-MA (Fig. 3C). These results suggest that the autophagy signaling pathway in the cytosol is functional in IRE1-deficient cells, whereas the signaling pathway that activates autophagy from the ER is disturbed in those cells. It is notable that the GFP-LC3 punctate formation induced by amino acid starvation was inhibited at approximately 50% in IRE1αβ-deficient cells compared with that in wild-type cells (Fig. 3C). To ascertain whether IRE1 is also required for starvation-induced autophagy, we examined the dot formation of GFP-LC3 in cells treated with IRE1 siRNA. Dot formation after amino acid starvation was induced as well as in the control cells, although autophagy induced by ER stress was inhibited in cells treated with IRE1 siRNA (Fig. 3D). The results indicated that IRE1 was not required for autophagosome formation after amino acid starvation. The reasons that IRE1-deficient cells showed a 50% reduction in starvation-induced autophagy are unknown, but clonal variation of IRE1-deficient cells could have a role.
FIG. 3.
Autophagosome formation is inhibited in IRE1-deficient cells. (A) Autofluorescence of GFP-LC3 in wild-type MEFs and IRE1α-deficient MEFs (left panels, whole fields; right panels, high magnification images). Cells were incubated in the presence or absence of 1 μM TG for 2 h. (B and C) Quantification of the GFP-LC3-labeled punctate areas in the cells. The values are the means ± standard deviations of three independent experiments. Asterisks indicate a significant difference from control cells (**, P < 0.01; *, P < 0.05) (Student's t test). WT, wild type. (D) Quantification of the GFP-LC3-labeled punctate areas in cells treated with IRE1α siRNA. Note that treatment with IRE1α siRNA inhibits the formation of autophagosomes induced by ER stress but not by amino acid starvation. The right panel shows IRE1α protein levels in HeLa cells treated with IRE1α or control siRNA (lamin A/C). In IRE1α siRNA-treated cells, the protein levels of IRE1α are reduced.
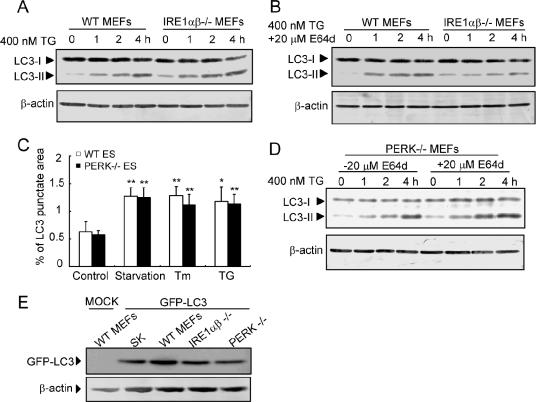
Endogenous LC3 processing was induced by ER stress in IRE1αβ-deficient cells in a manner similar to that in wild-type cells (Fig. 4A), whereas treatment of IRE1αβ-deficient cells with E64d attenuated the accumulation of LC3-II after ER stress (Fig. 4B). The findings indicate that LC3 is processed after ER stress but is not recruited on the membrane of autophagosome and not subject to degradation by the lysosome. These results raise the possibility that IRE1 signaling may be required at a point in the cascade that is later than the LC3 processing stage. However, it remains unclear how recruitment of LC3 to autophagosomes (GFP-LC3 dot formation) is inhibited despite the intact processing of LC3 in IRE1αβ-deficient cells. One possibility is that IRE1-independent signaling may be involved in the intact processing of LC3 during ER stress.
FIG. 4.
Processing of LC3 in IRE1- and PERK-deficient cells. (A) Processing of endogenous LC3 in MEFs exposed to 400 nM TG. Conversion of LC3-I to LC3-II is detected not only in wild-type (WT) MEFs but also in IRE1αβ−/− MEFs after ER stress. The lower panel shows the Western blotting analysis of β-actin used as an internal control. (B and D) Western blotting of endogenous LC3 in wild-type, IRE1αβ−/− (B), and PERK−/− (D) MEFs after exposure to TG in the presence or absence of E64d. (C) Quantification of the GFP-LC3-labeled punctate areas in the PERK-deficient cells. The values are the means ± standard deviations of three independent experiments. Asterisks indicate a significant difference from control cells (**, P < 0.01; *, P < 0.05 (Student's t test). (E) Expression levels of GFP-LC3 in the cells used in each experiment. Cell lysates from each experiment were subjected to Western blotting analyses with anti-GFP and anti-β-actin antibodies.
In PERK-deficient cells, GFP-LC3 dot formation was significantly increased after ER stress (Fig. 4C). Furthermore, treatment of PERK-deficient cells with E64d caused accumulation of LC3-II after ER stress similar to that in wild-type cells (Fig. 4D). ATF6αβ knockdown MEFs showed GFP-LC3 dot formation similar to that with control MEFs (data not shown). The expression levels of GFP-LC3 in cells used in each experiment were almost equivalent (Fig. 4E). Taken together, these results indicate that the PERK and ATF6 pathways do not play important roles in activation of autophagy after ER stress.
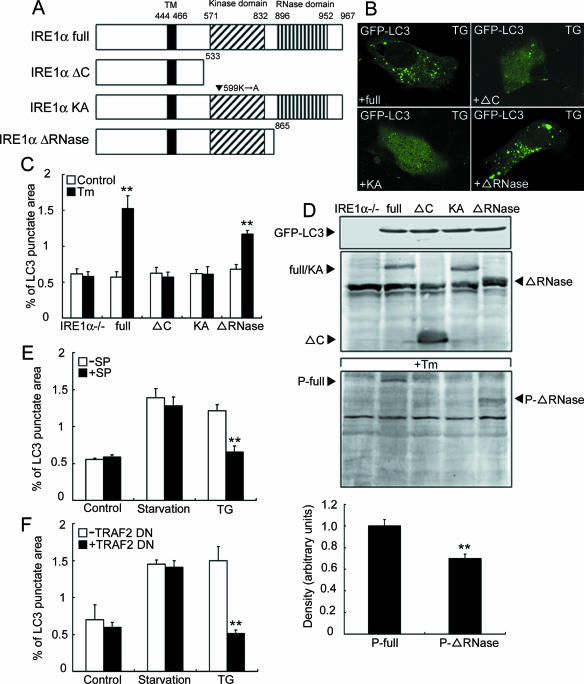
To analyze the detailed signaling pathway from IRE1 to the formation of autophagosomes, expression vectors for various mutants of IRE1α (Fig. 5A) and GFP-LC3 were cotransfected into IRE1α-deficient cells, and the formation of GFP-LC3 dots was examined. Transfection of full-length IRE1α into IRE1α-deficient cells recovered the GFP-LC3 dot formation (Fig. 5B and C). The same result was obtained when the RNase L domain-truncated mutant was introduced into the cells, although the level of recovery was lower than that for full-length IRE1α. Mutants with a C-terminally truncated cytoplasmic region (10) and a Lys599-to-Ala (K599A) mutation that abolishes the kinase activity (37) could not recover the GFP-LC3 dot formation, suggesting that the kinase domain in IRE1α is required for autophagy activation after ER stress. The phosphorylation level of the RNase L domain-truncated mutant was lower than that of full-length IRE1α after ER stress (Fig. 5D). Therefore, the lower level of phosphorylation of the RNase L domain-truncated mutant could attenuate the downstream signaling and partially reduce the formation of the GFP-LC3 dot structure compared with that for the full-length IRE1.
FIG. 5.
Autophagy after ER stress is activated by the IRE1-JNK pathway. (A) Schematic structures of full-length IRE1α (IRE1α full) as well as its carboxy-terminal deletion mutant (IRE1α ΔC), kinase-dead mutant (IRE1α KA), and RNase L domain-truncated mutant (IRE1α ΔRNase). The locations of the transmembrane (TM), kinase, and RNase L domains are indicated. (B) Autofluorescence of GFP-LC3 in IRE1α-deficient MEFs cotransfected with expression vectors for each IRE1α mutant construct and GFP-LC3. After transfection, the cells were exposed to 2 μg/ml Tm for 2 h. (C) Quantification of the GFP-LC3 punctate areas in cells treated with Tm. The values are the means ± standard deviations of three independent experiments. Asterisks indicate a significant difference from IRE1α−/− cells (**, P < 0.01) (Student's t test). (D) Expression levels of GFP-LC3, the IRE1α mutants, and their phosphorylated forms. Lysates from cells double transfected with GFP-LC3 and each IRE1α construct were subjected to Western blotting with anti-GFP, anti-IRE1α, and anti-phosphorylated IRE1α antibodies, respectively. The lower panel shows quantitative analysis of phosphorylated levels of each IRE1 mutant with arbitrary intensities of each band of the Western blotting. The values are the means ± standard deviations of three independent experiments. Asterisks indicate a significant difference from P-full (**, P < 0.01) (Student's t test). (E) Effects of a JNK inhibitor on autophagosome formation after ER stress. Wild-type MEFs were transfected with an expression vector for GFP-LC3, pretreated with a JNK inhibitor, SP600125 (SP) (10 μM), and then exposed to amino acid deprivation (Starvation) and 1 μM TG for 2 h. The values are the means ± standard deviations of three independent experiments. (F) Quantification of the GFP-LC3-labeled punctate areas in cells transfected with an expression vector for a TRAF2 dominant-negative mutant (TRAF2 DN). The values are the means ± standard deviations of three independent experiments.
It has been shown that activated IRE1 on the ER membrane recruits tumor necrosis factor receptor-associated factor 2 (TRAF2) and then activates JNK and that this JNK activation requires the kinase activity of IRE1 (37). That report and our present results raise the possibility that the autophagy activated during ER stress is mediated by JNK after activation by the IRE1 kinase domain. Therefore, we subjected MEFs transfected with the GFP-LC3 expression vector to treatment with a JNK inhibitor, SP600125. The punctate GFP-LC3 dot formation observed in the cells after ER stress was significantly reduced in cells treated with the JNK inhibitor, although the JNK inhibitor did not suppress autophagosome formation after amino acid starvation (Fig. 5E). To confirm that the IRE1-TRAF2-JNK pathway plays an important role in activation of autophagosome formation after ER stress, we cotransfected expression vectors for dominant-negative TRAF2 (40) and GFP-LC3 into SK-N-SH cells and then treated the cells with thapsigargin. The introduction of dominant-negative TRAF2 inhibited the GFP-LC3 dot formation (Fig. 5F). In contrast, dominant-negative TRAF2 did not affect activation of autophagy after amino acid starvation. Taken together, these results suggest that activation of JNK through the IRE1-TRAF2 pathway is indeed required and plays a role specifically in autophagy activation during ER stress.
Protective effects of autophagy on cell death induced by ER stress.
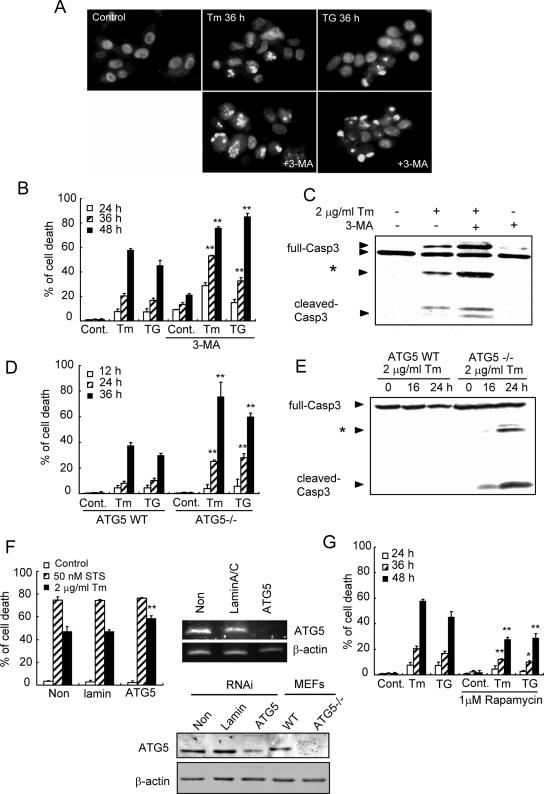
Paradoxically, autophagy can serve to protect cells (29, 41) but may also contribute to cell damage (5, 39). To examine whether the autophagy induced by ER stress plays roles in cell survival or cell death, cells in which autophagy was blocked by 3-MA were exposed to ER stress. The 3-MA-treated cells underwent dramatic cell death (Fig. 6A and B) and revealed more rapid cleavage of caspase-3 after ER stress compared to that for nontreated cells (Fig. 6C). We further analyzed the sensitivity of ATG5-deficient MEFs to ER stress. Cells exposed to tunicamycin or thapsigargin were examined for ER stress-dependent cell death at the indicated times (Fig. 6D). ATG5-deficient cells showed significantly increased vulnerability to ER stress, as well as more rapid activation of caspase-3, compared with wild-type cells (Fig. 6E). Furthermore, cells treated with an ATG5 siRNA showed more sensitivity to ER stress than control cells (Fig. 6F). These results indicate that inhibition of autophagy enhances the cell death induced by ER stress. In contrast, cells pretreated with rapamycin, which was previously shown to effectively inhibit mTOR function and induce autophagy (2, 26), were more resistant to ER stress-induced cell death than control cells (Fig. 6G). Taken together, we conclude that autophagy plays pivotal roles in protecting against the cell death induced by ER stress.
FIG. 6.
Autophagy plays pivotal roles in protecting against the cell death induced by ER stress. (A) Inhibition of autophagy increases vulnerability to ER stress. SK-N-SH cells were treated with 0.5 μg/ml Tm or 300 nM TG for 36 h in the presence or absence of 3-MA and then stained with Hoechst dye. Note that the cells treated with 3-MA show marked chromatin condensation and fragmentation. (B) Quantification of the cell death induced by ER stress in the cells shown in panel A. The values are the means ± standard deviations of three independent experiments. Asterisks indicate a significant difference from cells not treated with 3-MA (**, P < 0.01) (Student's t test). (C) Western blotting of caspase-3 in SK-N-SH cells after exposure to 2 μg/ml Tm for 24 h with or without 10 mM 3-MA. ER stress results in the cleavage of procaspase-3. Treatment with 3-MA accelerates its cleavage. (D) Quantification of cell death in ATG5-deficient MEFs after ER stress. Wild-type or ATG5-deficient MEFs were stimulated with 2 μg/ml Tm or 1 μM TG for the indicated times. The values are the means ± standard deviations of three independent experiments. Asterisks indicate a significant difference from wild-type MEFs (**, P < 0.01) (Student's t test). (E) Western blotting of caspase-3 in ATG5-deficient cells after ER stress. (F) Cell death assay of HeLa cells treated with an ATG5 siRNA (left panel). Cells transfected with each siRNA (final concentration, 10 nM) were incubated at 37°C for 12 h and then stimulated with 2 μg/ml Tm or 50 nM staurosporine (STS), which is an inducer of apoptosis but does not lead to ER stress, or were not stimulated (Control). Note that ATG5 siRNA accelerated cell death by Tm but not by STS. The right panel and lower panel show the effects of the ATG5 siRNA or control siRNA (lamin A/C) on ATG5 mRNA (right) or protein (lower) expression in HeLa cells. (G) Quantification of cell death after ER stress with or without 1 μM rapamycin. The values are the means ± standard deviations of three independent experiments. Asterisks indicate a significant difference from rapamycin-untreated cells (**, P < 0.01; *, P < 0.05) (Student's t test).
JNK activation in the early phase of ER stress is essential for autophagy.
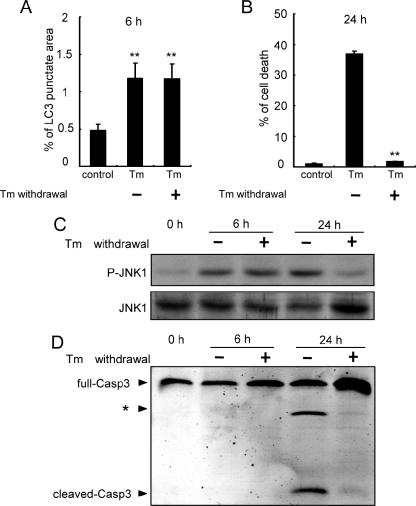
As demonstrated in the present study, activation of JNK is required for the autophagy induced by ER stress. In contrast, it is well established that JNK activation after ER stress is necessary for apoptosis induction (24, 37). Therefore, in order to separate the effect of ER stress on the induction of autophagy from its effect on the induction of apoptosis, we established experimental conditions under which ER stress activated autophagy but did not induce apoptosis. Specifically, the cells were treated with 0.5 μg/ml tunicamycin for 1 h, and the medium was then washed out and exchanged for fresh medium. These conditions led the cells toward autophagy as shown in Fig. 1C but not toward apoptosis (Fig. 7A and B). At the same time, phosphorylated forms of JNK were upregulated (Fig. 7C), indicating that JNK is indeed activated and correlated with the autophagosome formation. In contrast, pretreatment of the cells with a JNK inhibitor suppressed the autophagosome formation. When cells were treated with tunicamycin for 24 h, they underwent apoptosis involving sustained activation of JNK and activation of caspase-3 (Fig. 7B, C, and D). These results indicate that JNK activation in the early phase of ER stress is needed to activate autophagy but does not induce apoptosis. In contrast, sustained activation of JNK for 24 h by ER stress could cause apoptosis.
FIG. 7.
JNK activation and autophagosome formation after ER stress. Cells were treated with 0.5 μg/ml Tm for 1 h, and the medium was then washed out and exchanged for fresh medium. LC3 punctate area at 6 h (A), the number of dead cells at 24 h after ER stress (B), levels of phosphorylated JNK1 (C), and activation of caspase-3 (D) are shown. The values are the means ± standard deviations of three independent experiments. Asterisks indicate a significant difference from control (A) and Tm-treated (B) cells (**, P < 0.01) (Student's t test).
DISCUSSION
We found activation of the autophagy system during ER stress. Our hypothesis that autophagy is induced during ER stress is supported by the following points: first, all ER stressors we examined activated the autophagosome formation, which was shown in examinations of electron microscopy and LC3 dot formation; second, LC3 was convertedfrom LC3-I to LC3-II in response to ER stress; third, autophagy induced by ER stress was inhibited in IRE1-deficient cells. Unfolded or misfolded proteins in the ER lumen are retrotranslocated through the translocon to the cytoplasm, where they are usually ubiquitinated and degraded by the proteasome. Our present study has demonstrated the possibility that autophagy could act as a degradation system for unfolded proteins accumulated in the ER in addition to ERAD.
In Saccharomyces cerevisiae, both the ΔVps30/Atg6 strain, which is unable to degrade the Z variant of human α1 proteinase inhibitor (A1PiZ) by ERAD, and the ΔAtg14 strain, which is unable to induce autophagy, have shown less degradation of A1PiZ and resultant constitutive activation of UPR than the wild type (15). The findings support our notion that unfolded proteins are dealt with by both ERAD and autophagy systems and disturbance of these degradation systems damages cells by ER stress. Recently, autophagy was demonstrated to play a role in the maintenance of ATP production from catabolism of intracellular substrates for cell survival after growth factor withdrawal (18). It is conceivable that autophagy after ER stress also allows for maintenance of energy homeostasis to protect against cell death.
It has been reported that autophagy is induced via eIF2α phosphorylation during starvation in S. cerevisiae and during starvation and viral infection in mammalian cells (35). There are four eIF2α kinases (GCN2, PKR, PERK, and HRI) which are activated by amino acid starvation, viral infection, ER stress, and heme depletion, respectively (6). The previous findings raise the possibility that the PERK-eIF2α pathway may associate with activation of autophagy after ER stress. Surprisingly, our present data showed that the PERK-eIF2α pathway is not essential for ER stress-induced autophagy because autophagosome formation and LC3-II conversion was induced intact in PERK-deficient cells. Recently, Kouroku et al. reported that expanded polyglutamine (polyQ)-induced ER stress activates autophagosome formation with LC3 conversion from LC3-I to -II via the PERK-eIF2α pathway (14). However, in that paper, they did not examine whether agents that specifically induce ER stress such as tunicamycin or thapsigargin activate autophagosome formation via the PERK-eIF2α pathway and whether autophagy is induced in PERK-deficient cells. The cytoplasmic aggregates of malfolded proteins such as polyQ induce ER stress by the accumulation of unfolded proteins in the ER due to the inhibition of proteasome activity and retrotranslocation from ER to cytosol; that is, inhibited ERAD results in ER stress, so it is not the direct effect of polyQ. Therefore, polyQ aggregation could activate eIF2α via cytosolic eIF2α kinases independent of ER eIF2α kinase PERK.
We have also demonstrated that JNK activation mediated by IRE1 in the early phase of ER stress is required for autophagosome formation after ER stress but is not sufficient for the induction of apoptosis. In contrast, sustained activation of JNK for 24 h by ER stress could cause apoptosis. It remains unclear whether the activated JNK pathway after ER stress can affect the functions of mTOR, which occupies a central position in the signaling cascade of autophagy in eukaryotic cells (34), or can associate with a novel signaling pathway for the activation of autophagy. Furthermore, even in IRE1-deficient cells, LC3-II formation was intact after ER stress, indicating that a signaling pathway other than the IRE1-JNK pathway may also play important roles in the activation of autophagy signaling after ER stress. The detailed signaling pathway for activation of the autophagy induced by ER stress awaits further analysis. Both autophagy and ER stress have been implicated in certain human diseases, such as Parkinson's disease (38) and Huntington's disease (28, 30), and exploration of the novel signaling pathways relevant to ER stress and autophagy could lead to the development of new therapeutic strategies for these diseases.
Acknowledgments
We thank Noboru Mizushima for providing the ATG5-deficient cells, David Ron for providing the PERK-deficient MEFs and embryonic stem cells, Laurie H. Glimcher for the ATF6αβ-knockdown MEFs, Eeva-Liisa Eskelinen for the anti-ATG5 antibody, and Tamotsu Yoshimori for the GFP-LC3 plasmids.
This work was partly supported by grants from the JSPS KAKENHI (no. 17200026), the Mitsui Sumitomo Insurance Welfare Foundation, the NOVARTIS Foundation (Japan) for the Promotion of Science, and the Mitsubishi Pharma Research Foundation. Funding was also provided by the Japan Society for the Promotion of Science (M.O.).
Footnotes
Published ahead of print on 9 October 2006.
REFERENCES
- 1.Adham, I. M., T. J. Eck, K. Mierau, N. Müller, M. A. Sallam, I. Paprotta, S. Schubert, S. Hoyer-Fender, and W. Engel. 2005. Reduction of spermatogenesis but not fertility in Creb3l4-deficient mice. Mol. Cell. Biol. 25:7657-7664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blommaart, E. F., J. J. Luiken, P. J. Blommaart, G. M. van Woerkom, and A. J. Meijer. 1995. Phosphorylation of ribosomal protein S6 is inhibitory for autophagy in isolated rat hepatocytes. J. Biol. Chem. 270:2320-2326. [DOI] [PubMed] [Google Scholar]
- 3.Boya, P., R.-A. Gonzalez-Polo, N. Casares, J.-L. Perfettini, P. Dessen, N. Larochette, D. Metivier, D. Meley, S. Souquere, T. Yoshimori, G. Pierron, P. Codogno, and G. Kroemer. 2005. Inhibition of macroautophagy triggers apoptosis. Mol. Cell. Biol. 25:1025-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burry, R. W., D. D. Vandre, and D. M. Hayes. 1992. Silver enhancement of gold antibody probes in pre-embedding electron microscopic immunocytochemistry. J. Histochem. Cytochem. 40:1849-1956. [DOI] [PubMed] [Google Scholar]
- 5.Daido, S., T. Kanzawa, A. Yamamoto, H. Takeuchi, Y. Kondo, and S. Kondo. 2004. Pivotal role of the cell death factor BNIP3 in ceramide-induced autophagic cell death in malignant glioma cells. Cancer Res. 64:4286-4293. [DOI] [PubMed] [Google Scholar]
- 6.Dever, T. E. 2002. Gene-specific regulation by general translation factors. Cell 108:545-556. [DOI] [PubMed] [Google Scholar]
- 7.Imai, Y., M. Soda, H. Inoue, N. Hattori, Y. Mizuno, and R. Takahashi. 2001. An unfolded putative transmembrane polypeptide, which can lead to endoplasmic reticulum stress, is a substrate of Parkin. Cell 105:891-902. [DOI] [PubMed] [Google Scholar]
- 8.Kabeya, Y., N. Mizushima, T. Ueno, A. Yamamoto, T. Kirisako, T. Noda, E. Kominami, Y. Ohsumi, and T. Yoshimori. 2000. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19:5720-5728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kabeya, Y., N. Mizushima, A. Yamamoto, S. Oshitani-Okamoto, Y. Ohsumi, and T. Yoshimori. 2004. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J. Cell Sci. 117:2805-2812. [DOI] [PubMed] [Google Scholar]
- 10.Katayama, T., K. Imaizumi, N. Sato, K. Miyoshi, T. Kudo, J. Hitomi, T. Morihara, T. Yoneda, F. Gomi, Y. Mori, Y. Nakano, J. Takeda, T. Tsuda, Y. Itoyama, O. Murayama, A. Takashima, P. St. George-Hyslop, M. Takeda, and M. Tohyama. 1999. Presenilin-1 mutations downregulate the signalling pathway of the unfolded-protein response. Nat. Cell Biol. 1:479-485. [DOI] [PubMed] [Google Scholar]
- 11.Kaufman, R. J. 2002. Orchestrating the unfolded protein response in health and disease. J. Clin. Investig. 110:1389-1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim, K. S., V. J. Sapienza, and R. I. Carp. 1980. Antiviral activity of arildone on deoxyribonucleic acid and ribonucleic acid viruses. Antimicrob. Agents Chemother. 18:276-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kondo, S., T. Murakami, K. Tatsumi, M. Ogata, S. Kanemoto, K. Otori, K. Iseki, A. Wanaka, and K. Imaizumi. 2005. OASIS, a CREB/ATF-family member, modulates UPR signalling in astrocytes. Nat. Cell Biol. 7:186-194. [DOI] [PubMed] [Google Scholar]
- 14.Kouroku, Y., E. Fujita, I. Tanida, T. Ueno, A. Isoai, H. Kumagai, S. Ogawa, R. J. Kaufman, E. Kominami, and T. Momoi. ER stress (PERK/eIF2α phosphorylation) mediates the polyglutamine-induced LC3 conversion, an essential step for autophagy formation. Cell Death Diff., in press. [DOI] [PubMed]
- 15.Kruse, K. B., J. L. Brodsky, and A. A. McCracken. 2006. Characterization of an ERAD gene as VPS30/ATG6 reveals two alternative and functionally distinct protein quality control pathways: one for soluble Z variant of human α-1 proteinase inhibitor (A1PiZ) and another for aggregates of A1PiZ. Mol. Biol. Cell 17:203-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuma, A., M. Hatano, M. Matsui, A. Yamamoto, H. Nakaya, T. Yoshimori, Y. Ohsumi, T. Tokuhisa, and N. Mizushima. 2004. The role of autophagy during the early neonatal starvation period. Nature 432:1032-1036. [DOI] [PubMed] [Google Scholar]
- 17.Levine, B., and D. J. Klionsky. 2004. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev. Cell 6:463-477. [DOI] [PubMed] [Google Scholar]
- 18.Lum, J. J., D. E. Bauer, M. Kong, M. H. Harris, C. Li, T. Lindsten, and C. B. Thompson. 2005. Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell 120:237-248. [DOI] [PubMed] [Google Scholar]
- 19.Mann, S. S., and J. A. Hammarback. 1994. Molecular characterization of light chain 3. J. Biol. Chem. 269:11492-11497. [PubMed] [Google Scholar]
- 20.Mizushima, N., A. Yamamoto, M. Hatano, Y. Kobayashi, Y. Kabeya, K. Suzuki, T. Tokuhisa, Y. Ohsumi, and T. Yoshimori. 2001. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J. Cell Biol. 152:657-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mori, K. 2000. Tripartite management of unfolded proteins in the endoplasmic reticulum. Cell 101:451-454. [DOI] [PubMed] [Google Scholar]
- 22.Nakagawa, I., A. Amano, N. Mizushima, A. Yamamoto, H. Yamaguchi, T. Kamimoto, A. Nara, J. Funao, M. Nakata, K. Tsuda, S. Hamada, and T. Yoshimori. 2004. Autophagy defends cells against invading group A Streptococcus. Science 306:1037-1040. [DOI] [PubMed] [Google Scholar]
- 23.Nakagawa, T., H. Zhu, N. Morishima, E. Li, J. Xu, B. A. Yankner, and J. Yuan. 2000. Caspase-12 mediates endoplasmic-reticulum-specific apoptosis and cytotoxicity by amyloid-β. Nature 403:98-103. [DOI] [PubMed] [Google Scholar]
- 24.Nishitoh, H., A. Matsuzawa, K. Tobiume, K. Saegusa, K. Takeda, K. Inoue, S. Hori, A. Kakizuka, and H. Ichijo. 2002. ASK1 is essential for endoplasmic reticulum stress-induced neuronal cell death triggered by expanded polyglutamine repeats. Genes Dev. 16:1345-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nixon, R. A. 2006. Autophagy in neurodegenerative disease: friend, foe or turncoat? Trends Neurosci. 29:528-535. [DOI] [PubMed] [Google Scholar]
- 26.Noda, T., and Y. Ohsumi. 1998. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J. Biol. Chem. 273:3963-3966. [DOI] [PubMed] [Google Scholar]
- 27.Pedruzzi, E., C. Guichard, V. Ollivier, F. Driss, M. Fay, C. Prunet, J.-C. Marie, C. Pouzet, M. Samadi, C. Elbim, Y. O'Dowd, M. Bens, A. Vandewalle, M.-A. Gougerot-Pocidalo, G. Lizard, and E. Ogier-Denis. 2004. NAD(P)H oxidase Nox-4 mediates 7-ketocholesterol-induced endoplasmic reticulum stress and apoptosis in human aortic smooth muscle cells. Mol. Cell. Biol. 24:10703-10717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qin, Z.-H., Y. Wang, K. B. Kegel, A. Kazantsev, B. L. Apostol, L. M. Thompson, J. Yoder, N. Aronin, and M. DiFiglia. 2003. Autophagy regulates the processing of amino terminal huntingtin fragments. Hum. Mol. Gen. 12:3231-3244. [DOI] [PubMed] [Google Scholar]
- 29.Qu, X., J. Yu, G. Bhagat, N. Furuya, H. Hibshoosh, A. Troxel, J. Rosen, E.-L. Eskelinen, N. Mizushima, Y. Ohsumi, G. Cattoretti, and B. Levine. 2003. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J. Clin. Investig. 112:1809-1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ravikumar, B., C. Vacher, Z. Berger, J. E. Davies, S. Luo, L. G. Oroz, F. Scaravilli, D. F. Easton, R. Duden, C. J. O'Kane, and D. C. Rubinsztein. 2004. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nature Gen. 36:585-595. [DOI] [PubMed] [Google Scholar]
- 31.Ron, D. 2002. Translational control in the endoplasmic reticulum stress response. J. Clin. Investig. 110:1383-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rutkowski, D. T., and R. J. Kaufman. 2004. A trip to the ER: coping with stress. Trends Cell Biol. 14:20-28. [DOI] [PubMed] [Google Scholar]
- 33.Seglen, P. O., and P. B. Gordon. 1982. 3-Methyladenine: specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proc. Natl. Acad. Sci. USA 79:1889-1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shintani, T., and D. J. Klionsky. 2004. Autophagy in health and disease: a double-edged sword. Science 306:990-995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Talloczy, Z., W. Jiang, H. W. Virgin IV, D. A. Leib, D. Scheuner, R. J. Kaufman, E.-L. Eskelinen, and B. Levine. 2002. Regulation of starvation- and virus-induced autophagy by the eIF2α kinase signaling pathway. Proc. Natl. Acad. Sci. USA 99:190-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tanaka, Y., G. Guhde, A. Suter, E.-L. Eskelinen, D. Hartmann, R. Lullmann-Rauch, P. M. L. Janssen, J. Blanz, K. von Figura, and P. Saftig. 2000. Accumulation of autophagic vacuoles and cardiomyopathy in LAMP-2-deficient mice. Nature 406:902-906. [DOI] [PubMed] [Google Scholar]
- 37.Urano, F., X. Wang, A. Bertolotti, Y. Zhang, P. Chung, H. Harding, and D. Ron. 2000. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science 287:664-666. [DOI] [PubMed] [Google Scholar]
- 38.Webb, J. L., B. Ravikumar, J. Atkins, J. N. Skepper, and D. C. Rubinsztein. 2003. α-Synuclein is degraded by both autophagy and the proteasome. J. Biol. Chem. 278:25009-25013. [DOI] [PubMed] [Google Scholar]
- 39.Xue, L., G. C. Fletcher, and A. M. Tolkovsky. 1999. Autophagy is activated by apoptotic signalling in sympathetic neurons: an alternative mechanism of death execution. Mol. Cell. Neurosci. 14:180-198. [DOI] [PubMed] [Google Scholar]
- 40.Yoneda, T., K. Imaizumi, K. Oono, D. Yui, F. Gomi, T. Katayama, and M. Tohyama. 2001. Activation of caspase-12, an endoplasmic reticulum (ER) resident caspase, through tumor necrosis factor receptor-associated factor 2-dependent mechanism in response to the ER stress. J. Biol. Chem. 276:13935-13940. [DOI] [PubMed] [Google Scholar]
- 41.Yue, Z., S. Jin, C. Yang, A. J. Levine, and N. Heintz. 2003. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc. Natl. Acad. Sci. USA 100:15077-15082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang, K., X. Shen, J. Wu, K. Sakaki, T. Saunders, D. T. Rutkowski, S. H. Back, and R. J. Kaufman. 2006. Endoplasmic reticulum stress activates cleavage of CREBH to induce a systemic inflammatory response. Cell 10:587-599. [DOI] [PubMed] [Google Scholar]