Abstract
Localization and translational control of Drosophila melanogaster gurken and oskar mRNAs rely on the hnRNP proteins Squid and Hrp48, which are complexed with one another in the ovary. Imp, the Drosophila homolog of proteins acting in localization of mRNAs in other species, is also associated with Squid and Hrp48. Notably, Imp is concentrated at sites of gurken and oskar mRNA localization in the oocyte, and alteration of gurken localization also alters Imp distribution. Imp binds gurken mRNA with high affinity in vitro; thus, the colocalization with gurken mRNA in vivo is likely to be the result of direct binding. Imp mutants support apparently normal regulation of gurken and oskar mRNAs. However, loss of Imp activity partially suppresses a gurken misexpression phenotype, indicating that Imp does act in control of gurken expression but has a largely redundant role that is only revealed when normal gurken expression is perturbed. Overexpression of Imp disrupts localization of gurken mRNA as well as localization and translational regulation of oskar mRNA. The opposing effects of reduced and elevated Imp activity on gurken mRNA expression indicate a role in gurken mRNA regulation.
The restriction of proteins to discrete subcellular regions can be accomplished by a coordinated program of mRNA localization and translational control. These mechanisms are used prominently during oogenesis in Drosophila melanogaster, where several localized proteins direct body patterning. The dorsoventral axis of the oocyte and later the embryo are established by a process that involves the specific expression of Gurken (Grk) protein at a dorsal position near the anterior of the oocyte. Similarly, patterning along the anteroposterior axis relies on restricted expression of Bicoid (Bcd) and Oskar (Osk) proteins at the anterior and posterior poles, respectively, of the oocyte and embryo. In each case, the deployment of the protein is a consequence of localization of the mRNA to the appropriate region within the oocyte, coupled with translational controls to enhance accumulation of the protein at this destination (reviewed in reference 12).
These programs of posttranscriptional control of gene expression require RNA binding proteins that recognize regulatory elements within the mRNAs and mediate association with the localization or translational control machinery. Although it has proven difficult in most cases to demonstrate that a particular protein/RNA interaction contributes to regulation, multiple RNA binding proteins are required for correct expression of grk mRNA; these include Squid (Hrp40), Hrp48 (also known as Hrb27C), Bruno (Bru), Vasa, and Otu (8, 9, 13, 31, 41, 45, 49). Each of these proteins is also required for correct expression of osk (11, 14, 20, 30, 32, 40, 44, 47, 51), revealing substantial similarities in the control of grk and osk mRNAs.
Mutants defective for Sqd, Hrp48, and Otu have a common grk mRNA localization defect (9). Normally, grk mRNA is transiently localized to the anterior of the oocyte at stage 8 of oogenesis and then becomes restricted to the dorsal side of the anterior. In the mutants, grk mRNA persists along the anterior and fails to localize dorsally. Because localization of grk mRNA has been suggested to result from two vectorial movements—one toward the anterior and a second directed dorsally (19)—these genes could act specifically in the second movement. Sqd and Hrp48 have also been implicated in translational regulation and act to limit the translation of grk mRNA to the fraction of the mRNA that is properly localized at the dorsal side of the oocyte (9, 31). The mechanistic details of how these proteins contribute to localization and translational control remain to be determined, but it does appear that they function as part of a regulatory RNP complex, since Hrp48 interacts physically with both Sqd and Otu (9). Two components of the complex, Sqd and Hrp48, have been suggested to assemble with the mRNAs in the nucleus and associate with other factors in the cytoplasm (22-24, 31). It is likely that additional members of this complex have not yet been identified.
One candidate for another regulatory factor is Imp, the Drosophila homolog of a family of proteins that act in posttranscriptional regulation in a variety of animals (34, 50). One of the founding members of the family, ZBP-1, binds to a localization element in the chicken beta-actin mRNA (15) and appears to direct localization to the leading edge of embryonic fibroblasts (7). Another founding member, the Xenopus Vg1RBP/VERA protein, binds to signals directing localization of Vg1 and VegT mRNAs to the vegetal pole of the oocyte (4, 5, 10, 16). Mammalian homologs, the Imp proteins, have been suggested to act in mRNA localization (36), mRNA stability (6), and translational regulation (28). A recent report examined the RNA binding properties of Drosophila Imp protein, focusing specifically on the osk mRNA and its possible regulation by Imp (26). Although mutation of candidate Imp binding sites in the osk mRNA did block accumulation of Osk protein, loss of Imp activity did not cause a similar defect.
Here we also characterize the Drosophila Imp protein and show that it interacts with Sqd and Hrp48, two proteins that regulate expression of osk and grk mRNAs. Mutation of the Imp gene does not substantially alter grk or osk expression. Nevertheless, the Imp mutant partially suppresses a grk misexpression phenotype, arguing that it does contribute to grk regulation but may act redundantly and does not have an essential role. Consistent with this interpretation, overexpression of Imp interferes with localization of grk mRNA.
MATERIALS AND METHODS
Fly stocks and transgenics.
A full-length Imp cDNA (expressed sequence tag [EST] clone SD07045) was cloned into a UASp vector (33), and transgenic stocks were generated by standard methods. Multiple independent P[UAS-Imp] stocks produced similar phenotypes, with some differences in severity, when expressed from the P[matα4-GAL4VP16] V37 driver.
Fly stocks l(1)G0072 (now called ImpG0072), Df(1)HCl33, P[matα4-GAL4VP16] V37 and Dp(1;Y)v+y+ were obtained from the Bloomington stock center. Secondary mutations on the w67c23 ImpG0072 chromosome were removed by extensive backcrossing to w1118 flies. The kinesin-LacZ reporter (3) was obtained from David Stein, the fs(1)K101 and sqd1 flies were from Trudi Schupbach, and the TauGFP flies were from Daniel St Johnston.
Plasmid rescue was performed to confirm that the P element of ImpG0072 is inserted into the Imp gene. The lethality of ImpG0072 was confirmed to be due to the P element by isolation and characterization of revertants: five excision lines were obtained using the PΔ23 transposase, and none of them shows the lethality observed in ImpG0072.
Homozygous ImpG0072 flies were obtained by the following cross scheme. Df(1)v-L2/Dp(1;Y)v+y+ males were crossed with ImpG0072/FM7c females. Progeny ImpG0072/Dp(1;Y)v+y+ males were crossed with ImpG0072/FM7c females to get ImpG0072 homozygous flies. Homozygous fs(1)K101 ImpG0072 flies were obtained by the same strategy, using an fs(1)K101 ImpG0072 chromosome obtained by recombination.
Generation of Imp antibody and purification of His-Imp.
The coding region of the Imp gene was amplified by PCR and cloned into pET3b vector (Novagen). The Imp protein was expressed in Escherichia coli Codon-Plus (Stratagene) and partially purified. Polyclonal antibody against Imp was raised by Josman, LLC.
The Imp coding region was also cloned into PET15a vector (Novagen) to allow expression of Imp with an amino-terminal 6× His tag. The 6× His-Imp fusion protein was expressed in E. coli Codon-Plus RP (Stratagene) and purified using Probond resin (Invitrogen).
Immunodetection and in situ hybridization.
Ovaries were dissected and stained as described previously (17, 18). Primary antibodies were used at the following dilutions: rabbit anti-Imp, 1:600; rat anti-Vasa, 1:500; rabbit anti-Oskar, 1:4,000; rat anti-Staufen, 1:100; rabbit anti-Stau, 1:1,000; mouse anti-Gurken [1D12 from the Developmental Studies Hybridoma Bank], 1:10; mouse anti-beta-galactosidase (40-1a; Developmental Studies Hybridoma Bank), 1:40. Secondary antibodies were labeled with Cy5 (Jackson Immunoresearch Laboratories) or Alexa Fluor 488 (Molecular Probes). Stained ovaries were mounted in Vectashield medium (Vector Labs) and imaged with a Leica TCS-SP confocal microscope.
Live imaging of egg chambers was performed as described previously (39) using flies expressing TauGFP maternally to mark microtubules (25).
To quantitate the loss of dorsal localization of Imp in sqd mutant oocytes, images acquired by confocal microscopy were analyzed for signal intensity using the ImageJ software (NIH). For each of four oocytes of each genotype, four nonoverlapping boxes were drawn at random within the dorsal cortical region adjacent to the nucleus or along the cortical region near the posterior pole. The signal intensity of each region was measured, to yield an average value, and the ratios of the dorsal and posterior values were determined. For the wild-type oocytes, the ratios were 1.54, 1.79, 1.83, and 2.00. For the sqd mutant oocytes, the ratios were 1.07, 1.14, 1.14, and 1.24.
In situ hybridization was performed as described previously (42). Linearized plasmids containing the osk 3′ untranslated region (UTR) (pY107 cut by BamHI), the bcd 3′ UTR (p908 cut by MluI), and the grk 3′ UTR (p848 cut by BglII) were used as templates for synthesis of antisense RNA probes. The probes were labeled with digoxigenin-conjugated nucleotides (Roche Diagnostic GmbH).
Western blot analysis.
Protein samples were electrophoresed in a 10% sodium dodecyl sulfate-polyacrylamide gel and electroblotted to a polyvinylidene difluoride membrane. Proteins were detected by chemiluminescence (Western Light; Tropix). Primary antibodies were affinity-purified rabbit polyclonal anti-Imp at 1:3,000, mouse monoclonal anti-α-tubulin at 1:20,000 (gift from Tim Stearns), mouse monoclonal anti-Sqd at 1:100 (gift from Trudi Schubach), and rabbit polyclonal anti-Hrp48 at 1:20,000 (gift from Don Rio).
Immunoprecipitation.
Ovaries of w1118 flies were hand-dissected in phosphate-buffered saline buffer, washed with lysis buffer (50 mM Tris-Cl [pH 8.0], 150 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol, 0.1% NP-40, 2 mM Pefabloc, 5 mM benzamidine. 2 μg/ml pepstatin, and 2 μg/ml leupeptin) three times, homogenized, and cleared by centrifugation at 13,000 rpm for 25 min at 4°C. Aliquots of the extract (300 μl; equivalent to 50 ovary pairs) were incubated with primary antibody at 4°C for 1 h. Subsequently, 20 μl of protein A/G PLUS agarose beads (Santa Cruz Biotechnology) preequilibrated with lysis buffer were added to the extract and incubated at 4°C for 30 min. Agarose beads were spun down and washed three times with lysis buffer. Next, they were incubated with lysis buffer with or without 50 ng/ml RNase A/T1 (Ambion) for 15 min at 4°C. Finally, beads were recovered by centrifugation and washed with lysis buffer three times. Sodium dodecyl sulfate loading buffer (2×, 50 μl) was added to the beads and boiled at 100°C for 5 min. Samples were assayed by Western blotting.
Filter binding assay.
Probes were generated by in vitro transcription in the presence of [32P]UTP and gel purified. Details of the plasmids used to prepare the osk and grk RNAs described in the legend to Fig. 4 are available on request. Twenty microliters of reaction mix containing labeled probe (<0.1 nM in final concentration) and various amounts of purified Imp protein in filter binding buffer (10 mM Tris-Cl [pH 8.0], 25 mM NaCl, 0.2 mM EDTA, 0.1 mg/ml tRNA, 5 mg/ml heparin, 1 mM dithiothreitol) were incubated on ice for 1 h. Filter binding buffer (80 μl) was added to each reaction mixture, and the samples were filtered though nitrocellulose membrane filters (Millipore) preequilibrated with filter binding buffer at 4°C for at least 1 h. The membrane filters were washed three times with 1 ml filter binding buffer and assayed for radioactivity by scintillation spectrometry. Dissociation constants (Kd) were calculated using Kaleidagraph (Synergy Software).
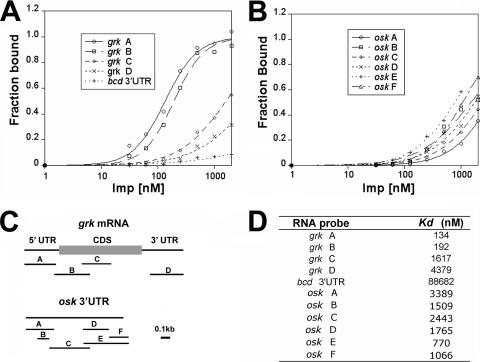
FIG. 4.
Imp binds with high affinity to grk mRNA. (A and B) Filter binding assays of Imp binding to portions of the grk mRNA (A) and osk mRNA 3′ UTR (B). The binding assays were performed with various concentrations of Imp to allow calculation of dissociation constants. (C) Diagram of the grk mRNA and osk mRNA 3′ UTR, indicating the regions used as probes. (D) Summary of dissociation constants obtained from the binding assays.
RESULTS
Imp associates with hnRNP proteins Sqd and Hrp48.
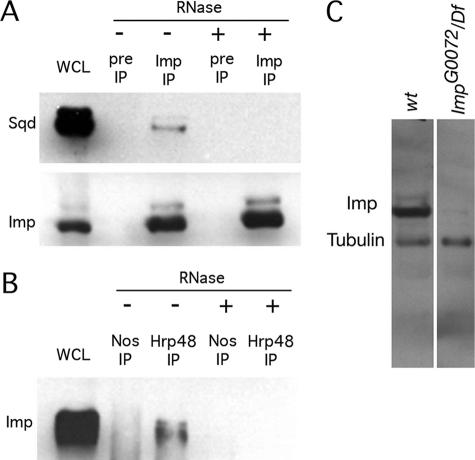
Coimmunoprecipitation experiments with ovary extracts were performed to test for association of Imp with proteins known to act in posttranscriptional regulation. We found that Imp coimmunoprecipitates with Sqd and with Hrp48 (Fig. 1). Both Sqd and Hrp48 are, like Imp, RNA binding proteins, and their association with Imp could involve only protein/protein contacts or could depend on RNA binding. The coimmunoprecipitations were also performed after treatment with RNase, and in each case, the interaction is disrupted. An additional RNA binding protein, Nanos, was also tested by the same assay but did not coimmunoprecipitate with Imp. Thus, the RNA-dependent association of Imp with Sqd and Hrp48 is specific and is not a common property of all RNA binding proteins.
FIG. 1.
Imp is coimmunoprecipitated with Sqd and Hrp48. (A) Western blot of immunoprecipitation (IP) from ovary extract with anti-Imp antibodies or preimmune serum (Pre) with (+) or without (−) RNase A/T1 treatment. Proteins on the blot were detected with anti-Imp and anti-Sqd. An amount of ovary extract equal to 5% of that used for the immunoprecipitations was loaded in lane WCL. (B) Western blot of immunoprecipitation from ovary extract with Hrp48 or Nanos (Nos) antibodies with or without RNase A/T1 treatment. The blot was probed with anti-Imp. (C) Western blot analysis of Imp protein. Similar amounts of ovarian protein from w1118 or ImpG0072/Df(1)HC133 mutant females were probed for Imp and for α-tubulin as a loading control. The ImpG0072 hemizygous mutant has dramatically reduced levels of the two immunoreactive bands of Imp protein (one prominent and indicated as Imp, the other less abundant and slightly larger). The identities of the bands were confirmed in blots probed separately for Imp or Tubulin. wt, wild type.
Imp is concentrated at the site of grk mRNA localization.
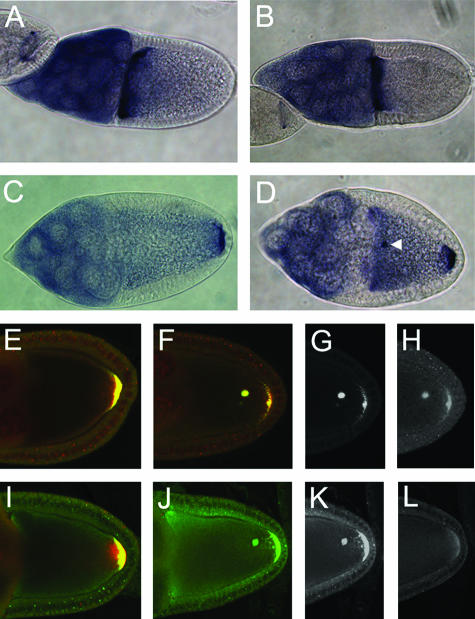
Imp protein is cytoplasmic and present in essentially all cells of the ovary, both somatic follicle cells and the germ line nurse cells and oocyte (see Fig. S1 in the supplemental material). Within the germ line, Imp displays a changing pattern of abundance in different cells. At the earliest stages of oogenesis, Imp is initially uniform in the dividing germ line cells of each cyst but becomes rapidly concentrated in the oocyte (see Fig. S1 in the supplemental material). This enrichment is lost by stage 7, after which the level of Imp in the oocyte is noticeably reduced. Although the uniform level of Imp in the oocyte decreases, Imp levels become elevated in during stages 8 and 9 in the narrow zone between the nucleus and the anterior and dorsolateral margins of the oocyte (Fig. 2A). This is precisely the region in which grk mRNA and protein accumulate, with Grk protein then trafficking to the follicle cells to provide a localized signal in the pathway that specifies dorsal fates (29). The enrichment of Imp could be specific and perhaps related to the localization or translation of grk mRNA. Alternatively, all cytoplasmic proteins might display a concentration in this restricted region of the ooplasm. To distinguish between these options, we examined other cytoplasmic proteins that also appear in the oocyte. Unlike Imp, neither Vasa (Fig. 2D and E) nor Spindle E (data not shown) proteins were enriched between the nucleus and the oocyte margins, despite being present throughout the ooplasm. Thus, the regional concentration of Imp is specific.
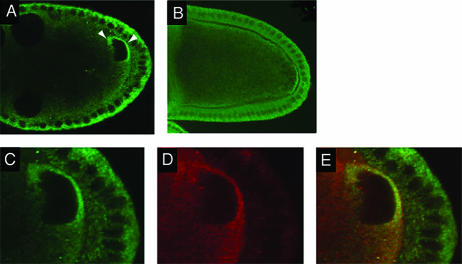
FIG. 2.
Distribution of Imp protein in the ovary. (A) Imp is transiently concentrated in an anterodorsal zone flanking the oocyte nucleus on the anterior and lateral sides (arrowheads). This localization can be detected as soon as the nucleus migrates to the anterior of the oocyte and is largely lost by mid-stage 9. (B) Imp is concentrated in a crescent at the posterior pole of the oocyte. (C to E) The anterodorsal concentration of Imp in the oocyte is specific. Egg chambers were double labeled for Imp (C) or Vas (D). Both proteins can be detected throughout the oocyte cytoplasm. In the overlay of the two signals (E), the ratio of green to red is substantially greater in the lateral and anterior sides of the nucleus than elsewhere in the cytoplasm, indicating that Imp is specifically concentrated at these regions of the oocyte.
To determine if the concentration of Imp correlates with localization of grk mRNA, we examined Imp protein distribution under conditions when the anterodorsal localization of grk mRNA is altered (Fig. 3). In sqd mutant ovaries, grk mRNA remains concentrated at the anterior of the oocyte but is no longer restricted to the dorsal region (31). The concentration of Imp at the dorsal side of the nucleus is substantially reduced in the mutant ovaries, and the degree of residual localization correlates well with the level of residual localized Grk protein expression. Furthermore, Imp still appears concentrated along the anterior of the oocyte, just as does grk mRNA. To quantitate the loss of dorsal localization, anti-Imp fluorescence intensity levels were measured in the dorsal cortical region and in a more posterior cortical region for multiple wild-type and sqd mutant oocytes (see Materials and Methods). The dorsal/posterior ratio was 1.54 to 2.00 for the wild type, reflecting the dorsal concentration, and 1.07 to 1.24 for the sqd mutant, confirming that dorsal localization of Imp is reduced (Fig. 3). Thus, Imp not only colocalizes with grk mRNA but may rely on the grk mRNA localization machinery or grk mRNA itself for that distribution. Consequently, Imp could bind directly to grk mRNA and act in regulation of its expression.
FIG. 3.
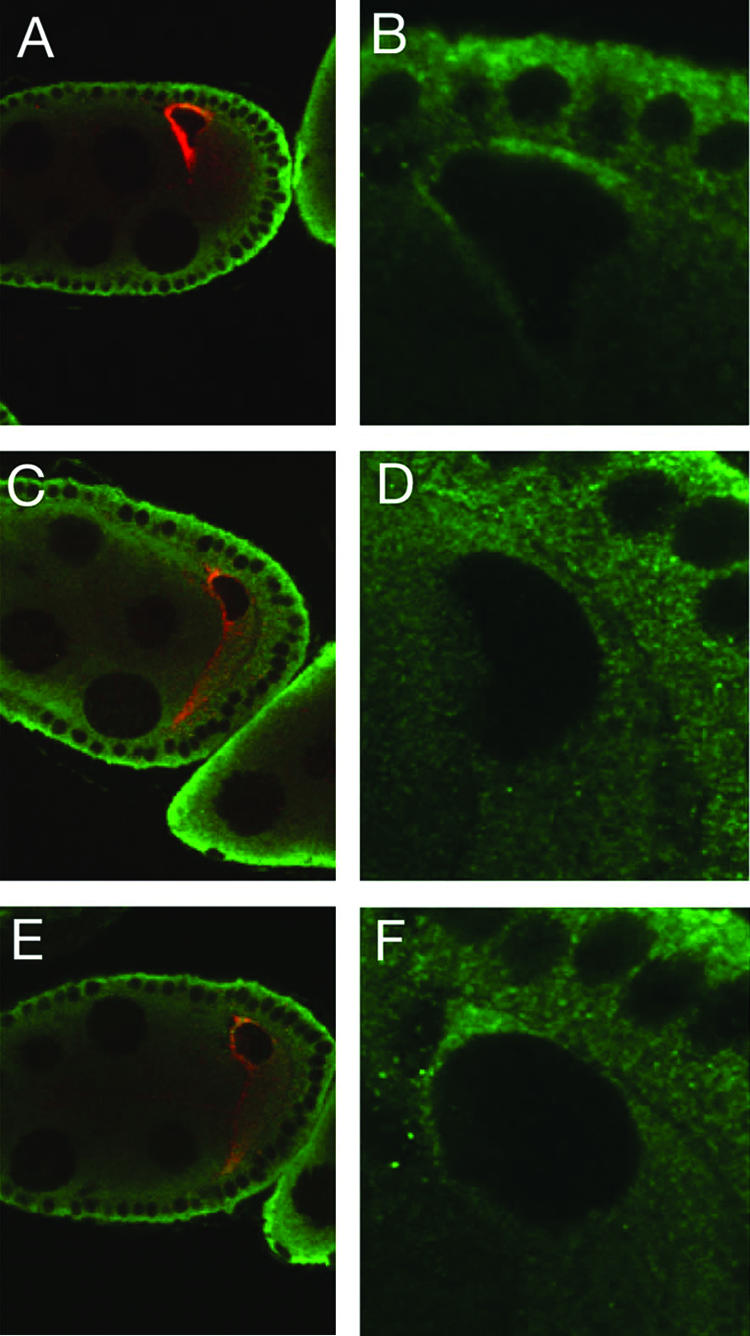
Anterodorsal localization of Imp in the oocyte is dependent on Sqd. Panels A, C, and E are stained for both Grk protein (red) and Imp protein (green). Panels B, D, and F are magnifications, with only the Imp signal shown. In a wild-type oocyte (A), Grk is highly concentrated adjacent to the nucleus, and Imp is concentrated in a similar pattern. In sqd mutant oocytes (B and C) Grk protein remains at the anterior of the oocyte but now at both dorsal and ventral positions in the optical sections. The Imp localization is greatly reduced at the dorsal surface, primarily along the lateral cortex. The residual concentration of Imp between the oocyte nucleus and the nurse cells parallels the distribution of grk mRNA.
Imp protein also colocalizes with osk mRNA at the posterior pole of the oocyte (Fig. 2B), raising the possibility that it is associated with osk mRNA. Munro et al. (26) examined this association in detail and found that Imp remained colocalized with osk mRNA that had been mispositioned in various mutants, very strongly arguing that Imp is bound, directly or indirectly, to osk mRNA.
Imp binds to grk mRNA.
The RNA binding activity of purified Imp was monitored using a quantitative nitrocellulose filter binding assay. RNA probes for the assay were prepared from different parts of the grk and osk mRNAs (Fig. 4C) as well as from bcd mRNA 3′ UTR, which serves as a negative control (there is no indication of any role for Imp in bcd expression). Imp binds with highest affinity to the grk mRNA 5′ UTR (Kd is 134 nM) and the 5′ part of the coding sequence (Kd of 192 nM) (Fig. 4A and D). Other parts of the grk mRNA, including the 3′ coding region and 3′ UTR, bind with much lower affinity (Kd of 1.6 and 4.3 μM, respectively). Weak binding is also observed for the osk 3′ UTR (various segments bind with a Kd of 0.77 to 3.4 μM) (Fig. 4B and D). The negative control RNA is largely unbound at similar Imp protein concentrations. The high-affinity binding of Imp to grk mRNA, taken together with the colocalization of the mRNA and protein in the oocyte, strongly suggests that Imp binds grk mRNA in vivo. It also appears possible that Imp binds directly to osk mRNA in vivo. However, the lower affinity of the in vitro interaction with osk mRNA raises the possibility that binding occurs in the context of an RNP complex in which multiple proteins contact the mRNA and a high affinity is achieved through multiple contacts, much as has been suggested for the binding of a localization complex to the bcd mRNA (1). Obvious candidates for additional complex components are the Imp-associated proteins Sqd and Hrp48, both of which are concentrated with Imp and osk mRNA at the posterior pole of the oocyte (11, 30, 51).
Imp mutants have no overt ovarian phenotype but suppress the dorsalization of fs(1)K10.
For genetic analysis of Imp function, we used a P element insertion mutant, ImpG0072, in which the transposon is inserted into the Imp gene. ImpG0072 is semilethal, with rare escapers surviving as adults for up to several days. Two forms of Imp protein are detected in wild-type ovaries by Western blot analysis (Fig. 1). The most abundant form is about 70 kDa, consistent with the reported structure of the Imp protein. Both protein forms are present at greatly reduced levels in ImpG0072/Df(1)H133 females. The ImpG0072 chromosome was extensively backcrossed to the wild type to remove secondary mutations. Reversion of the mutation by excision of the P element restored full viability and expression of Imp protein.
Homozygous ImpG0072 females, although unhealthy and prone to getting stuck in the growth medium, can produce eggs before they die. The eggs appear phenotypically normal and, if fertilized by wild-type sperm, form viable and fertile adults. Not surprisingly then, the Imp mutants have no substantial defects in distribution or activity of localized mRNAs that contribute to embryonic body patterning (data not shown). Nevertheless, Imp could play a redundant role, perhaps in regulation of grk or osk, given its concentration at the sites where these mRNAs are localized and its association with Hrp48 and Sqd.
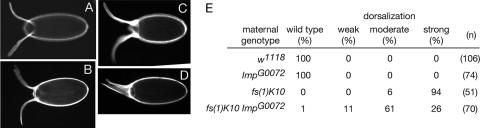
As a more sensitive assay for a role in the dorsoventral pathway, we sought to determine whether the Imp mutation could modify an existing dorsoventral patterning defect. Mutation of fs(1)k10, which encodes a transcriptional factor, results in mislocalization of grk mRNA and protein along the anterior margin of the oocyte during midoogenesis, instead of restriction to the anterodorsal corner. Consequently, eggs laid by mutant fs(1)K10 mothers are strongly dorsalized and display expansion and fusion of the two dorsal appendages that normally lie near the dorsal midline (27, 35, 38, 48) (Fig. 5D). When fs(1)k10 females are also homozygous for the ImpG0072 mutation, the dorsalization phenotype is partially suppressed (Fig. 5C and D). In keeping with the absence of a detectable ImpG0072 mutant phenotype, the fs(1)K10 eggshell phenotype is only fully suppressed in very rare cases. More commonly, the embryos from the Imp fs(1)K10 mothers display some degree of reduced dorsalization, as scored by the effects on the dorsal appendages. No substantial change in the grk mRNA distribution of the fs(1)K10 mutant accompanies the partial suppression of the eggshell phenotype (data not shown). This is not surprising, given the normal appearance of grk mRNA distribution in Imp mutants. However, because the eggshell phenotype provides a very sensitive measure of grk patterning activity, it can presumably reveal defects not discernible by the in situ hybridization assay. We conclude that reduction of Imp activity has a very weak effect on dorsoventral patterning.
FIG. 5.
Suppression of the fs(1)K10 phenotype by reduction of Imp activity. Panels A to D are eggshells from wild-type (A) or fs(1)K10 ImpG0072 (B to D) mothers showing weak (B), moderate (C), or strong dorsalization (D). The dorsal appendages are well separated in the wild type (A) and fused ventrally in the strongly dorsalized eggshells (D). Quantitation of the phenotypes is provided in panel E.
Overexpression of Imp alters dorsoventral polarity and expression of grk.
As an alternate assay for Imp activity in dorsoventral patterning, we overexpressed the protein in the germ line cells of the ovary using the GAL4/UAS expression system (2, 33). Overexpression of Imp produces a strong and highly penetrant effect: dorsalization of the eggshell. In the wild type, embryos have an eggshell in which the two anterior dorsal appendages lie close to the dorsal midline and are separated from one another (Fig. 6E). Only 17% of the embryos from Imp overexpression mothers (n = 255) have the wild-type dorsal appendages. The remainder of the embryos show various degrees of dorsalization, in which the dorsal appendages fuse and form an anterior ring around the embryo (Fig. 6F to H). The Imp overexpression also causes a partial dumpless phenotype (a defect in transfer of nurse cell contents to the oocyte), such that the embryos are smaller than normal (data not shown).
FIG. 6.
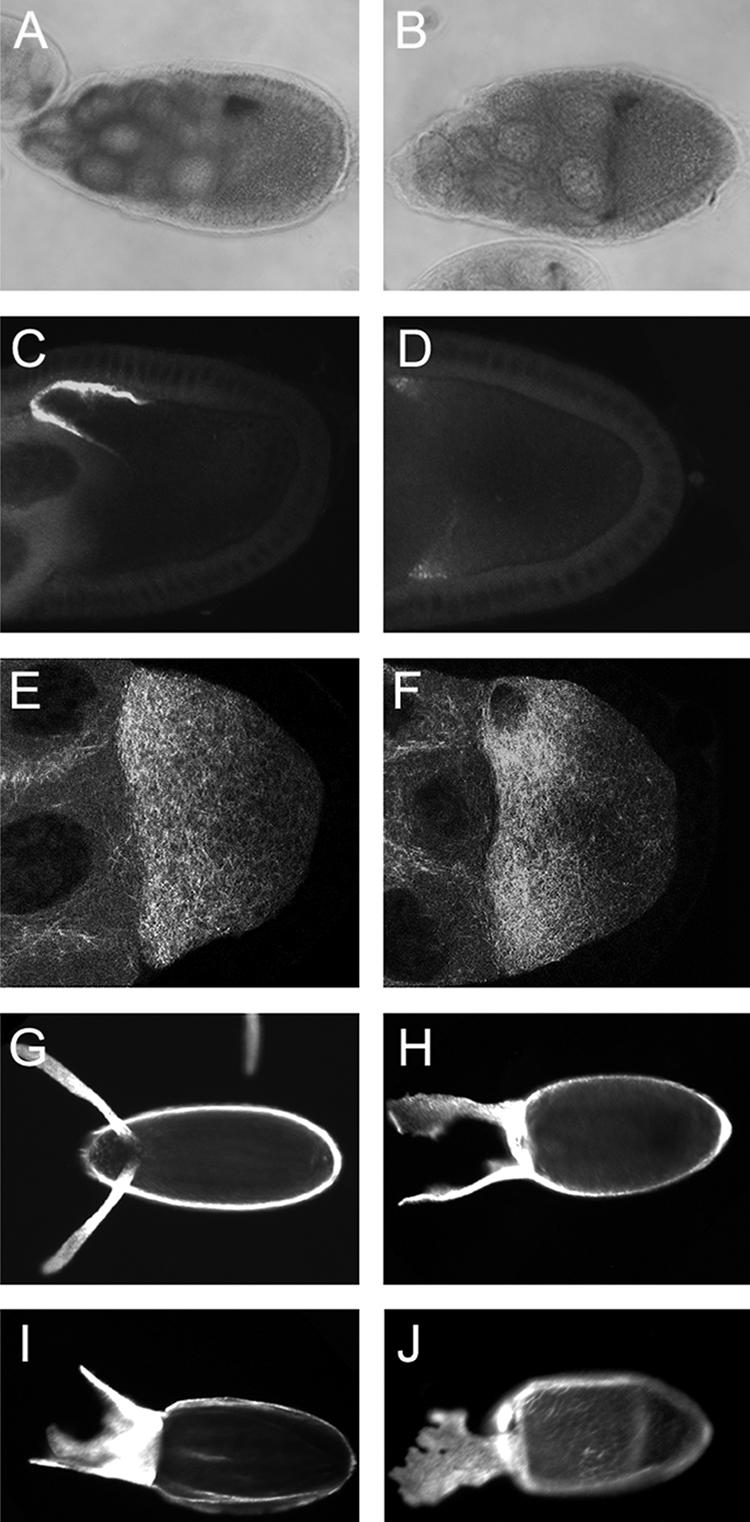
Imp overexpression alters dorsoventral patterning and regulation of grk mRNA. Both grk mRNA (A and B) and protein (C and D) are misexpressed in Imp overexpression egg chambers. Panels A and C are the wild type, while panels B and D express one copy of P[UAS-Imp] under the control of the matα4-GAL4VP16 driver. Panels E and F show microtubule organization in live wild type (E) and Imp overexpression (F) oocytes, as detected by TauGFP. Both show a gradient of microtubule density, highest at the anterior. There are no substantial differences between the mutant and wild type. Panels G to J show eggshells (anterior to the left) of wild-type or Imp overexpression oocytes (one copy of P[UAS-Imp] with the matα4-GAL4VP16 driver). All eggshells of wild-type oocytes are normal (G) (n = 105). Imp overexpression produces eggshells (n = 255) of which 17% are wild type, 20% are moderately dorsalized (H), and 58% are strongly dorsalized (I). The remaining 4% have fused but slightly expanded dorsal appendages (J).
Consistent with the eggshell dorsalization phenotype, grk mRNA and protein fail to be tightly restricted to the anterodorsal corner of the oocyte and are instead dispersed along the anterior margin. At stage 9, 97% of Imp overexpression oocytes display an abnormal circular ring of grk mRNA (Fig. 6B), with only 3% having the normal anterodorsal restriction (n = 31) (Fig. 6A). By stage 10, localization has improved, with 52% showing the ring and the remainder appear wild type (n = 33). For Grk protein, 79% of stage 9 oocytes have an anterior ring of Grk (Fig. 6D), with 21% displaying the normal wild-type anterodorsal distribution (n = 53) (Fig. 6C). Much as for the grk mRNA, the localization improves by stage 10, with 56% having an anterior ring and 44% being wild type (n = 45). Wild-type controls show normal anterodorsal localization of both grk mRNA and protein at both stages. To address the possibility that the grk mRNA localization defect is an indirect consequence of altered microtubule organization, microtubules were monitored by imaging TauGFP in live egg chambers (25). No substantial differences were observed in comparison of wild-type and Imp overexpression oocytes (Fig. 6E and F, respectively). Similar results were obtained when microtubules were imaged by immunodetection in fixed samples (data not shown).
In wild-type oocytes, grk mRNA is initially spread across the anterior of the oocyte and becomes tightly restricted to the dorsal anterior before stage 9. Because localization improves in the Imp overexpression oocytes as development of the oocyte advances, it is possible that the actions that restrict the mRNA dorsally are active but less effective or retarded. Alternatively, there may be mechanistically distinct phases in localization—one acting earlier and one later—with overexpression of Imp primarily or exclusively disrupting the early phase. Localization during the later phase would gradually restore the wild-type distribution of grk mRNA and protein.
Imp overexpression disrupts oocyte polarity and expression of osk.
osk mRNA is tightly localized to the posterior pole of wild-type oocytes from stage 9 throughout the remainder of oogenesis, and Osk protein only accumulates after localization of the mRNA (14, 20, 32). In Imp overexpression ovaries, osk mRNA (as well as Stau protein, which typically marks the distribution of osk mRNA) (21) appears at two positions within individual oocytes: some of the osk mRNA is localized in a crescent at the posterior pole, the normal site of localization, and some of the osk mRNA appears in a discrete body in the ooplasm (Fig. 7D and G). These bodies, which are never seen in the wild type (Fig. 7C and E), are present in 79% of stage 9 egg chambers (n = 113) and 55% of stage 10 egg chambers (n = 99). Osk protein is present at both sites of osk mRNA concentration (Fig. 7H), demonstrating that translation of osk mRNA is not negatively affected by Imp overexpression. Moreover, the accumulation of Osk is no longer dependent on the posterior localization of the mRNA, revealing a loss of the regulation that normally prevents accumulation of Osk from unlocalized mRNA.
FIG. 7.
Imp overexpression disrupts polarization of the oocyte along the anteroposterior axis. The distribution of bcd mRNA in wild-type (A) and Imp overexpression (B) egg chambers is indistinguishable. In contrast, the posterior localization of osk mRNA in the wild type (C) is disrupted by overexpression of Imp, with the mRNA often present in a discrete body (or rarely, bodies) in the ooplasm (arrowhead) as well as at the posterior pole (D). A similar effect is observed for Stau protein (green signal in panels E and F, wt and Imp overexpression, respectively), which also serves to mark the distribution of osk mRNA. Notably, Osk protein (red signal in panels E and F) also appears in the ooplasmic bodies containing osk mRNA and Stau protein. The Stau and Osk signals are also shown separately in panels G (Stau) and H (Osk) for clarity. The kinesin-β-gal marker (red signal in panels I and J), which is heavily concentrated at the posterior pole in the wild-type and colocalized with Stau (green signal), is dispersed in Imp overexpression oocytes (J), with no evidence of concentration in a central zone. The Stau and kinesin-β-gal signals are also shown separately in panels K (Stau) and L (kinesin-β-gal) for clarity.
Imp is concentrated at the site of osk mRNA localization and is associated with osk mRNA (26). Thus, the osk mRNA localization defect arising from overexpression of Imp may involve a direct effect on osk mRNA. However, another consequence of Imp overexpression suggests that osk mRNA mislocalization may result, at least in part, from a more primary defect in microtubule organization. In wild-type stage 9 oocytes, the microtubule polarity marker Kin-β-gal (3) is concentrated at the posterior pole (Fig. 7I). When Imp is overexpressed, posterior localization of Kin-β-gal is greatly reduced (Fig. 7L). However, the effects on microtubule organization must be subtle, as there are no obvious differences in microtubules imaged in wild-type and Imp overexpression oocytes (Fig. 6E and F), and bcd mRNA remains normally localized at the anterior of Imp overexpression oocytes (Fig. 7B).
DISCUSSION
Deployment of proteins that control patterning in the oocyte relies on coordinated programs of mRNA localization and translational control. Many RNA binding proteins contribute to these programs, and some interact with one another in regulatory RNPs. Here we have shown that Imp is associated in an RNA-dependent manner with Sqd and Hrp48 and is thus part of a complex whose other members have clearly established roles in control of grk and osk expression. Imp does not have an essential role in regulation of either grk or osk mRNAs, as both mRNAs are expressed with no obvious defects in Imp mutant ovaries. However, loss of Imp activity does partially suppress the grk misexpression defect in fs(1)K10 mutant oocytes, providing strong evidence that Imp contributes to regulation of grk. This view is reinforced by the colocalization of Imp with grk mRNA in vivo. Imp's role must be largely redundant, only becoming detectable when grk expression is perturbed. Overexpression of Imp has a much more dramatic effect, transiently blocking the dorsal localization of grk mRNA and disrupting localization and translational control of osk mRNA.
The evidence that Imp, Sqd, and Hrp48 interact physically is complemented by striking similarities in grk and osk expression defects that arise from loss of sqd or Hrp48 activity or from overexpression of Imp. In each case, grk mRNA accumulates at the anterior of the oocyte, fails to become dorsally localized, and leads to misexpression of Grk protein. The defects of sqd and Hrp48 mutants in osk expression may result from both direct and indirect effects: a direct effect via binding to osk mRNA and an indirect effect owing to alterations in microtubule organization (11, 26, 30, 51). The same is true for Imp overexpression. In Imp overexpression oocytes, the posterior localization of Kin-β-gal is disrupted, indicating some degree of microtubule defects. In addition, Imp, like Sqd and Hrp48, colocalizes with osk mRNA to the posterior pole of the oocyte.
The correlations between the consequences of excess Imp activity and loss of sqd or Hrp48 activity may be significant and suggest that Imp competes with these proteins at some level, either for binding to a common substrate or by exerting opposing effects on such a substrate. Alternatively, Imp could inactivate Sqd or Hrp48. Imp overexpression does not substantially alter the amount of Sqd or Hrp48 (data not shown), ruling out one form of inactivation. In addition, sqd and Hrp48 mutants display one phenotype, altered polytenization of nurse cell nuclei (9), which does not occur when Imp is overexpressed, arguing against any simple model in which Sqd and Hrp48 are inhibited by Imp.
Sqd and Hrp48 could compete with Imp at the level of RNA binding: excess Imp would displace Sqd or Hrp48 from shared or closely positioned binding sites on regulated mRNAs, yielding the same phenotype as if Sqd or Hrp48 were eliminated by mutation and thus not available for binding. This model seems unlikely, since Imp binds best to the 5′ UTR and 5′ coding regions of grk mRNA (regions implicated in grk mRNA localization) (43, 46), while Sqd and Hrp48 bind to the 3′ UTR (9, 31) (osk mRNA is considered below). However, Imp does bind with lower affinity to the grk mRNA 3′ UTR, and the assays with Sqd and Hrp48 have not tested for binding to the grk mRNA 5′ UTR.
Competition could also occur for events that transpire after RNA binding, with bound Imp promoting one outcome for the mRNA and bound Sqd and Hrp48 promoting another. For example, localization of grk mRNA has been suggested to involve two vectorial movements within the oocyte, one directed anteriorly and one directed dorsally (19). In this model, Imp could promote the anterior movement, while Sqd and Hrp48 could contribute to the dorsal movement (a role in keeping with known phenotypes). Increasing the number of copies of one protein that become bound to the mRNA, even without a reduction in the binding of other proteins, could enhance association with the machinery that drives one vectorial movement and thus alter the balance between the two movements. This type of interpretation would explain the partial suppression of the fs(1)K10 ventralization phenotype by the Imp mutant. In the absence of K10, the competition would be skewed in favor of the Imp-promoted outcome. Removing Imp, even if it acts redundantly, could shift the competition back toward the balance normally achieved in wild-type ovaries. This model might appear to be at odds with the known distributions of Imp, Hrp48, and Sqd. Specifically, Imp is colocalized with grk mRNA even after the proposed second vectorial movement of localization, while Hrp48 and Sqd are never detectably colocalized with the mRNA. However, the proposed competition would not require displacement of Imp from the mRNA, and Hrp48 and Sqd might act very early in the localization process (perhaps beginning in the nucleus where the proteins are concentrated) to orchestrate events that only occur later. Thus, the positions of the proteins in the ovary only rule out the possibility that they are all persistently associated with one another but do not argue against the models described here.
The defect in grk mRNA localization caused by overexpression of Imp is accompanied by ectopic accumulation of Grk protein, whose distribution mirrors that of grk mRNA along the anterior of the oocyte. In wild-type ovaries, grk mRNA is transiently concentrated along the anterior of the oocyte at stages 7 and 8, but there is no corresponding anterior ring of Grk protein (37). Thus, the anterior accumulation of Grk when Imp is overexpressed reveals a defect in the control of grk mRNA translation, as well as localization. The premature translation could be an indirect consequence of derailing grk mRNA localization, or it could indicate a more direct effect of excess Imp on translation.
Does Imp act in regulation of osk mRNA?
Our discussion of Imp has focused on regulation of grk mRNA, since this role is supported by multiple lines of evidence. Overexpression of Imp also dramatically alters osk expression, acting indirectly by altering microtubule organization and perhaps acting directly through binding to osk mRNA. The data implicating Imp in osk regulation, whether direct or indirect, are substantially less compelling than for regulation of grk. Most importantly, we have no loss-of-function evidence that implicates Imp in osk mRNA regulation or in control of microtubule organization. Second, the binding of Imp to the osk mRNA 3′ UTR is relatively weak, with Kd values near or above 1 μM.
Munro et al. (26) specifically explored the possible regulation of osk by Imp. They identified sequences (Imp binding elements [IBEs]) in the osk mRNA as Imp binding sites. Inactivation of the IBEs eliminates accumulation of Osk protein. The osk mRNA initially localizes normally to the posterior of the oocyte but is later delocalized and dispersed in the ooplasm, apparently an indirect consequence of a failure to accumulate Osk protein, which is required for anchoring of osk mRNA (26). Because loss of Imp activity did not cause similar defects, they concluded that another factor (factor X) must bind the IBEs for osk mRNA translation. Factor X could act redundantly with Imp, or factor X alone could mediate the action of the IBEs. Munro et al. argue for the latter option and propose a regulatory interplay between Imp and factor X, in which they compete for binding. By that model, overexpression of Imp would be expected to have consequences similar to mutation of the IBEs. The consequences of Imp overexpression differ when comparing our work to that of Munro et al. They present evidence that Imp overexpression reduces the level of Osk at stage 10 and do not report on osk mRNA localization. We also find a reduction in the accumulation of Osk protein at the posterior pole, but this is accompanied by mislocalization of a fraction of Osk protein to a discrete body in the ooplasm, a feature not observed for the IBE mutants. This body also contains osk mRNA, a localization defect that is clearly different from the dispersal of osk mRNA caused by mutation of the IBEs. To consider the possibility that the difference between our results and those of Munro et al. may reflect different levels of Imp overexpression, we varied the dosage of the P[UAS-Imp] transgene: increasing from one to two copies greatly enhanced the shift of osk mRNA and protein to multiple discrete bodies in the ooplasm but did not eliminate Osk accumulation (data not shown). Thus, in our extensive analysis, the effects of Imp overexpression on osk mRNA localization and translation are markedly different from the IBE inactivation phenotype. We cannot explain why our results differ from those of Munro et al.; further characterization of their Imp overexpression mutant might provide insight.
Imp mutants do not have an osk misexpression phenotype, but the in vitro binding properties of Imp and the consequences of Imp overexpression suggest that Imp plays a redundant role, much as we have argued in the case of grk mRNA. It would not be surprising for Imp to act in regulation of osk, as well as grk, since Imp is associated with proteins known to regulate both mRNAs.
Supplementary Material
Acknowledgments
This work was supported by grants GM42612 and GM54409 from the National Institutes of Health.
Members of the Macdonald lab, especially Mark Snee, Nan Yan, and Eric Arn, provided useful discussion and comments on the manuscript. We thank the Bloomington Stock Center, Trudi Schupbach, Daniel St Johnston, Don Rio, Tim Stearns, and Tom Hays for fly stocks, cDNAs, or antibodies. Some antibodies were obtained from the Developmental Studies Hybridoma Bank, which was developed under the auspices of the NICHD and maintained by The University of Iowa, Department of Biological Sciences, Iowa City, Iowa.
Footnotes
Published ahead of print on 9 October 2006.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Arn, E. A., B. J. Cha, W. E. Theurkauf, and P. M. Macdonald. 2003. Recognition of a bicoid mRNA localization signal by a protein complex containing Swallow, Nod, and RNA binding proteins. Dev. Cell 4:41-51. [DOI] [PubMed] [Google Scholar]
- 2.Brand, A. H., and N. Perrimon. 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118:401-415. [DOI] [PubMed] [Google Scholar]
- 3.Clark, I., E. Giniger, H. Ruohola-Baker, L. Y. Jan, and Y. N. Jan. 1994. Transient posterior localization of a kinesin fusion protein reflects anteroposterior polarity of the Drosophila oocyte. Curr. Biol. 4:289-300. [DOI] [PubMed] [Google Scholar]
- 4.Deshler, J. O., M. I. Highett, T. Abramson, and B. J. Schnapp. 1998. A highly conserved RNA-binding protein for cytoplasmic mRNA localization in vertebrates. Curr. Biol. 8:489-496. [DOI] [PubMed] [Google Scholar]
- 5.Deshler, J. O., M. I. Highett, and B. J. Schnapp. 1997. Localization of Xenopus Vg1 mRNA by vera protein and the endoplasmic reticulum. Science 276:1128-1131. [DOI] [PubMed] [Google Scholar]
- 6.Doyle, G. A., N. A. Betz, P. F. Leeds, A. J. Fleisig, R. D. Prokipcak, and J. Ross. 1998. The c-myc coding region determinant-binding protein: a member of a family of KH domain RNA-binding proteins. Nucleic Acids Res. 26:5036-5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Farina, K. L., S. Huttelmaier, K. Musunuru, R. Darnell, and R. H. Singer. 2003. Two ZBP1 KH domains facilitate beta-actin mRNA localization, granule formation, and cytoskeletal attachment. J. Cell Biol. 160:77-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Filardo, P., and A. Ephrussi. 2003. Bruno regulates gurken during Drosophila oogenesis. Mech. Dev. 120:289-297. [DOI] [PubMed] [Google Scholar]
- 9.Goodrich, J. S., K. N. Clouse, and T. Schupbach. 2004. Hrb27C, Sqd and Otu cooperatively regulate gurken RNA localization and mediate nurse cell chromosome dispersion in Drosophila oogenesis. Development 131:1949-1958. [DOI] [PubMed] [Google Scholar]
- 10.Havin, L., A. Git, Z. Elisha, F. Oberman, K. Yaniv, S. P. Schwartz, N. Standart, and J. K. Yisraeli. 1998. RNA-binding protein conserved in both microtubule- and microfilament- based RNA localization. Genes Dev. 12:1593-1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huynh, J. R., T. P. Munro, K. Smith-Litiere, J. A. Lepesant, and D. St. Johnston. 2004. The Drosophila hnRNPA/B homolog, Hrp48, is specifically required for a distinct step in osk mRNA localization. Dev. Cell 6:625-635. [DOI] [PubMed] [Google Scholar]
- 12.Johnstone, O., and P. Lasko. 2001. Translational regulation and RNA localization in Drosophila oocytes and embryos. Annu. Rev. Genet. 35:365-406. [DOI] [PubMed] [Google Scholar]
- 13.Kelley, R. L. 1993. Initial organization of the Drosophila dorsoventral axis depends on an RNA-binding protein encoded by the squid gene. Genes Dev. 7:948-960. [DOI] [PubMed] [Google Scholar]
- 14.Kim-Ha, J., K. Kerr, and P. M. Macdonald. 1995. Translational regulation of oskar mRNA by bruno, an ovarian RNA-binding protein, is essential. Cell 81:403-412. [DOI] [PubMed] [Google Scholar]
- 15.Kislauskis, E. H., X. Zhu, and R. H. Singer. 1994. Sequences responsible for intracellular localization of β-actin messenger RNA also affect cell phenotype. J. Cell Biol. 127:441-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kwon, S., T. Abramson, T. P. Munro, C. M. John, M. Kohrmann, and B. J. Schnapp. 2002. UUCAC- and Vera-dependent localization of VegT RNA in Xenopus oocytes. Curr. Biol. 12:558-564. [DOI] [PubMed] [Google Scholar]
- 17.Macdonald, P. M., S. K.-S. Luk, and M. Kilpatrick. 1991. Protein encoded by the exuperantia gene is concentrated at sites of bicoid mRNA accumulation in Drosophila nurse cells but not in oocytes or embryos. Genes Dev. 5:2455-2466. [DOI] [PubMed] [Google Scholar]
- 18.Macdonald, P. M., and G. Struhl. 1986. A molecular gradient in early Drosophila embryos and its role in specifying the body pattern. Nature 324:537-545. [DOI] [PubMed] [Google Scholar]
- 19.MacDougall, N., A. Clark, E. MacDougall, and I. Davis. 2003. Drosophila gurken (TGFalpha) mRNA localizes as particles that move within the oocyte in two dynein-dependent steps. Dev. Cell 4:307-319. [DOI] [PubMed] [Google Scholar]
- 20.Markussen, F.-H., A.-M. Michon, W. Breitwieser, and A. Ephrussi. 1995. Translational control of oskar generates Short OSK, the isoform that induces pole plasm assembly. Development 121:3723-3732. [DOI] [PubMed] [Google Scholar]
- 21.Martin, S. G., and D. St. Johnston. 2003. A role for Drosophila LKB1 in anterior-posterior axis formation and epithelial polarity. Nature 421:379-384. [DOI] [PubMed] [Google Scholar]
- 22.Matunis, E. L., M. J. Matunis, and G. Dreyfuss. 1992. Characterization of the major hnRNP proteins from Drosophila melanogaster. J. Cell Biol. 116:257-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matunis, M. J., E. L. Matunis, and G. Dreyfuss. 1992. Isolation of hnRNP complexes from Drosophila melanogaster. J. Cell Biol. 116:245-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matunis, M. J., E. L. Matunis, and G. Dreyfuss. 1993. PUB1: a major yeast poly(A)+ RNA-binding protein. Mol. Cell. Biol. 13:6114-6123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Micklem, D. R., R. Dasgupta, H. Elliott, F. Gergely, C. Davidson, A. Brand, A. González-Reyes, and D. St Johnston. 1997. The mago nashi gene is required for the polarisation of the oocyte and the formation of perpendicular axes in Drosophila. Curr. Biol. 7:468-478. [DOI] [PubMed] [Google Scholar]
- 26.Munro, T. P., S. Kwon, B. J. Schnapp, and D. St. Johnston. 2006. A repeated IMP-binding motif controls oskar mRNA translation and anchoring independently of Drosophila melanogaster IMP. J. Cell Biol. 172:577-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neuman-Silberberg, F. S., and T. Schüpbach. 1996. The Drosophila TGF-α-like protein Gurken: expression and cellular localization during Drosophila oogenesis. Mech. Dev. 59:105-113. [DOI] [PubMed] [Google Scholar]
- 28.Nielsen, J., J. Christiansen, J. Lykke-Andersen, A. H. Johnsen, U. M. Wewer, and F. C. Nielsen. 1999. A family of insulin-like growth factor II mRNA-binding proteins represses translation in late development. Mol. Cell. Biol. 19:1262-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nilson, L. A., and T. Schupbach. 1999. EGF receptor signaling in Drosophila oogenesis. Curr. Top. Dev. Biol. 44:203-243. [DOI] [PubMed] [Google Scholar]
- 30.Norvell, A., A. Debec, D. Finch, L. Gibson, and B. Thoma. 2005. Squid is required for efficient posterior localization of oskar mRNA during Drosophila oogenesis. Dev. Genes Evol. 215:340-349. [DOI] [PubMed] [Google Scholar]
- 31.Norvell, A., R. L. Kelley, K. Wehr, and T. Schupbach. 1999. Specific isoforms of squid, a Drosophila hnRNP, perform distinct roles in Gurken localization during oogenesis. Genes Dev. 13:864-876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rongo, C., E. R. Gavis, and R. Lehmann. 1995. Localization of oskar RNA regulates oskar translation and requires Oskar protein. Development 121:2737-2746. [DOI] [PubMed] [Google Scholar]
- 33.Rorth, P. 1998. Gal4 in the Drosophila female germline. Mech. Dev. 78:113-118. [DOI] [PubMed] [Google Scholar]
- 34.Ross, A. F., Y. Oleynikov, E. H. Kislauskis, K. L. Taneja, and R. H. Singer. 1997. Characterization of a β-actin mRNA zipcode-binding protein. Mol. Cell. Biol. 17:2158-2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roth, S., and T. Schupbach. 1994. The relationship between ovarian and embryonic dorsoventral patterning in Drosophila. Development 120:2245-2257. [DOI] [PubMed] [Google Scholar]
- 36.Runge, S., F. C. Nielsen, J. Nielsen, J. Lykke-Andersen, U. M. Wewer, and J. Christiansen. 2000. H19 RNA binds four molecules of insulin-like growth factor II mRNA-binding protein. J. Biol. Chem. 275:29562-29569. [DOI] [PubMed] [Google Scholar]
- 37.Saunders, C., and R. S. Cohen. 1999. The role of oocyte transcription, the 5′UTR, and translation repression and derepression in Drosophila gurken mRNA and protein localization. Mol. Cell 3:43-54. [DOI] [PubMed] [Google Scholar]
- 38.Serano, T. L., M. Karlin-McGinness, and R. S. Cohen. 1995. The role of fs(1) K10 in the localization of the mRNA of the TGFa homolog gurken within the Drosophila oocyte. Mech. Dev. 51:183-192. [DOI] [PubMed] [Google Scholar]
- 39.Snee, M. J., and P. M. Macdonald. 2004. Live imaging of nuage and polar granules: evidence against a precursor-product relationship and a novel role for Oskar in stabilization of polar granule components. J. Cell Sci. 117:2109-2120. [DOI] [PubMed] [Google Scholar]
- 40.Steinhauer, J., and D. Kalderon. 2005. The RNA-binding protein Squid is required for the establishment of anteroposterior polarity in the Drosophila oocyte. Development 132:5515-5525. [DOI] [PubMed] [Google Scholar]
- 41.Styhler, S., A. Nakamura, A. Swan, B. Suter, and P. Lasko. 1998. vasa is required for GURKEN accumulation in the oocyte, and is involved in oocyte differentiation and germline cyst development. Development 125:1569-1578. [DOI] [PubMed] [Google Scholar]
- 42.Tautz, D., and C. Pfeifle. 1989. A non radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals a translational control of the segementation gene hunchback. Chromosoma 98:81-85. [DOI] [PubMed] [Google Scholar]
- 43.Thio, G. L., R. P. Ray, G. Barcelo, and T. Schupbach. 2000. Localization of gurken RNA in Drosophila oogenesis requires elements in the 5′ and 3′ regions of the transcript. Dev. Biol. 221:435-446. [DOI] [PubMed] [Google Scholar]
- 44.Tirronen, M., V.-P. Lahti, T. I. Heino, and C. Roos. 1995. Two otu transcripts are selectively localised in Drosophila oogenesis by a mechanism that requires a function of the otu protein. Mech. Dev. 52:65-75. [DOI] [PubMed] [Google Scholar]
- 45.Tomancak, P., A. Guichet, P. Zavorszky, and A. Ephrussi. 1998. Oocyte polarity depends on regulation of gurken by Vasa. Development 125:1723-1732. [DOI] [PubMed] [Google Scholar]
- 46.Van De Bor, V., E. Hartswood, C. Jones, D. Finnegan, and I. Davis. 2005. gurken and the I factor retrotransposon RNAs share common localization signals and machinery. Dev. Cell 9:51-62. [DOI] [PubMed] [Google Scholar]
- 47.Webster, P. J., L. Liang, C. A. Berg, P. lasko, and P. M. Macdonald. 1997. Translational repressor bruno plays multiple roles in development and is widely conserved. Genes Dev. 11:2510-2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wieschaus, E., J. L. Marsh, and W. Gehring. 1978. fs(1)K10, a germline-dependent female sterile mutation causing abnormal chorion morphology in Drosophila melanogaster. Roux's Arch. Dev. Biol. 184:75-82. [DOI] [PubMed] [Google Scholar]
- 49.Yan, N., and P. M. Macdonald. 2004. Genetic interactions of Drosophila melanogaster arrest reveal roles for translational repressor Bruno in accumulation of Gurken and activity of Delta. Genetics 168:1433-1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yaniv, K., and J. K. Yisraeli. 2002. The involvement of a conserved family of RNA binding proteins in embryonic development and carcinogenesis. Gene 287:49-54. [DOI] [PubMed] [Google Scholar]
- 51.Yano, T., S. L. de Quinto, Y. Matsui, A. Shevchenko, and A. Ephrussi. 2004. Hrp48, a Drosophila hnRNPA/B homolog, binds and regulates translation of oskar mRNA. Dev. Cell 6:637-648. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.