Abstract
The DNA and the spindle assembly checkpoints play key roles in maintaining genomic integrity by coordinating cell responses to DNA lesions and spindle dysfunctions, respectively. These two surveillance pathways seem to operate mostly independently of one another, and little is known about their potential physiological connections. Here, we show that in Saccharomyces cerevisiae, the activation of the spindle assembly checkpoint triggers phosphorylation changes in two components of the DNA checkpoint, Rad53 and Rad9. These modifications are independent of the other DNA checkpoint proteins and are abolished in spindle checkpoint-defective mutants, hinting at specific functions for Rad53 and Rad9 in the spindle damage response. Moreover, we found that after UV irradiation, Rad9 phosphorylation is altered and Rad53 inactivation is accelerated when the spindle checkpoint is activated, which suggests the implication of the spindle checkpoint in the regulation of the DNA damage response.
The DNA and the spindle assembly checkpoints are surveillance pathways in charge of a common task: the proper transmission of genetic material. This task involves checking for (i) the complete and accurate replication of nuclear DNA, (ii) the absence of DNA lesions, and (iii) the equal repartition of the sister chromatids in the daughter cells. The DNA checkpoints are activated in cases of DNA damage or replication defects. They do not seem to be sensitive to incomplete DNA replication per se but rather to pathological DNA structures resulting from stalled replication forks or DNA lesions. They comprise different proteins able to sense these pathological structures and/or to transduce a signal, mostly via phosphorylation cascades. In Saccharomyces cerevisiae, their main components include the kinases Mec1, Tel1, Rad53, Dun1, and Chk1, along with adaptors (Rad9 and Mrc1) mediating their interactions (for a review, see reference 39).
The primary defect that activates the spindle assembly checkpoint remains unclear (48). This checkpoint seems to check for the accurate segregation of the chromatids by monitoring either the attachment of microtubules to kinetochores (the protein complexes that assemble at the centromeres) or the tension that is exerted at kinetochores upon bipolar attachment (for reviews, see references 26, 33, and 43). The spindle assembly checkpoint is thus sensitive to defects altering all aspects of the spindle function. Drugs or mutations affecting microtubule polymerization, spindle pole body duplication, microtubule motors, kinetochore components, or DNA centromeric sequences arrest cells in mitosis by spindle assembly checkpoint-dependent mechanisms or are synthetically lethal with mutations of spindle assembly checkpoint genes (19, 21, 34, 45). Its core components were first identified through genetic screens in yeast (21, 34) and consist of the Mad1, Mad2, Mad3, Bub1, and Bub3 proteins and the Mps1 kinase (70).
The DNA checkpoint can be activated at any point of the cell cycle depending on the nature of DNA lesions and on the phase when DNA damage occurs, whereas the activation of the spindle assembly checkpoint is restricted to G2/mitosis (G2/M). The two checkpoints have in common the ability to block both the metaphase/anaphase transition and the exit from mitosis. The progression from metaphase to anaphase is triggered by the degradation of the securin Pds1, which depends on the anaphase promoting complex (APC) (for a review, see reference 44). Upon checkpoint activation, Pds1 is stabilized and sequesters the separin Esp1 into an inactive complex, thus precluding the release of the cohesin Mcd1/Scc1 from the chromosomes and sister chromatid separation (2, 3, 14, 50). The stabilization of Pds1 also concurs to inhibit mitotic exit (8, 56, 62). Exit from mitosis is regulated in S. cerevisiae by the mitotic exit network (MEN), which includes the phosphatase Cdc14; the kinases Cdc5, Cdc15, Dbf2, and Dbf20; the GTPase Tem1; and the two-component GTPase-activating protein Bub2/Bfa1 (4, 24, 41). The Bub2/Bfa1 complex inhibits Tem1, whose activation promotes APC-dependent destruction of B-type cyclins, activation of the Cdk1 inhibitor Sic1, and mitotic exit (5, 47, 53, 66). BFA1 and BUB2 are required to prevent mitotic exit after activation of the DNA or the spindle assembly checkpoints (21, 67).
Whereas the DNA checkpoint was characterized from the beginning as a pathway responding to DNA damage (69), the fact that the spindle assembly checkpoint contributes to the cell response to DNA damage and replication defect was only lately acknowledged. Stern and Murray have elegantly demonstrated that Pds1 is stabilized in a Mad2-dependent manner in a cdc6 mutant undergoing anaphase without prior DNA replication (57), which suggests that the spindle assembly checkpoint can respond to the lack of tension at kinetochores induced by defective DNA replication. Mutations in centromeric DNA that impair kinetochore attachment were shown to induce a spindle assembly checkpoint-dependent mitotic delay (45, 55), which suggested that lesions affecting centromeric DNA could also be detected by the spindle assembly checkpoint via their effects on kinetochore structure. Several studies have subsequently shown that the spindle assembly checkpoint is involved in cell cycle arrest following DNA damage in yeast, Drosophila, and mammal cells (7, 10, 13, 15, 36, 40, 49, 58). Thus, Collura et al. have demonstrated that in Schizosaccharomyces pombe, the Chk1 pathway causes a Mad2-dependent delay at the metaphase-to-anaphase transition, when cells enter mitosis after camptothecin treatment (10). In budding yeast cells with a defective DNA checkpoint, Mad2 contributes to the residual G2/M arrests induced by the DNA-alkylating agent methyl methanesulfonate or by mutations in genes encoding DNA polymerase subunits or components of the prereplicative complex (15). Similarly, Clerici et al. have shown that the absence of Mad2 results in advanced nuclear division of rad53Δ chk1Δ cells and in a partial suppression of the metaphase arrest of mec1Δ cells after UV irradiation (7). In S. cerevisiae, Mad2 seems to have an effect on the DNA damage response mostly in the absence of a functional DNA checkpoint and therefore independently from it. Mad2 influences in cases of DNA damage could be explained in two ways. First, the spindle assembly checkpoint could be activated as a consequence of DNA lesions (DNA lesions affecting centromeric sequences could lead to defective kinetochore structures, or DNA damage could prevent replication of the centromeres). Second, Mad2 could have a constitutive, inhibitory activity regarding the APC that would be efficiently offset by APC activation during an unperturbed cell cycle but not after DNA damage, even in the absence of core components of the DNA checkpoint. In the latter case, the deletion of MAD2 would suppress Mad2-constitutive inhibition of the APC and the G2/M arrest observed in DNA checkpoint-deficient cells upon DNA damage. However, this hypothesis implies that DNA lesions affect APC activation independently of the known components of the DNA checkpoint, so it appears more logical to presume that the spindle assembly checkpoint is activated as a result of DNA lesions. This assumption is also in agreement with observations made for mammalian and in Drosophila cells (40, 49).
Reciprocally, several studies have demonstrated strong connections between the DNA checkpoint and the spindle apparatus. In syncytial Drosophila embryos, DmChk2 (the Rad53 homolog in Drosophila melanogaster) localizes to centrosomes and spindle microtubules. Furthermore, localization of DmChk2 to these structures increases upon DNA damage and a DmChk2-dependent pathway disrupts spindle assembly and chromosome segregation in response to DNA lesions, eliminating defective nuclei from the embryonic precursor pool (61). Similarly, subpopulations of Chk2 phosphorylated at Thr-68 and Thr-26 or Ser-28 localize to centrosomes throughout the cell cycle and to midbodies in human cells in the absence of DNA damage and independently of ATM, the human homolog of Mec1/Tel1 (63). 53BP1, presumably a mammalian homolog of Rad9, localizes to kinetochores specifically in mitotic cells and is hyperphosphorylated during mitosis under conditions where the spindle checkpoint is activated (25). This observation also suggests cross talks between the DNA and the spindle checkpoint in mammalian cells. ATM was shown to be involved in centrosome amplification induced by DNA damage in chicken DT40 cells (12). This mechanism was interpreted as promoting mitotic catastrophe and ensuring the death of cells evading the DNA checkpoint. Finally, Krishnan and collaborators have demonstrated that in budding yeast, Rad53- and Mec1-dependent pathways directly modulate spindle dynamics in response to stalled replication forks by controlling the levels of microtubule-associated proteins (29). These examples show that in many cases, proteins of the DNA checkpoint colocalize with or can act on spindle components.
Both the DNA and the spindle assembly checkpoints thus seem able to detect DNA damage and replication defects by monitoring distinct parameters (the presence of pathological DNA structures and the absence of bipolar attachment, respectively), both checkpoints, when activated, block the metaphase/anaphase transition and the mitotic exit, and both checkpoints have physiological connections with the spindle apparatus. Given these common features, interregulations of the DNA and the spindle assembly checkpoint were conceivable. Here, we show that the activation of the spindle assembly checkpoint induces changes in Rad53 and Rad9 phosphorylation states. These modifications are independent of all the DNA checkpoint proteins that we tested and are abolished by the deletion of MAD2, which suggests that Rad53 and Rad9 could perform specific functions in the spindle damage response. We tested whether the influence of the spindle assembly checkpoint on the phosphorylation states of Rad9 and Rad53 could play a part in the DNA damage response, and we found that in cases of DNA damage, spindle checkpoint activation modifies the phosphorylation of Rad9 and significantly accelerates the inactivation of Rad53.
MATERIALS AND METHODS
Strains, plasmids, and media.
Strains used in this study are listed in Table 1. All strains (except MCM336 and L208) are congenic with YPH499 (54). Standard molecular biology and molecular genetic techniques, such as gene disruption, tetrad dissection, and PCR-based tagging or disruption of chromosomal genes, were used to construct the strains. PCR-based genotyping was used to confirm gene disruption. The mps1-1 strain L300 was obtained by crossing L125 (as YPH500, bar1Δ::LEU2 RAD9-myc::his5+) with strain 290 (MATa ura3-52 his3-Δ200 trp1-Δ1 mps1-1), kindly provided by Mark Winey. Further details are available upon request.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| MCM001 (YPH499) | MATaura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 | 54 |
| MCM181 | rad17Δ::kanMXa | This study |
| MCM182 | mec1Δ::HIS3 + pBAD70 (2μm TRP1 RNR1)a | This study |
| MCM185 | bar1Δ::LEU2a | 32 |
| MCM215 | bar1Δ::LEU2 rad9Δ::kanMXa | 32 |
| MCM275 | rad17Δ::kanMX rad9Δ::KlURA3a | This study |
| MCM403 | bar1Δ::LEU2 RAD9-myc::his5+a | This study |
| L48 | bar1Δ::LEU2 mad2Δ::kanMXa | This study |
| L183 | bar1Δ::LEU2 mad2Δ::his5+a | This study |
| L78 | bar1Δ::LEU2 RAD9-myc::his5+mad2Δ::kanMXa | This study |
| L108 | bar1Δ::LEU2 RAD9-myc::his5+bub2Δ::kanMXa | This study |
| L138 | Matα bar1Δ::LEU2 RAD9-myc::his5+mec1Δ::HIS3 tel1Δ::KlURA3 + pBAD70 (2μm TRP1 RNR1)a | This study |
| L140 | bar1Δ::LEU2 RAD9-myc::his5+mec1Δ::HIS3 tel1Δ::KlURA3 + pBAD70 (2μm TRP1 RNR1)a | This study |
| L141 | bar1Δ::LEU2 RAD9-myc::his5+mec1Δ::HIS3 + pBAD70 (2μm TRP1 RNR1)a | This study |
| L145 | bar1Δ::LEU2 rad53Δ::kanMX RAD9-myc::his5+ + pBAD70 (2μm TRP1 RNR1)a | This study |
| L149 | bar1Δ::LEU2 RAD9-myc::his5+tel1Δ::KlURA3a | This study |
| L153 | bar1Δ::LEU2 rad53Δ::kanMX + pBAD70 (2μm TRP1 RNR1)a | This study |
| L305 | bar1Δ::LEU2 rad53Δ::kanMX + pBAD70 (2μm TRP1 RNR1)a | This study |
| L157 | bar1Δ::LEU2 rad9Δ::kanMXa | This study |
| L256 | bar1Δ::LEU2 rad9Δ::kanMXa | This study |
| L199 | bar1Δ::LEU2 pds1Δ::KlURA3a | This study |
| L241 | bar1Δ::LEU2 bub1Δ::kanMXa | This study |
| L281 | bar1Δ::LEU2 ptc2Δ::KlURA3 ptc3Δ::kanMXa | This study |
| L286 | bar1Δ::LEU2 mec1Δ::HIS3 tel1Δ::KlURA3 sml1Δ::kanMXa | This study |
| L289 | bar1Δ::LEU2 pph3Δ::KlURA3a | This study |
| L300 | mps1-1 bar1Δ::TRP1 RAD9-myc::his5+ | This study |
| L208 | MATacan1-100 ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 bar1Δ::TRP1 | This study |
| MCM336 (JKM139) | MATahoΔ hmlΔ::ADE1 hmrΔ::ADE1 ade3::pGAL-HO lys5 leu2-3,112 ura3-52 trp1::hisG | 30 |
As YPH499.
p416GAL1/MPS1, a URA3-marked, centromeric plasmid harboring the MPS1 gene under the control of a galactose-inducible promoter, was constructed by cloning the PCR-amplified coding sequence of MPS1 into the XmaI and XhoI sites of the p416GAL1 vector (42). pBAD70 is a generous gift from Steve Elledge and is described in reference 11. The pOC57-HA plasmid (CEN URA3 pGAL1-pds1-mdb-HA), described in reference 1, was kindly provided by Orna Cohen-Fix.
Yeast cells were grown in YP medium (1% yeast extract, 2% Bacto peptone) supplemented with 2% glucose unless indicated otherwise. For the overexpression of MPS1 and pds1-mdb, cells were grown in selective medium containing 2% raffinose or in YP medium supplemented with 2% galactose.
Other techniques.
Nocodazole and α-factor were used at final concentrations of 15 μg/ml and 0.5 μM, respectively. Analysis of Rad53 and Rad9 phosphorylation was performed as described previously (31, 37), except that gels were run for a longer time in order to clearly observe Rad53 nocodazole-induced modification. The in situ Rad53 autophosphorylation assay was carried out as described in reference 46.
For flow cytometric DNA analysis, cells were fixed in 70% ethanol-30% 1 M sorbitol. Cells were then washed in phosphate-buffered saline (PBS; Sigma), incubated with 0.25 mg/ml RNase for 1 hour at 50°C, washed again, and resuspended in PBS containing 50 μg/ml propidium iodide. Cells were diluted 1:10 into PBS, and DNA analysis was performed on a Becton Dickinson FACScan.
For the Rad53 dephosphorylation assay, total protein extracts (about 20 μg proteins) were incubated at 37°C for 1 hour in the presence of 10 units of calf intestine phosphatase (Biolabs) according to the manufacturer's instructions and in the presence or absence of a set of phosphatase inhibitors (1 mM sodium orthovanadate, 1.5 mM p-nitrophenyl phosphate, 5 mM beta-glycerophosphate).
The amount of cyclobutane pyrimidine dimers after UV irradiation was assessed as follows. Genomic DNA (0.2 μg) was slot blotted onto a nylon transfer membrane (Hybond N+; Amersham). Thymidine dimers were detected with the TDM-2 monoclonal antibody (MBL) as the primary antibody and with anti-mouse antibody (Promega) as the secondary antibody. The blot was revealed by chemiluminescence (Amersham).
RESULTS
Activation of the spindle assembly checkpoint triggers changes in the phosphorylation states of Rad53 and Rad9 that do not occur during an unperturbed cell cycle.
The core components of the budding yeast DNA checkpoint include the phosphatidylinositol 3-kinase-related protein kinases Mec1 and Tel1 and two central transducers, the Chk1 and Rad53 kinases, that phosphorylate downstream effectors. Chk1 and Rad53 themselves get phosphorylated upon activation of the DNA checkpoint, and their phosphorylation states, detected as electrophoretic mobility shifts, are commonly used as markers to monitor the activation of this transduction pathway. The phosphorylations of Chk1 and Rad53 depend on Mec1 and Tel1 and also on so-called “adaptors,” Rad9 in cases of DNA damage and Mrc1 in cases of replication blocks and DNA lesions during S phase. When DNA damage occurs, Rad9 also becomes highly phosphorylated on putative Mec1 consensus sites (52, 59, 65).
In order to reveal a potential influence of the spindle assembly checkpoint on the DNA checkpoint, we analyzed the electrophoretic behaviors of Rad53, Chk1, and Rad9 upon nocodazole treatment. Nocodazole is a microtubule-depolymerizing chemical that binds to β-tubulin with high affinity. Nocodazole treatment activates the spindle assembly checkpoint but has not been reported to have any DNA-damaging effect. As shown in Fig. 1A, nocodazole treatment of exponentially growing cells led to modifications in the electrophoretic mobilities of Rad53 and Rad9. These modifications became visible after 1 hour of nocodazole treatment, at the time when cells were mostly synchronized in G2/M phase. Rad53 then appeared as a doublet while Rad9 forms were completely shifted upwards. In contrast, Chk1 migration seemed unaltered by nocodazole treatment (data not shown), which does not rule out the possibility that some nocodazole-induced modifications of Chk1 could be undetected by our electrophoretic assay.
FIG. 1.
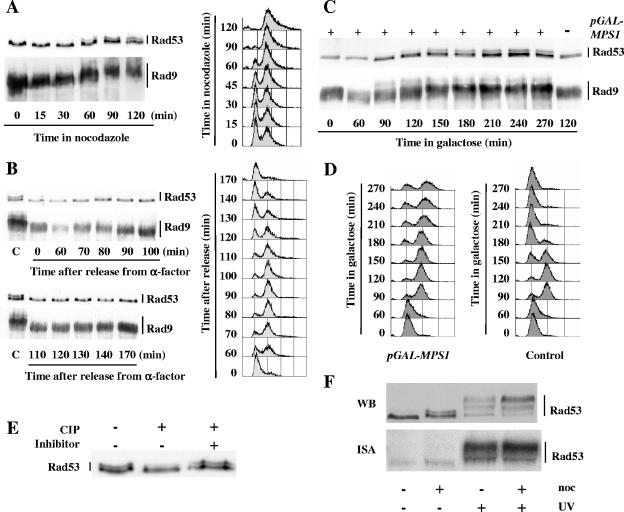
Activation of the spindle assembly checkpoint triggers changes in the phosphorylation states of Rad53 and Rad9. (A) Nocodazole (15 μg/ml) was added at time zero to an asynchronous culture of wild-type cells containing a RAD9::myc allele (MCM403). Aliquots were taken at the indicated times after addition of nocodazole and analyzed by FACS and by Western blotting using anti-Rad53 and anti-myc antibodies. (B) Exponentially growing wild-type cells harboring a RAD9::myc allele (MCM403) were synchronized in G1 phase with α-factor, washed thrice, and released into fresh medium. When the majority of the cells had rebudded after release, α-factor was added back to the culture to prevent a second cell cycle. Samples were collected at the indicated times after release from the α-factor block and analyzed as described above by FACS and by Western blotting. Lanes C, extracts from MCM403 cells after a 2-hour nocodazole treatment. (C and D) Wild-type cells harboring a RAD9::myc allele (MCM403) and containing either the empty vector p416GAL1 (Control) or the p416GAL1/MPS1 plasmid expressing MPS1 under the control of a galactose-inducible promoter were grown to the exponential phase in raffinose medium, synchronized in G1 with α-factor, washed thrice, and resuspended at time zero in galactose medium to induce MPS1 expression. At 135 min, α-factor was added back to the culture to prevent extra cell cycling. Aliquots were taken at the indicated times after galactose induction and analyzed by Western blotting (C) and by FACS (D) as described above. (E) Western blot analysis of total protein extracts from nocodazole-treated wild-type cells (MCM336) incubated in the presence (+) or absence (−) of calf intestine phosphatase (CIP) and in the presence (+) or absence (−) of phosphatase inhibitors. (F) Wild-type cells (MCM185) either treated with nocodazole (noc; 15 μg/ml) for 90 min or left untreated were either immediately collected or UV irradiated (40 J/m2) and subsequently harvested. Whole-cell extracts were then analyzed by Western blotting (WB), and Rad53 autophosphorylation activity was assessed by an in situ renaturation assay (ISA).
A modification of Rad9 electrophoretic mobility upon nocodazole treatment has been reported by Vialard et al., who have demonstrated that it corresponds to a phosphorylation change (65). This phosphorylated state of Rad9 was considered (without further examination) a G2/M phase-specific alteration, independent of the nocodazole treatment that was then used to synchronize the cells (65). In contrast, when we analyzed α-factor-synchronized cells progressing through an entire cycle, we found that Rad9 and Rad53 nocodazole-induced modifications do not occur during an unperturbed cell cycle (Fig. 1B).
In order to exclude the possibility of previously unnoticed DNA-damaging effects of nocodazole, we artificially activated the spindle assembly checkpoint by overexpressing Mps1, an essential kinase required for spindle function (20, 70). Wild-type cells were grown overnight on raffinose medium and synchronized with α-factor, and Mps1 overexpression was induced from a pGAL-MPS1 construct by addition of galactose. As shown in Fig. 1C and D, electrophoretic mobility changes for Rad53 and Rad9 similar to the ones observed after nocodazole treatment appeared about 2 h after galactose addition, at the time when most cells were in G2/M.
We investigated the nature and activity of the modified form of Rad53 induced by nocodazole treatment. Incubating protein extract from nocodazole-treated wild-type cells with calf intestine phosphatase resulted in the disappearance of the slower-migrating form of Rad53, establishing that it corresponds to a phosphorylation variant (Fig. 1E). Upon DNA damage, a large proportion of Rad53 phosphorylation changes are due to the autophosphorylation activity of Rad53 (46). In contrast, an in situ assay revealed similar, low autophosphorylation activities for Rad53 with or without nocodazole treatment (Fig. 1F). Figure 1F also illustrates that the modification of Rad53 induced by nocodazole treatment is quite different from the ones observed after DNA damage. Vialard and collaborators have also reported differences in Rad9 phosphorylations resulting from nocodazole treatment or UV irradiation (65).
Nocodazole-induced phosphorylations of Rad53 and Rad9 are independent of other components of the DNA checkpoints.
Rad17, Rad9, Tel1, and the Mec1/Ddc2 complex are upstream components of the DNA checkpoint that are required for maximal phosphorylation of Rad53 after DNA damage. Mec1/Ddc2 has a predominant function in the S. cerevisiae DNA checkpoint and has been proposed to detect processed DNA lesions via an interaction with replication protein A-coated single-strand DNA (71). Tel1 plays only a minor part in the budding yeast DNA checkpoint, and its functions, mostly redundant with those of Mec1/Ddc2, appear only in the absence of Mec1 (7, 51, 64). Rad17 is part of the Rad17/Mec3/Ddc1 complex, which presents structural similarities to PCNA and is loaded onto DNA double-strand breaks (DSBs), and presumably other kinds of DNA lesions, independently of Mec1 (for a review, see reference 39). It is generally assumed that the Rad17/Mec3/Ddc1 complex is involved in the activation of Mec1 at the sites of DNA lesions. As previously stated, Rad9 is an activator of Rad53. Upon DNA damage, Rad9 becomes highly phosphorylated in a Mec1/Tel1-dependent way, which triggers its binding to Rad53 via Rad53 FHA (forkhead-associated) domains (52, 59, 65). The resulting increase in the local Rad53 concentration on the Rad9 surface has been proposed to induce trans autophosphorylation and catalytic activation of Rad53 (17). Sweeney and collaborators also recently proposed that binding of Rad53 to Rad9 promotes recruitment of Rad53 to DNA lesions and its direct phosphorylation by Mec1, which would contribute to Rad53 activation and subsequent autophosphorylation (60).
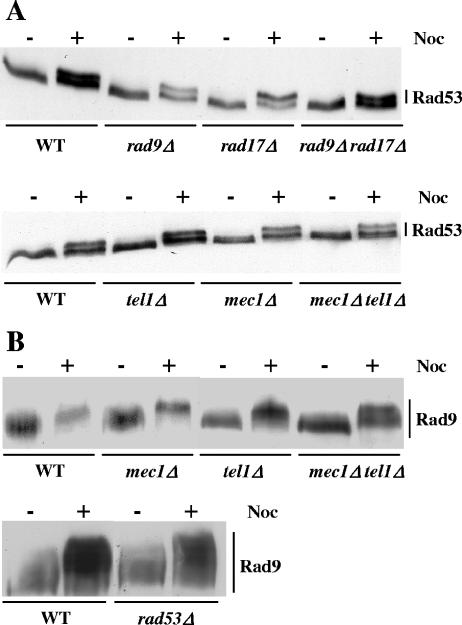
Whereas the absence of RAD17, RAD9, or MEC1 severely affects Rad53 phosphorylation after DNA damage (see, for example, reference 65), the phosphorylation of Rad53 after a 2-hour nocodazole treatment was similar in wild-type cells and in rad9Δ, rad17Δ, rad9Δ rad17Δ, mec1Δ, tel1Δ, and mec1Δ tel1Δ mutants (Fig. 2A). The effector kinase Chk1 functions in parallel with Rad53 and does not influence Rad53 phosphorylation upon DNA damage (50). We found that the nocodazole-induced phosphorylation of Rad53 was also independent of CHK1 (data not shown). Since Rad53 phosphorylation upon DNA damage is completely abolished in mec1Δ tel1Δ cells, we analyzed more thoroughly nocodazole-induced Rad53 phosphorylation in these cells. Exponentially growing wild-type and mec1Δ tel1Δ cells were synchronized in G1 with α-factor and subsequently released into nocodazole-containing medium. As shown in Fig. 3A, Rad53 phosphorylation appeared in wild-type cells about 90 min after release from α-factor arrest and was maintained until the end of the experiment. We confirmed that Rad53 phosphorylation in mec1Δ tel1Δ mutants was comparable to that in the wild type.
FIG. 2.
Nocodazole-induced phosphorylations of Rad53 and Rad9 are independent of other components of the DNA checkpoints. (A) Asynchronous cultures of wild-type (WT) (MCM001) and rad9Δ (MCM215), rad17Δ (MCM181), rad9Δ rad17Δ (MCM275), mec1Δ (L141), tel1Δ (L149), and mec1Δ tel1Δ (L140) mutant strains were collected before (−) or after (+) a 2-h treatment with nocodazole (Noc; 15 μg/ml). Total protein extracts were analyzed by Western blotting using anti-Rad53 antibodies. (B) Asynchronous cultures of wild-type (MCM403) and mec1Δ (L141), tel1Δ (L149), mec1Δ tel1Δ (L138), and rad53Δ (L145) mutant strains harboring the RAD9::myc allele were harvested before (−) or after (+) a 2-h treatment with nocodazole (Noc; 15 μg/ml). Total protein extracts were analyzed by Western blotting using anti-myc antibodies.
FIG. 3.
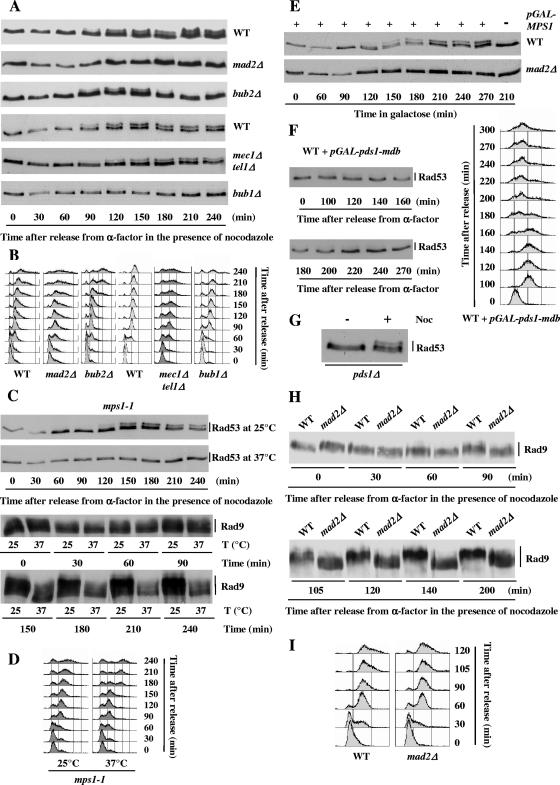
Nocodazole-induced phosphorylations of Rad53 and Rad9 depend on the components of the spindle assembly checkpoint. (A and B) Wild-type (WT) (MCM185) and mad2Δ (L78), bub1Δ (L241), bub2Δ (L108), and mec1Δ tel1Δ (L286) mutant cells were arrested in G1 with α-factor, washed thrice, and released into nocodazole-containing medium. Cell aliquots were taken at the indicated times after release from the α-factor block and analyzed by Western blotting using anti-Rad53 antibodies (A) and by FACS (B). (C and D) mps1-1 (L300) cells were synchronized in G1 with α-factor at the permissive temperature (25°C), washed thrice, and released into nocodazole-containing medium at either the permissive (25°C) or the restrictive (37°C) temperature. Cell aliquots were taken at the indicated times after release from the α-factor block and analyzed by Western blotting (C) and by FACS (D). (E) Wild-type (MCM403) and mad2Δ (L78) cells containing the p416GAL1/MPS1 plasmid expressing MPS1 under the control of a galactose-inducible promoter were grown to the exponential phase in raffinose medium, synchronized in G1 with α-factor, washed thrice, and resuspended at time zero in galactose medium to induce MPS1 expression. At 135 min, α-factor was added back to the culture to prevent extra cell cycling. Rad53 phosphorylation was analyzed by Western blotting using anti-Rad53 antibodies. (F) Wild-type (L208) cells containing the pOC57-HA plasmid expressing pds1-mdb under the control of a galactose-inducible promoter were grown to the exponential phase in raffinose medium, synchronized in G1 with α-factor, washed thrice, and resuspended at time zero in galactose medium to induce pds1-mdb expression. At 100 min, α-factor was added back to the culture to prevent extra cell cycling. Rad53 phosphorylation was analyzed by Western blotting using anti-Rad53 antibodies. (G) An asynchronous culture of pds1Δ (L199) cells was collected before (−) or after (+) a 3-h treatment with nocodazole (Noc; 15 μg/ml). Total protein extracts were analyzed by Western blotting using anti-Rad53 antibodies. (H and I) Wild-type (MCM403) and mad2Δ (L78) mutant cells carrying the RAD9::myc allele were arrested in G1 with α-factor, washed thrice, and released into nocodazole-containing medium. Samples were retrieved at the indicated times after release from the α-factor block and analyzed by Western blotting using anti-myc antibodies (H) and by FACS (I).
Upon DNA damage, Rad9 becomes phosphorylated in a Mec1/Tel1-dependent but Rad53-independent manner (65). In contrast, we found that the phosphorylation states of Rad9 after 2-hour nocodazole treatments were similar in wild-type cells and in mec1Δ, tel1Δ, and mec1Δ tel1Δ mutants (Fig. 2B).
In conclusion, we have shown that the phosphorylations of Rad53 and Rad9 after nocodazole treatment are independent of the major components of the DNA damage checkpoint. Interestingly, Rad9, although strongly phosphorylated, is not required for nocodazole-induced Rad53 modification, and Rad9 phosphorylation does not bring about a hyperphosphorylated state for Rad53 comparable to the one observed after DNA damage.
Nocodazole-induced phosphorylations of Rad53 and Rad9 depend on the components of the spindle assembly checkpoint.
Nocodazole-induced Rad53 phosphorylation in mutants affected in components of the spindle assembly checkpoint (mad2Δ and bub1Δ) or in the MEN pathway (bub2Δ) was analyzed as described above. Wild-type, mad2Δ, bub1Δ, and bub2Δ cells were synchronized with α-factor and released into nocodazole-containing medium. As shown in Fig. 3A, mad2Δ and bub1Δ cells, although momentarily arrested in G2/M by nocodazole (Fig. 3B), were completely defective in inducing Rad53 phosphorylation change. Nocodazole-induced Rad53 phosphorylation in bub2Δ mutants appeared at the same time as that in wild-type cells but was more transient and disappeared about 180 min after release from α-factor arrest, at the time when cells were escaping the mitotic arrest (Fig. 3B). The disappearance of phosphorylated forms of Rad53 was most probably due to exit from anaphase. Indeed, we found that the Rad53 phosphorylation state was unaffected when cdc15-2 mutants blocked in telophase at the restrictive temperature were treated with nocodazole (data not shown).
The spindle checkpoint kinase Mps1 could conceivably phosphorylate Rad53 upon nocodazole treatment. We thus tested whether nocodazole-induced Rad53 phosphorylation was dependent upon Mps1 functionality. mps1-1 cells were synchronized with α-factor at the permissive temperature and released into nocodazole-containing medium at the permissive (25°C) or restrictive (37°C) temperature. As shown in Fig. 3C, mps1-1 cells were completely defective in inducing Rad53 phosphorylation change at the restrictive temperature, although they were transiently delayed in G2/M (Fig. 3D). If Rad53 were a direct substrate of Mps1, one could expect that Rad53 phosphorylation induced by MPS1 overexpression should be independent of the other spindle checkpoint proteins. However, we found that mad2Δ cells were defective in inducing Rad53 phosphorylation change upon MPS1 overexpression (Fig. 3E). Although we cannot rule out the hypothesis that a Mad2-dependent modification of Mps1 could be required for Mps1 activity toward Rad53, this result suggests that Rad53 is not a direct substrate of Mps1.
We have thus shown that the Rad53 phosphorylation changes observed upon nocodazole treatment or Mps1 overexpression depend on spindle assembly checkpoint components but are independent of the MEN protein Bub2. To further test the hypothesis that Rad53 modification results directly from spindle checkpoint activation and is not the indirect consequence of a persistent cell cycle arrest, we verified that a spindle checkpoint-independent G2/M arrest induced by the overexpression of an indestructible form of Pds1, Pds1-mdb (9), did not bring about similar changes in Rad53 electrophoretic mobility (Fig. 3F). We also observed that Pds1 is not required for Rad53 nocodazole-induced modification (Fig. 3G). Taken together, our data indicate that Rad53 modification upon nocodazole treatment is strictly dependent on and specific to the activation of the spindle assembly checkpoint.
Similar experiments showed that, whereas Rad9 phosphorylation appeared in wild-type cells about 90 min after the cells were released from α-factor arrest into the nocodazole-containing medium and was maintained until the end of the experiment, mad2Δ cells were completely defective in inducing Rad9 phosphorylation in response to nocodazole (Fig. 3H and I). The fact that Rad53 and Rad9 are not modified upon nocodazole treatment in mad2Δ cells also confirmed our previous observations that the nocodazole-induced modifications of Rad53 and Rad9 do not occur during an unperturbed cell cycle, in the absence of spindle checkpoint activation (Fig. 1B).
The Pph3 phosphatase modulates Rad53 nocodazole-induced phosphorylation.
Three phosphatases have been shown to regulate the Rad53 phosphorylation state upon DNA damage. Leroy et al. have provided evidence that the PP2C phosphatases Ptc2 and Ptc3 bind the Rad53 FHA1 domain and directly dephosphorylate Rad53 after an HO-induced DSB (31). More recently, Keogh et al. have demonstrated that a three-protein complex including the Pph3 phosphatase dephosphorylates histone H2A, whose phosphorylation on Ser-129 is an early mark of DSB (27). They also showed that Pph3 indirectly affects Rad53 phosphorylation, as Rad53 inactivation following an HO-induced DSB is strongly delayed in pph3Δ cells in a way that is dependent on histone H2A Ser-129.
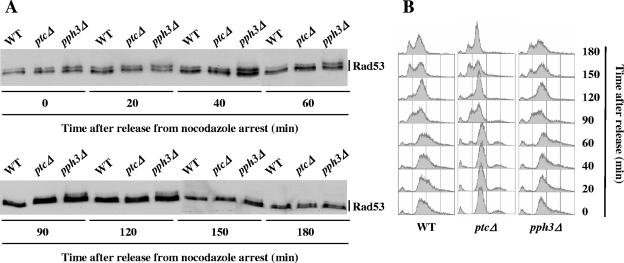
To test the potential regulation of Rad53 nocodazole-induced phosphorylation by Ptc2, Ptc3, and Pph3, we analyzed the reversion of Rad53 phosphorylation in wild-type, ptc2Δ ptc3Δ, and pph3Δ cells after nocodazole treatment. Wild-type and mutant cells were treated with nocodazole for 2.5 h and subsequently released from nocodazole arrest. We found that in wild-type and ptc2Δ ptc3Δ cells, nocodazole-induced Rad53 phosphorylation disappeared about 90 min after release but persisted, although diminished, in pph3Δ cells until the end of the experiment (Fig. 4A). Fluorescence-activated cell sorter (FACS) analysis showed that the wild-type, ptc2Δ ptc3Δ, and pph3Δ cells exited G2/M arrest with similar kinetics, ruling out the possibility of cell cycle effects on Rad53 phosphorylation (Fig. 4B). Since, to our knowledge, nocodazole treatment has never been described as inducing histone H2A phosphorylation, our data suggest that Rad53 might be a direct substrate of Pph3 upon spindle checkpoint activation.
FIG. 4.
Rad53 dephosphorylation upon nocodazole treatment is delayed in pph3Δ cells. Asynchronous cultures of wild-type (WT) (MCM185) and ptc2Δ ptc3Δ (ptcΔ, L281) and pph3Δ (L289) mutant strains were treated with nocodazole for 2.5 h and subsequently released from nocodazole arrest at time zero. Aliquots were harvested at the indicated time points and analyzed by Western blotting using anti-Rad53 antibodies (A) and by FACS (B).
Rad53 inactivation after DNA damage is accelerated when the spindle checkpoint is activated.
Having established that the activation of the spindle assembly checkpoint can modify the phosphorylation of a subset of DNA checkpoint proteins, we looked for the physiological functions of these modifications. We envisioned two hypotheses: (i) since the spindle assembly checkpoint seems to respond to some kinds of DNA damage (either directly affecting the DNA centromeric sequence or indirectly resulting in the lack of centromeric sequence replication), its activation under genotoxic conditions could represent an extra way to monitor the extent of DNA damage and to finely tune the DNA damage response by modifying the phosphorylation states of Rad9 and Rad53, and (ii) Rad53 and Rad9 could be involved in some aspect of the spindle damage response.
In order to explore the first hypothesis, asynchronous cultures of wild-type and mad2Δ cells were synchronized in G1 by α-factor treatment, released into S phase, and submitted to UV irradiation when cells were in G2/M. We found no reproducible difference between the kinetics of Rad53 and Rad9 phosphorylation in wild-type and in mad2Δ cells in response to UV irradiation (Fig. 5A), confirming previous observations on UV-irradiated, synchronized wild-type and mad2Δ cells (7). The kinds of DNA lesions that activate the spindle checkpoint in S. cerevisiae are not well known, so in order to get the spindle checkpoint unequivocally activated in the presence of DNA damage, we pretreated wild-type and mad2Δ cells with nocodazole. Nocodazole-treated wild-type and mad2Δ cells were thus UV irradiated and further grown in nocodazole-containing medium. As shown in Fig. 5B, the extents of Rad53 phosphorylation were similar in wild-type and in mad2Δ cells during the first 40 min following UV irradiation. However, after 60 min, the amount of phosphorylated forms of Rad53 decreased more rapidly in wild-type than in mad2Δ cells, whereas FACS analysis indicates that in both strains, the majority of the cells were blocked in G2/M (Fig. 5C). As expected, weaker Rad53 phosphorylation in wild-type cells was associated with a decrease in Rad53 autophosphorylation activity (data not shown). In contrast to that of Rad53, Rad9 hyperphosphorylation induced by UV irradiation was more intense in wild-type than in mad2Δ cells but disappeared in both strains around 90 min after irradiation (Fig. 5B).
FIG. 5.
Nocodazole treatment accelerates Rad53 inactivation after DNA damage. (A to D) Wild-type (WT) (MCM403) and mad2Δ (L78) mutant cells were arrested in G1 with α-factor, washed thrice, and released into fresh medium in the absence (untreated cells) or in the presence (nocodazole-treated cells) of nocodazole (15 μg/ml). After 90 min, both nocodazole-treated and untreated cells were synchronized in G2/M. They were then UV irradiated (40 J/m2) at time zero and further grown in fresh medium in the presence or absence of nocodazole as before. Aliquots were taken at the indicated times after UV irradiation and analyzed by Western blotting (A and B) and by FACS (C). The asterisks mark the presence of hyperphosphorylated Rad9 forms. For each time point, the extent of DNA repair was monitored by immunoblotting genomic DNA with the TDM-2 antibody against cyclobutane pyrimidine dimers (D). (E and F) Wild-type (MCM185) and mad2Δ (L48) mutant cells were arrested in G1 with α-factor, washed thrice, and released into fresh medium in the presence of nocodazole. After 2 h of nocodazole treatment, when the cells were synchronized in G2/M, phleomycin (10 μg/ml) was added to the culture (time zero). At 60 min, the cells were washed again and resuspended in fresh medium with nocodazole but without phleomycin. Samples were analyzed by Western blotting using anti-Rad53 antibodies (E) and by FACS (F).
In order to exclude the possibility that the slower inactivation of Rad53 in mad2Δ cells upon nocodazole treatment could be due to a defect in DNA repair, we verified that the extents and the kinetics of DNA repair were similar in wild-type and in mad2Δ cells by using an antibody against cyclobutane pyrimidine dimers (Fig. 5D). Nocodazole treatment thus accelerated Rad53 inactivation after UV irradiation in wild-type cells compared to that in mad2Δ mutants, although both types of cells were blocked in mitosis, with comparable amounts of DNA lesions. We performed similar experiments using Mps1 overexpression to activate the spindle checkpoint prior to UV irradiation, and we also observed an acceleration of Rad53 inactivation in wild-type cells compared to that in mad2Δ mutants (data not shown).
In order to test whether the effect of the spindle checkpoint activation on Rad53 phosphorylation was specific to UV-induced DNA lesions, we performed the same analyses using phleomycin, a radiomimetic drug that produces DNA double-strand breaks. Wild-type and mad2Δ cells were synchronized with α-factor and released into nocodazole-containing medium and, when synchronized in G2/M, were treated with phleomycin for 1 hour (phleomycin was added at time zero). As shown in Fig. 5E, Rad53 phosphorylation levels were comparable in wild-type and mad2Δ cells during phleomycin treatment. However, removal of phleomycin at 60 min led to a rapid decrease in phosphorylated forms of Rad53 in wild-type cells, whereas Rad53 phosphorylation was maintained in mad2Δ mutants until the end of the experiment. These results are less conclusive than those for the UV experiment because the mad2Δ cells did not keep the same degree of synchronization as the wild-type cells (Fig. 5F). However, they suggest that activation of the spindle assembly checkpoint accelerates the inactivation of Rad53 following different kinds of DNA damage.
We tested the influence of the Ptc2, Ptc3, and Pph3 phosphatases on Rad53 dephosphorylation under conditions of simultaneous DNA and spindle checkpoint activation. Wild-type, mad2Δ, ptc2Δ ptc3Δ, and pph3Δ cells were pretreated with nocodazole and UV irradiated as described above. As shown in Fig. 6A, the deletion of PTC2 and PTC3 had no visible effects on Rad53 dephosphorylation under these conditions. In contrast, Rad53 dephosphorylation was significantly delayed in pph3Δ cells (Fig. 6C). However, a delay in Rad53 dephosphorylation was also observed in pph3Δ cells that were synchronized with α-factor treatment, released, and UV irradiated in G2/M without nocodazole treatment (Fig. 6E). The absence of Pph3 leads to a sustained phosphorylation of histone H2A following DSBs, which was shown to induce a persistent phosphorylation of Rad53 (27). Since UV irradiation was demonstrated to trigger the phosphorylation of histone H2A in mammal cells (18, 35), the simplest interpretation of our data is that the deletion of PPH3 indirectly modifies Rad53 phosphorylation with or without nocodazole treatment, via its effect on histone H2A phosphorylation.
FIG. 6.
PPH3 deletion strongly affects Rad53 dephosphorylation after UV irradiation. (A to D) Asynchronous cultures of wild-type (WT) (MCM185), mad2Δ (L48), ptc2Δ ptc3Δ (ptcΔ, L281), and pph3Δ (L289) cells were synchronized in G1 with α-factor, washed thrice, and released into fresh medium in the presence of nocodazole. After 90 min, G2/M-arrested cells were UV irradiated (40 J/m2) at time zero and further grown in fresh medium in the presence of nocodazole. Aliquots were taken at the indicated times after UV irradiation and analyzed by Western blotting using anti-Rad53 antibodies (A and C) and by FACS (B and D). (E and F) Asynchronous cultures of wild-type (MCM185), mad2Δ (L48), and pph3Δ (L289) cells were synchronized in G1 with α-factor, washed thrice, and released into fresh medium (without nocodazole). After 90 min, the G2/M-synchronized cells were UV irradiated (40 J/m2) at time zero. Samples were retrieved at the indicated times after UV irradiation and analyzed by Western blotting using anti-Rad53 antibodies (E) and by FACS (F).
We have shown that Rad53 inactivation after DNA damage is accelerated when the spindle checkpoint is activated. One physiological interpretation of this effect is that, since Rad53 hyperactivation is deleterious (29, 37), an accelerated inactivation of Rad53 could be advantageous if G2/M arrest can be maintained in a redundant way by the spindle assembly checkpoint. If this were the case, we could expect nocodazole treatment to induce a Rad9-, Rad53-, and Mad2-dependent increase in UV resistance. RAD53 being essential, we analyzed rad53Δ cells in which the lethality of RAD53 deletion was suppressed by overexpressing RNR1 (which encodes the largest subunit of ribonucleotide reductase) (11). Asynchronous cultures of wild-type, rad9Δ, rad53Δ, and mad2Δ cells were synchronized in G1 with α-factor, released into fresh medium in the presence or in the absence of nocodazole, and UV irradiated when cells were in G2/M. The results for a representative experiment are shown in Table 2, with the number of surviving colonies after UV irradiation normalized by the number of surviving colonies in the absence of irradiation. Surprisingly, nocodazole treatment improved cell viability after UV irradiation in wild-type cells as well as in DNA checkpoint- and in spindle checkpoint-defective cells. The causes of this nocodazole effect on UV resistance were not investigated further, but no potential effect of the accelerated inactivation of Rad53 on cell viability after DNA damage could be demonstrated under these conditions.
TABLE 2.
Nocodazole treatment improves UV resistance of wild-type, DNA checkpoint-, and spindle checkpoint-deficient cellsa
| Strain | Presence of nocodazole | Cell viability (%) after indicated UV irradiation (J/m2)
|
||
|---|---|---|---|---|
| 0 | 120 | 120 (normalized) | ||
| Wild type | − | 97.5 ± 0.7 | 47.0 ± 0.0 | 48.2 ± 0.3 |
| + | 96.5 ± 0.7 | 85.2 ± 4.7 | 88.3 ± 5.6 | |
| mad2Δ | − | 97.5 ± 0.7 | 38.4 ± 0.8 | 39.4 ± 1.2 |
| + | 48.5 ± 2.7 | 35.8 ± 6.4 | 73.6 ± 9.0 | |
| rad9Δ | − | 96.0 ± 1.4 | 0.9 ± 0.6 | 1.0 ± 0.6 |
| + | 97.5 ± 0.7 | 4.8 ± 0.8 | 4.9 ± 0.9 | |
| rad53Δ | − | 79.1 ± 3.8 | 1.0 ± 0.0 | 1.3 ± 0.1 |
| + | 77.6 ± 0.4 | 4.8 ± 2.7 | 6.2 ± 3.4 | |
Two independent cultures of wild-type (MCM185), mad2Δ (L48 and L183), rad9Δ (L157 and L256), and rad53Δ (L153 and L305) strains were synchronized with α-factor, released into fresh medium in the presence (+) or in the absence (−) of nocodazole, plated, and either UV irradiated when cells were in G2/M or left untreated. Plates were incubated at 30°C, and the percentages of surviving colonies (means ± standard deviations) were determined after 24 h (200 colonies were examined for each point). In order to take into account UV irradiation-unrelated effects on cell viability, the percentages of surviving colonies after UV irradiation were divided by the proportions of surviving colonies in the absence of UV irradiation [in the “120 (normalized)” column].
rad9Δ and rad53Δ mutants exhibit wild-type viabilities under conditions of spindle dysfunction.
In order to investigate whether Rad53 or Rad9 could be involved in the spindle damage response, we looked for spindle-damaging conditions to which rad9Δ or rad53Δ mutants would be hypersensitive or hyperresistant. We tested the sensitivities of rad9Δ, rad53Δ, and rad9Δ rad53Δ mutants to benomyl (another microtubule-depolymerizing drug) or to the overexpression of Mps1, which is highly deleterious to mad1 and mad2 mutants (20). Neither rad9Δ, rad53Δ, nor rad9Δ rad53Δ cells showed any specific hypersensitivity to these stresses compared to wild-type cells (data not shown). Moreover, we found that upon nocodazole treatment, Pds1 was similarly stabilized in wild-type and in rad9Δ cells (data not shown). We also analyzed cbf1Δ and gim3Δ mutants that are affected in different spindle components and are hypersensitive to microtubule-depolymerizing drugs (19). CBF1 encodes a kinetochore protein that binds to centromeric DNA (6), whereas GIM3 codes for a subunit of the prefoldin complex that promotes the formation of functional tubulins (16). We found that the deletion of RAD9 or RAD53 had no reproducible effect on the viability of the cbf1Δ or gim3Δ mutant in the presence or in the absence of benomyl (data not shown).
Since Chk2 and 53BP1, the mammalian homologs of Rad53 and Rad9, have been shown to localize to centrosomes (63) and to kinetochores (25), respectively, we analyzed the localizations of Rad53-green fluorescent protein (GFP) and Rad9-GFP fusion proteins after nocodazole treatment. We observed diffuse nuclear localizations for both Rad53-GFP and Rad9-GFP in exponentially growing and in nocodazole-treated cells (consistent with previously reported results [22]), without any evidence for Rad53-GFP or Rad9-GFP accumulation at the spindle pole bodies or at the kinetochores (data not shown).
DISCUSSION
The activation of the spindle assembly checkpoint modulates Rad53 and Rad9 phosphorylation states.
Our results have established that the activation of the spindle assembly checkpoint alters the phosphorylation states of two components of the DNA checkpoint, Rad53 and Rad9, in the absence and in the presence of DNA damage. In the absence of DNA damage, Rad53 and Rad9 modifications induced by nocodazole treatment or Mps1 overexpression are independent of all the DNA checkpoint components that we tested (Fig. 2) but are abolished in spindle checkpoint-defective mutants (Fig. 3). Remarkably, Rad9, although highly phosphorylated, has no influence on Rad53 phosphorylation, which indicates that the physiological relations between Rad53 and Rad9 in this case are quite different from those observed upon DNA lesions. The nocodazole-induced phosphorylation of Rad53 is dependent on the spindle checkpoint components Mad1, Mad2, Bub1, and Mps1 but is independent of spindle checkpoint downstream targets, like the securin Pds1 or the MEN protein Bub2 (Fig. 3 and data not shown). Moreover, the observation that the Rad53 phosphorylation state is unaffected by the overexpression of an indestructible form of Pds1 that induces a metaphase/anaphase block further supports the assumption that Rad53 and Rad9 modifications result directly from spindle checkpoint activation.
We have found evidence suggesting that Rad53 might be a substrate of the Pph3 phosphatase upon spindle checkpoint activation, but the identity of the kinase(s) that phosphorylates Rad53 and Rad9 under these conditions remains undefined. Two protein kinases are part of the S. cerevisiae spindle assembly checkpoint: Mps1 and Bub1. We have shown that Mps1 functionality is required for Rad53 modification upon nocodazole treatment and that overexpression of Mps1 induces a modification of Rad53 similar to the one observed after nocodazole treatment (Fig. 1C and 3C and D). However, Rad53 modification induced by MPS1 overexpression depends on Mad2 (Fig. 3E), which suggests that Rad53 may not be a direct substrate of Mps1. Moreover, coimmunoprecipitation experiments failed to demonstrate any specific interaction between Rad53 and Mps1 or between Rad53 and Bub1 (data not shown), so the question has not been settled.
We have shown that the activation of the spindle checkpoint also modulates Rad53 phosphorylation in cases of DNA damage. This observation can be compared with recent data indicating that TTK/hMps1, the human homolog of yeast Mps1, contributes to the activation of Chk2, the human homolog of Rad53, upon DNA damage (68). hMps1 phosphorylates Chk2 on Thr-68 in vitro, and inhibition of hMps1 activity in vivo results in reduced Chk2 Thr-68 phosphorylation accompanied by a defective cell cycle arrest and a decreased checkpoint response to DNA damage. However, inactivating the spindle assembly checkpoint by transfection of small interfering RNA targeting hMad2 had no effect on Chk2 Thr-68 phosphorylation or on the DNA checkpoint response, which indicates that hMps1 participates in the regulation of the DNA checkpoint independently of its functions in the spindle checkpoint. Our results thus differ from those reported by Wei et al. (68) in that (i) the phosphorylation state of Rad53 is modified both in the absence and in the presence of DNA damage and (ii) this modification is dependent on the spindle assembly checkpoint as a whole and not on just one of its components.
Our observations both hint at some functions for Rad53 and Rad9 in the spindle damage response and suggest an influence of the spindle checkpoint on DNA checkpoint inactivation, two issues that we will address sequentially.
Potential implication of Rad53 and Rad9 in the spindle damage response.
The phosphorylations of Rad53 and Rad9 induced by the activation of the spindle checkpoint could indicate that Rad53 and Rad9 are involved in the spindle damage response and perform biochemical functions either identical to or different from the ones that they carry out upon DNA damage. If Rad53 and Rad9 were effectors of the spindle checkpoint, they should be required under some circumstances for an optimal response to spindle dysfunction and RAD53 and RAD9 deletions should affect cell viability upon spindle damage. However, we and others (e.g., references 23, 38, and 45) have found little evidence for it. This could mean that the parts played by Rad9 and Rad53 become limiting for cell viability only in very specific cases. Alternatively, Rad53 and Rad9 could represent so-far-unidentified targets of the spindle checkpoint. If this were the case, the implication of Rad9 and Rad53 in the spindle damage response might be demonstrated more readily by identifying and mutating the Rad9 or Rad53 sites that get phosphorylated following spindle checkpoint activation than by deleting RAD9 or RAD53. The fact that rad9 and rad53 sml1-1 mutants exhibit significantly higher rates of spontaneous chromosome loss than other DNA checkpoint mutants, like the dun1 and chk1 mutants (28), may be another indication of specific parts played by Rad53 and Rad9 in the spindle damage response. These functions could be conserved in mammal cells, which would explain the localizations of Chk2 and 53PB1 and the hyperphosphorylation of 53BP1 upon spindle damage (25, 61, 63).
Rad53 inactivation after DNA damage is accelerated when the spindle assembly checkpoint is activated.
As briefly reviewed in the introduction, many studies have shown that the spindle assembly checkpoint is involved in some DNA damage responses. The data regarding S. cerevisiae (7, 15, 36) have mostly established that Mad2-dependent pathways contribute to G2/M arrest upon DNA damage independently of the DNA checkpoint. These data also suggest that the spindle assembly checkpoint is activated by DNA lesions or replication defects.
Looking for interregulations of the DNA and the spindle checkpoints, we found that the activation of the spindle assembly checkpoint modifies the phosphorylation of both Rad53 and Rad9: Rad9 phosphorylation is more intense following UV irradiation, whereas Rad53 inactivation is accelerated (although the possibility of other effects on Rad53 activity and/or specificity cannot be excluded). Nocodazole treatment triggers several responses, including cell cycle arrest in mitosis, in spindle checkpoint-proficient cells. The accelerated inactivation of Rad53 after DNA damage in the presence of nocodazole could be due either to a direct influence of the spindle checkpoint on the DNA checkpoint or to an indirect effect of G2/M arrest on Rad53 activation. We verified that wild-type and mad2Δ cells exhibit similar kinetics of DNA repair and similar FACS profiles during the UV irradiation experiment (Fig. 5). Moreover, Clerici et al. demonstrated that the kinetics of Rad53 phosphorylation in UV-irradiated mec1Δ and mec1Δ mad2Δ cells were identical whereas mec1Δ cells remained blocked in G2/M and mec1Δ mad2Δ cells resumed their division cycle (7). Although we cannot completely rule out that Rad53 could be affected by secondary consequences of spindle checkpoint activation, our results suggest that the spindle checkpoint, when activated, could directly influence Rad53 inactivation after DNA damage.
Since Mad2-dependent pathways are presumed to be activated by some kinds of DNA lesions, the question as to why Mad2 has no effect on Rad53 inactivation in the absence of nocodazole arises. We propose that only a subset of DNA lesions is able to activate the spindle assembly checkpoint and that the DNA damage generated by UV irradiations in DNA checkpoint-proficient cells is not included in this subset. Another question concerns the physiological meaning of the influence of the spindle checkpoint on Rad53 inactivation after DNA damage. One possibility is that an accelerated inactivation of Rad53 could be beneficial if G2/M arrest can be maintained by the spindle assembly checkpoint. Although we could not demonstrate that nocodazole treatment induces a spindle checkpoint- and DNA checkpoint-dependent increase in UV resistance, we propose that upon some kinds of DNA damage, both the DNA checkpoint and the spindle checkpoint are activated and that the activation of the spindle checkpoint could contribute to adaptation to or recovery from DNA lesions.
As a conclusion, our study points to a direct regulation of some components of the DNA checkpoint (Rad53 and Rad9) by the spindle assembly checkpoint. This regulation suggests both specific functions for Rad53 and Rad9 in the spindle damage response and the implication of the spindle checkpoint in DNA checkpoint inactivation. Nocodazole is regularly used to synchronize cells in DNA checkpoint experiments, and our results should stand as a caveat for their interpretation.
Acknowledgments
We thank Mark Winey, David Lydall, Orna Cohen-Fix, and Steve Elledge for the generous gifts of plasmids and strains. We are grateful to the members of the MCMK laboratory, particularly to Anne Peyroche for discussions and to Emilie Ma for analyzing the localization of Rad53-GFP and Rad9-GFP.
This work was financed in part by the Association pour la Recherche sur le Cancer.
Footnotes
Published ahead of print on 23 October 2006.
REFERENCES
- 1.Agarwal, R., and O. Cohen-Fix. 2002. Phosphorylation of the mitotic regulator Pds1/securin by Cdc28 is required for efficient nuclear localization of Esp1/separase. Genes Dev. 16:1371-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agarwal, R., Z. Tang, H. Yu, and O. Cohen-Fix. 2003. Two distinct pathways for inhibiting Pds1 ubiquitination in response to DNA damage. J. Biol. Chem. 278:45027-45033. [DOI] [PubMed] [Google Scholar]
- 3.Alexandru, G., W. Zachariae, A. Schleiffer, and K. Nasmyth. 1999. Sister chromatid separation and chromosome re-duplication are regulated by different mechanisms in response to spindle damage. EMBO J. 18:2707-2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bardin, A. J., and A. Amon. 2001. Men and sin: what's the difference? Nat. Rev. Mol. Cell Biol. 2:815-826. [DOI] [PubMed] [Google Scholar]
- 5.Bardin, A. J., R. Visintin, and A. Amon. 2000. A mechanism for coupling exit from mitosis to partitioning of the nucleus. Cell 102:21-31. [DOI] [PubMed] [Google Scholar]
- 6.Cai, M., and R. W. Davis. 1990. Yeast centromere binding protein CBF1, of the helix-loop-helix protein family, is required for chromosome stability and methionine prototrophy. Cell 61:437-446. [DOI] [PubMed] [Google Scholar]
- 7.Clerici, M., V. Baldo, D. Mantiero, F. Lottersberger, G. Lucchini, and M. P. Longhese. 2004. A Tel1/MRX-dependent checkpoint inhibits the metaphase-to-anaphase transition after UV irradiation in the absence of Mec1. Mol. Cell. Biol. 24:10126-10144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cohen-Fix, O., and D. Koshland. 1999. Pds1p of budding yeast has dual roles: inhibition of anaphase initiation and regulation of mitotic exit. Genes Dev. 13:1950-1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cohen-Fix, O., J. M. Peters, M. W. Kirschner, and D. Koshland. 1996. Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes Dev. 10:3081-3093. [DOI] [PubMed] [Google Scholar]
- 10.Collura, A., J. Blaisonneau, G. Baldacci, and S. Francesconi. 2005. The fission yeast Crb2/Chk1 pathway coordinates the DNA damage and spindle checkpoint in response to replication stress induced by topoisomerase I inhibitor. Mol. Cell. Biol. 25:7889-7899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Desany, B. A., A. A. Alcasabas, J. B. Bachant, and S. J. Elledge. 1998. Recovery from DNA replicational stress is the essential function of the S-phase checkpoint pathway. Genes Dev. 12:2956-2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dodson, H., E. Bourke, L. J. Jeffers, P. Vagnarelli, E. Sonoda, S. Takeda, W. C. Earnshaw, A. Merdes, and C. Morrison. 2004. Centrosome amplification induced by DNA damage occurs during a prolonged G2 phase and involves ATM. EMBO J. 23:3864-3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fang, Y., T. Liu, X. Wang, Y. M. Yang, H. Deng, J. Kunicki, F. Traganos, Z. Darzynkiewicz, L. Lu, and W. Dai. 2006. BubR1 is involved in regulation of DNA damage responses. Oncogene 25:3598-3605. [DOI] [PubMed] [Google Scholar]
- 14.Fraschini, R., E. Formenti, G. Lucchini, and S. Piatti. 1999. Budding yeast Bub2 is localized at spindle pole bodies and activates the mitotic checkpoint via a different pathway from Mad2. J. Cell Biol. 145:979-991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garber, P. M., and J. Rine. 2002. Overlapping roles of the spindle assembly and DNA damage checkpoints in the cell-cycle response to altered chromosomes in Saccharomyces cerevisiae. Genetics 161:521-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geissler, S., K. Siegers, and E. Schiebel. 1998. A novel protein complex promoting formation of functional alpha- and gamma-tubulin. EMBO J. 17:952-966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gilbert, C. S., C. M. Green, and N. F. Lowndes. 2001. Budding yeast Rad9 is an ATP-dependent Rad53 activating machine. Mol. Cell 8:129-136. [DOI] [PubMed] [Google Scholar]
- 18.Halicka, H. D., X. Huang, F. Traganos, M. A. King, W. Dai, and Z. Darzynkiewicz. 2005. Histone H2AX phosphorylation after cell irradiation with UV-B: relationship to cell cycle phase and induction of apoptosis. Cell Cycle 4:339-345. [PubMed] [Google Scholar]
- 19.Hardwick, K. G., R. Li, C. Mistrot, R. H. Chen, P. Dann, A. Rudner, and A. W. Murray. 1999. Lesions in many different spindle components activate the spindle checkpoint in the budding yeast Saccharomyces cerevisiae. Genetics 152:509-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hardwick, K. G., E. Weiss, F. C. Luca, M. Winey, and A. W. Murray. 1996. Activation of the budding yeast spindle assembly checkpoint without mitotic spindle disruption. Science 273:953-956. [DOI] [PubMed] [Google Scholar]
- 21.Hoyt, M. A., L. Totis, and B. T. Roberts. 1991. S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell 66:507-517. [DOI] [PubMed] [Google Scholar]
- 22.Huh, W. K., J. V. Falvo, L. C. Gerke, A. S. Carroll, R. W. Howson, J. S. Weissman, and E. K. O'Shea. 2003. Global analysis of protein localization in budding yeast. Nature 425:686-691. [DOI] [PubMed] [Google Scholar]
- 23.Hyland, K. M., J. Kingsbury, D. Koshland, and P. Hieter. 1999. Ctf19p: a novel kinetochore protein in Saccharomyces cerevisiae and a potential link between the kinetochore and mitotic spindle. J. Cell Biol. 145:15-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jaspersen, S. L., J. F. Charles, R. L. Tinker-Kulberg, and D. O. Morgan. 1998. A late mitotic regulatory network controlling cyclin destruction in Saccharomyces cerevisiae. Mol. Biol. Cell 9:2803-2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jullien, D., P. Vagnarelli, W. C. Earnshaw, and Y. Adachi. 2002. Kinetochore localisation of the DNA damage response component 53BP1 during mitosis. J. Cell Sci. 115:71-79. [DOI] [PubMed] [Google Scholar]
- 26.Kadura, S., and S. Sazer. 2005. SAC-ing mitotic errors: how the spindle assembly checkpoint (SAC) plays defense against chromosome mis-segregation. Cell Motil. Cytoskeleton 61:145-160. [DOI] [PubMed] [Google Scholar]
- 27.Keogh, M. C., J. A. Kim, M. Downey, J. Fillingham, D. Chowdhury, J. C. Harrison, M. Onishi, N. Datta, S. Galicia, A. Emili, J. Lieberman, X. Shen, S. Buratowski, J. E. Haber, D. Durocher, J. F. Greenblatt, and N. J. Krogan. 2006. A phosphatase complex that dephosphorylates gammaH2AX regulates DNA damage checkpoint recovery. Nature 439:497-501. [DOI] [PubMed] [Google Scholar]
- 28.Klein, H. L. 2001. Spontaneous chromosome loss in Saccharomyces cerevisiae is suppressed by DNA damage checkpoint functions. Genetics 159:1501-1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krishnan, V., S. Nirantar, K. Crasta, A. Y. Cheng, and U. Surana. 2004. DNA replication checkpoint prevents precocious chromosome segregation by regulating spindle behavior. Mol. Cell 16:687-700. [DOI] [PubMed] [Google Scholar]
- 30.Lee, S. E., J. K. Moore, A. Holmes, K. Umezu, R. D. Kolodner, and J. E. Haber. 1998. Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell 94:399-409. [DOI] [PubMed] [Google Scholar]
- 31.Leroy, C., S. E. Lee, M. B. Vaze, F. Ochsenbien, R. Guerois, J. E. Haber, and M. C. Marsolier-Kergoat. 2003. PP2C phosphatases Ptc2 and Ptc3 are required for DNA checkpoint inactivation after a double-strand break. Mol. Cell 11:827-835. [DOI] [PubMed] [Google Scholar]
- 32.Leroy, C., C. Mann, and M. C. Marsolier. 2001. Silent repair accounts for cell cycle specificity in the signaling of oxidative DNA lesions. EMBO J. 20:2896-2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lew, D. J., and D. J. Burke. 2003. The spindle assembly and spindle position checkpoints. Annu. Rev. Genet. 37:251-282. [DOI] [PubMed] [Google Scholar]
- 34.Li, R., and A. W. Murray. 1991. Feedback control of mitosis in budding yeast. Cell 66:519-531. [DOI] [PubMed] [Google Scholar]
- 35.Limoli, C. L., E. Giedzinski, W. M. Bonner, and J. E. Cleaver. 2002. UV-induced replication arrest in the xeroderma pigmentosum variant leads to DNA double-strand breaks, gamma-H2AX formation, and Mre11 relocalization. Proc. Natl. Acad. Sci. USA 99:233-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maringele, L., and D. Lydall. 2002. EXO1-dependent single-stranded DNA at telomeres activates subsets of DNA damage and spindle checkpoint pathways in budding yeast yku70Delta mutants. Genes Dev. 16:1919-1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marsolier, M. C., P. Roussel, C. Leroy, and C. Mann. 2000. Involvement of the PP2C-like phosphatase Ptc2p in the DNA checkpoint pathways of Saccharomyces cerevisiae. Genetics 154:1523-1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mayer, M. L., S. P. Gygi, R. Aebersold, and P. Hieter. 2001. Identification of RFC(Ctf18p, Ctf8p, Dcc1p): an alternative RFC complex required for sister chromatid cohesion in S. cerevisiae. Mol. Cell 7:959-970. [DOI] [PubMed] [Google Scholar]
- 39.Melo, J., and D. Toczyski. 2002. A unified view of the DNA-damage checkpoint. Curr. Opin. Cell Biol. 14:237-245. [DOI] [PubMed] [Google Scholar]
- 40.Mikhailov, A., R. W. Cole, and C. L. Rieder. 2002. DNA damage during mitosis in human cells delays the metaphase/anaphase transition via the spindle-assembly checkpoint. Curr. Biol. 12:1797-1806. [DOI] [PubMed] [Google Scholar]
- 41.Morgan, D. O. 1999. Regulation of the APC and the exit from mitosis. Nat. Cell Biol. 1:E47-E53. [DOI] [PubMed] [Google Scholar]
- 42.Mumberg, D., R. Muller, and M. Funk. 1994. Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 22:5767-5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Musacchio, A., and K. G. Hardwick. 2002. The spindle checkpoint: structural insights into dynamic signalling. Nat. Rev. Mol. Cell Biol. 3:731-741. [DOI] [PubMed] [Google Scholar]
- 44.Nasmyth, K. 2001. Disseminating the genome: joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu. Rev. Genet. 35:673-745. [DOI] [PubMed] [Google Scholar]
- 45.Pangilinan, F., and F. Spencer. 1996. Abnormal kinetochore structure activates the spindle assembly checkpoint in budding yeast. Mol. Biol. Cell 7:1195-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pellicioli, A., C. Lucca, G. Liberi, F. Marini, M. Lopes, P. Plevani, A. Romano, P. P. Di Fiore, and M. Foiani. 1999. Activation of Rad53 kinase in response to DNA damage and its effect in modulating phosphorylation of the lagging strand DNA polymerase. EMBO J. 18:6561-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pereira, G., T. Hofken, J. Grindlay, C. Manson, and E. Schiebel. 2000. The Bub2p spindle checkpoint links nuclear migration with mitotic exit. Mol. Cell 6:1-10. [PubMed] [Google Scholar]
- 48.Pinsky, B. A., and S. Biggins. 2005. The spindle checkpoint: tension versus attachment. Trends Cell Biol. 15:486-493. [DOI] [PubMed] [Google Scholar]
- 49.Royou, A., H. Macias, and W. Sullivan. 2005. The Drosophila Grp/Chk1 DNA damage checkpoint controls entry into anaphase. Curr. Biol. 15:334-339. [DOI] [PubMed] [Google Scholar]
- 50.Sanchez, Y., J. Bachant, H. Wang, F. Hu, D. Liu, M. Tetzlaff, and S. J. Elledge. 1999. Control of the DNA damage checkpoint by chk1 and rad53 protein kinases through distinct mechanisms. Science 286:1166-1171. [DOI] [PubMed] [Google Scholar]
- 51.Sanchez, Y., B. A. Desany, W. J. Jones, Q. Liu, B. Wang, and S. J. Elledge. 1996. Regulation of RAD53 by the ATM-like kinases MEC1 and TEL1 in yeast cell cycle checkpoint pathways. Science 271:357-360. [DOI] [PubMed] [Google Scholar]
- 52.Schwartz, M. F., J. K. Duong, Z. Sun, J. S. Morrow, D. Pradhan, and D. F. Stern. 2002. Rad9 phosphorylation sites couple Rad53 to the Saccharomyces cerevisiae DNA damage checkpoint. Mol. Cell 9:1055-1065. [DOI] [PubMed] [Google Scholar]
- 53.Shou, W., J. H. Seol, A. Shevchenko, C. Baskerville, D. Moazed, Z. W. Chen, J. Jang, H. Charbonneau, and R. J. Deshaies. 1999. Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell 97:233-244. [DOI] [PubMed] [Google Scholar]
- 54.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Spencer, F., and P. Hieter. 1992. Centromere DNA mutations induce a mitotic delay in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 89:8908-8912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stegmeier, F., R. Visintin, and A. Amon. 2002. Separase, polo kinase, the kinetochore protein Slk19, and Spo12 function in a network that controls Cdc14 localization during early anaphase. Cell 108:207-220. [DOI] [PubMed] [Google Scholar]
- 57.Stern, B. M., and A. W. Murray. 2001. Lack of tension at kinetochores activates the spindle checkpoint in budding yeast. Curr. Biol. 11:1462-1467. [DOI] [PubMed] [Google Scholar]
- 58.Sugimoto, I., H. Murakami, Y. Tonami, A. Moriyama, and M. Nakanishi. 2004. DNA replication checkpoint control mediated by the spindle checkpoint protein Mad2p in fission yeast. J. Biol. Chem. 279:47372-47378. [DOI] [PubMed] [Google Scholar]
- 59.Sun, Z., J. Hsiao, D. S. Fay, and D. F. Stern. 1998. Rad53 FHA domain associated with phosphorylated Rad9 in the DNA damage checkpoint. Science 281:272-274. [DOI] [PubMed] [Google Scholar]
- 60.Sweeney, F. D., F. Yang, A. Chi, J. Shabanowitz, D. F. Hunt, and D. Durocher. 2005. Saccharomyces cerevisiae Rad9 acts as a Mec1 adaptor to allow Rad53 activation. Curr. Biol. 15:1364-1375. [DOI] [PubMed] [Google Scholar]
- 61.Takada, S., A. Kelkar, and W. E. Theurkauf. 2003. Drosophila checkpoint kinase 2 couples centrosome function and spindle assembly to genomic integrity. Cell 113:87-99. [DOI] [PubMed] [Google Scholar]
- 62.Tinker-Kulberg, R. L., and D. O. Morgan. 1999. Pds1 and Esp1 control both anaphase and mitotic exit in normal cells and after DNA damage. Genes Dev. 13:1936-1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tsvetkov, L., X. Xu, J. Li, and D. F. Stern. 2003. Polo-like kinase 1 and Chk2 interact and co-localize to centrosomes and the midbody. J. Biol. Chem. 278:8468-8475. [DOI] [PubMed] [Google Scholar]
- 64.Usui, T., H. Ogawa, and J. H. Petrini. 2001. A DNA damage response pathway controlled by Tel1 and the Mre11 complex. Mol. Cell 7:1255-1266. [DOI] [PubMed] [Google Scholar]
- 65.Vialard, J. E., C. S. Gilbert, C. M. Green, and N. F. Lowndes. 1998. The budding yeast Rad9 checkpoint protein is subjected to Mec1/Tel1-dependent hyperphosphorylation and interacts with Rad53 after DNA damage. EMBO J. 17:5679-5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Visintin, R., K. Craig, E. S. Hwang, S. Prinz, M. Tyers, and A. Amon. 1998. The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk-dependent phosphorylation. Mol. Cell 2:709-718. [DOI] [PubMed] [Google Scholar]
- 67.Wang, Y., F. Hu, and S. J. Elledge. 2000. The Bfa1/Bub2 GAP complex comprises a universal checkpoint required to prevent mitotic exit. Curr. Biol. 10:1379-1382. [DOI] [PubMed] [Google Scholar]
- 68.Wei, J. H., Y. F. Chou, Y. H. Ou, Y. H. Yeh, S. W. Tyan, T. P. Sun, C. Y. Shen, and S. Y. Shieh. 2005. TTK/hMps1 participates in the regulation of DNA damage checkpoint response by phosphorylating CHK2 on threonine 68. J. Biol. Chem. 280:7748-7757. [DOI] [PubMed] [Google Scholar]
- 69.Weinert, T. A., and L. H. Hartwell. 1988. The RAD9 gene controls the cell cycle response to DNA damage in Saccharomyces cerevisiae. Science 241:317-322. [DOI] [PubMed] [Google Scholar]
- 70.Weiss, E., and M. Winey. 1996. The Saccharomyces cerevisiae spindle pole body duplication gene MPS1 is part of a mitotic checkpoint. J. Cell Biol. 132:111-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zou, L., and S. J. Elledge. 2003. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300:1542-1548. [DOI] [PubMed] [Google Scholar]