Abstract
Cripto is a developmental oncoprotein and a member of the epidermal growth factor-Cripto, FRL-1, Cryptic family of extracellular signaling molecules. In addition to having essential functions during embryogenesis, Cripto is highly expressed in tumors and promotes tumorigenesis. During development, Cripto acts as an obligate coreceptor for transforming growth factor β (TGF-β) ligands, including nodals, growth and differentiation factor 1 (GDF1), and GDF3. As an oncogene, Cripto is thought to promote tumor growth via mechanisms including activation of mitogenic signaling pathways and antagonism of activin signaling. Here, we provide evidence supporting a novel mechanism in which Cripto inhibits the tumor suppressor function of TGF-β. Cripto bound TGF-β and reduced the association of TGF-β with its type I receptor, TβRI. Consistent with its ability to block receptor assembly, Cripto suppressed TGF-β signaling in multiple cell types and diminished the cytostatic effects of TGF-β in mammary epithelial cells. Furthermore, targeted disruption of Cripto expression by use of small inhibitory RNA enhanced TGF-β signaling, indicating that endogenous Cripto plays a role in restraining TGF-β responses.
Members of the transforming growth factor β (TGF-β) superfamily of ligands, which includes the TGF-β, activin, and bone morphogenetic protein (BMP) families, control a myriad of cellular processes, including proliferation and differentiation during development and in adult tissues (20, 29, 38, 53). Given the wide-ranging roles of these ligands, it is not surprising that disruption or dysregulation of their activities is associated with multiple pathological states, including tumorigenesis (43).
TGF-β ligands exert their biological effects by binding and assembling cell surface type I and type II transmembrane serine/threonine kinase receptors. Receptor assembly allows the constitutively active type II receptor kinase to phosphorylate the type I receptor, thereby activating the type I receptor kinase (42). This receptor activation mechanism was initially established for TGF-β, which first binds its type II receptor, TβRII, to allow subsequent recruitment and phosphorylation of its type I receptor, TβRI (55, 56). Intracellular signals emanating from active TGF-β receptor complexes include those mediated by the cytoplasmic receptor-activated Smad proteins Smad2 and Smad3, which are target substrates for the TβRI kinase (42). Following phosphorylation by TβRI, Smad2 and Smad3 bind the common mediator Smad, Smad4, and the resulting hetero-oligomeric Smad complexes migrate into the nucleus to regulate transcription of target genes (42).
The ability of TGF-β superfamily members to bind and assemble their signaling receptors is regulated in a complex manner by membrane-bound coreceptors and accessory proteins (23). One example is betaglycan (TβRIII), a multifunctional transmembrane proteoglycan that binds TGF-β isoforms and enhances their ability to bind and assemble signaling receptors (27, 28, 50). Betaglycan also binds inhibins (25) and facilitates the ability of these inhibitory TGF-β ligands to bind activin type II receptors and block signaling by activins (25) and BMPs (54). As another example, endoglin also binds multiple TGF-β ligands and, interestingly, determines whether TGF-β signals via TβRI as opposed to the functionally distinct type I receptor ALK1 (24, 35). Finally, the accessory protein BMP and activin receptor membrane-bound inhibitor (BAMBI) resembles a type I receptor but lacks a cytoplasmic kinase domain and inhibits BMP, activin, and TGF-β signaling by forming inactive complexes with these ligands and their respective signaling receptors (34). As illustrated by these three cell surface proteins, accessory receptors control the ability of TGF-β ligands to access signaling receptors and generate cellular responses.
Cripto (Cripto-1, TDGF1) is a multifunctional signaling protein and an accessory receptor based on its ability to bind TGF-β ligands and modulate their signaling properties (19, 40, 46). Under normal physiological conditions, Cripto is selectively expressed during developmental processes and it has been shown to have activity both as a soluble factor and as a glycosylphosphatidylinositol (GPI)-anchored cell surface protein (46). In the developing embryo, Cripto and other epidermal growth factor-Cripto, FRL-1, Cryptic (EGF-CFC) proteins have essential roles as necessary coreceptors for TGF-β ligands, including nodals (38, 41), growth and differentiation factor 1 (GDF1) (8), and GDF3 (7). In this capacity, EGF-CFC proteins are required for processes such as mesendoderm induction (38), correct positioning of the body axes (4, 12, 38), and cardiac development (58). Cripto has also been identified as a marker for embryonic stem cells (5), suggesting it facilitates maintenance of the stem cell phenotype (46). Although generally absent from adult tissues, Cripto is expressed at high levels in human tumors and several molecular mechanisms that may contribute to its ability to promote tumorigenesis have been proposed previously (46). For example, Cripto can activate ras/mitogen-activated protein kinase (22) and phosphatidylinositol 3-kinase (13) pathways associated with mitogenesis and cell survival and it also inhibits signaling by activins (1, 15). Determining the extent to which these and other oncogenic Cripto actions contribute to the tumor phenotype remains an important area of investigation.
Here, we identify a new molecular mechanism in which Cripto inhibits TGF-β signaling. Our results show that Cripto binds TGF-β and reduces TGF-β binding to TβRI, suggesting that it interferes with receptor assembly. Consistent with this interpretation, we demonstrate that Cripto attenuates TGF-β signaling in multiple cell types and signaling assays. Importantly, Cripto substantially diminished the cytostatic effects of TGF-β on mammary epithelial cells grown under either anchorage-dependent or anchorage-independent conditions. We have also measured the activity of Cripto deletion mutants lacking either the EGF-like domain or the CFC domain, and our results suggest that the EGF-like domain is both necessary and sufficient for TGF-β binding and inhibition of TGF-β signaling. Finally, we have shown that knockdown of endogenous Cripto expression by use of small interfering RNA (siRNA) enhances TGF-β responsiveness. Since Cripto is highly expressed in tumors (46) and since loss of the antiproliferative effects of TGF-β is frequently associated with tumorigenesis (43), we propose that Cripto antagonism of TGF-β signaling may contribute to tumor initiation and progression.
MATERIALS AND METHODS
Materials.
NuPAGE gels, molecular weight standards, and a Cyquant cell proliferation assay kit were from Invitrogen (San Diego, CA). TGF-β1 was purchased from R&D Systems (Minneapolis, MN). 125I-TGF-β1 was purchased from Perkin-Elmer Life and Analytical Sciences, Inc. (Boston, MA). Anti-His antibody and protein G-agarose were purchased from Calbiochem (San Diego, CA). Anti-FLAG (M2) and antihemagglutinin (anti-HA) antibodies were purchased from Sigma-Aldrich (St. Louis, MO). Tetramethylbenzidine peroxidase substrate was purchased from Pierce (Rockford, IL). Anti-phospho-Smad2 antibody was purchased from Cell Signaling (Danvers, MA). Rabbit polyclonal antibodies used to measure Smad2 levels were raised against a peptide conserved between Smad2 and Smad3 spanning amino acids 159 to 175 of human Smad3 and were produced by Joan Vaughan (Peptide Biology Laboratories, Salk Institute).
Expression constructs.
We used overlapping PCR (14) to generate constructs in which a single FLAG epitope tag sequence (DYKDDDDK) was inserted upstream of the EGF-like domain between glycine 55 and isoleucine 56 of mouse Cripto. The Cripto deletion mutants were generated using a previously described PCR strategy (17) in which either the EGF-like domain or the CFC domain was replaced with an in-frame BamHI site (GGATCC) encoding the amino acids Gly-Ser. For transient transfection into HepG2 and 293T cells, Cripto constructs were subcloned into pcDNA3 (Invitrogen, Carlsbad, CA). Cripto constructs were also subcloned into the lentiviral vector pCSC (31) for production of lentivirus. Lentiviral vectors used in this study were a gift from Inder Verma (Salk Institute). The TβRI-HA and TβRII-His expression constructs have been described previously (56) and were a gift from Joan Massagué (Memorial Sloan-Kettering Cancer Center, New York).
Transfection and infection of cell lines.
293T cells and HeLa cells were grown in Dulbecco's modified Eagle's medium, and HepG2 cells were grown in alpha minimal essential medium. Both media were supplemented with 10% fetal calf serum, penicillin, streptomycin, and l-glutamine. MCF10A cells were grown in Dulbecco's modified Eagle's medium-F-12 supplemented with 5% donor horse serum, 20 ng/ml EGF, 10 μg/ml insulin, 0.5 μg/ml hydrocortisone, and 100 ng/ml cholera toxin.
For transient transfection, cells were plated at densities between 40 and 60% confluence and GenePorter 2 (Gene Therapy Systems) was used for HepG2 cells and Perfectin (Gene Therapy Systems) was used for 293T cells according to the manufacturer's instructions. For viral transduction, lentivirus was produced essentially as previously described (31). An appropriate dilution of virus-containing media to obtain a multiplicity of infection of 3 to 5 was used to generate pools of cells containing the vector, and the efficiency of viral infection was determined by monitoring green fluorescent protein expression in infected cells by use of fluorescence microscopy.
Measurement of cell surface expression of Cripto constructs in 293T cells.
Detection of Cripto expressed at the surface of intact cells via cell surface enzyme-linked immunosorbent assay (ELISA) was performed essentially as previously described (18). 293T cells were plated on 24-well polylysine-coated plates at a density of 100,000 cells per well, transfected 24 h later with 0.5 μg vector or Cripto DNA per well, and then assayed for cell surface expression 48 h after transfection. Cells were rinsed in HEPES dissociation buffer (HDB) (12.5 mM HEPES [pH 7.4], 140 mM NaCl, and 5 mM KCl) and then incubated in 3% bovine serum albumin (BSA) in HDB for 30 min at room temperature (RT). Cells were then incubated for 2 h with 2 μg/ml anti-FLAG (M2) antibody in 3% BSA in HDB, rinsed with HDB, and incubated with peroxidase-conjugated anti-mouse immunoglobulin G in 3% BSA in HDB for 1 h at room temperature. Wells were rinsed with HDB, and then 100 μl of tetramethylbenzidine peroxidase substrate was added to each well. Plates were incubated at RT until the solutions turned visibly blue. Peroxidase activity was stopped by adding 100 μl of 0.18 M H2SO4 to each well, and peroxidase activity was quantified by measuring the absorbance of the resulting yellow solutions at 450 nm.
Covalent cross-linking.
293T cells were plated on six-well plates coated with polylysine at a density of 400,000 cells per well and then transfected approximately 24 h later. Cells were transfected with 4 μg DNA per well with a ratio of 0.5 μg TβRII/0.5 μg TβRI/3 μg Cripto unless otherwise indicated. As necessary, empty vector was used to keep the amount of DNA transfected constant at 4 μg. Covalent cross-linking was performed approximately 48 h after transfection by first rinsing cells in HDB and then incubating them with 125I-TGF-β1 in binding buffer (HDB containing 0.1% BSA, 5 mM MgSO4, and 1.5 mM CaCl2) at RT for approximately 4 h. Cells were rinsed in HDB, incubated in HDB containing 0.5 mM disuccinylsuberate for 30 min on ice, rinsed in HDB, and then solubilized in lysis buffer (Tris-buffered saline containing 1% NP-40, 0.5% deoxycholate, and 2 mM EDTA) supplemented with standard protease inhibitors for 1 h on ice. Solubilized, cross-linked complexes were incubated for approximately 24 h at 4°C with anti-His, anti-Flag, or anti-HA antibody, and then immune complexes were precipitated using protein G-agarose and analyzed using sodium dodecyl sulfate-polyacrylamide gel electrophoresis and autoradiography as previously described (14).
Luciferase assays.
Luciferase assays were carried out using TGF-β-responsive 3TP-lux and A3-luciferase reporters essentially as previously described (15). The 3TP-lux luciferase reporter contains three consecutive tetradecanoylphorbol acetate response elements and a portion of the plasminogen activator inhibitor 1 (PAI-1) promoter region, while the A3-luciferase construct contains three copies of the ARE (activin response element) from the Xenopus laevis Mix.2 promoter linked to a basic TATA box and a luciferase reporter gene. HepG2 cells were plated at 150,000 cells per well in 24-well plates and transfected in triplicate approximately 24 h later with 1 μg DNA per well, with a ratio of 800 ng Cripto/100 ng 3TP-lux/100 ng cytomegalovirus-β-galactosidase (CMV-β-Gal). Cells were treated with TGF-β1 approximately 30 h after the transfection and harvested 16 h following treatment. Cells were incubated in solubilization buffer (1% Triton X-100, 25 mM glycylglycine [pH 7.8], 15 mM MgSO4, 4 mM EGTA, and 1 mM dithiothreitol) for 30 min on ice, and luciferase reporter activity was measured and normalized relative to CMV-β-Gal activities. 293T cells were plated on 24-well plates treated with polylysine at 100,000 cells per well and transfected in triplicate approximately 24 h later with 0.5 μg DNA per well by use of 400 ng Cripto/50 ng FAST2 (FoxH1)/25 ng A3-luciferase/25 ng CMV-β-galactosidase per well. Cells were treated approximately 24 h following transfection and then harvested approximately 16 h following treatment. Luciferase assays were performed as described above for HepG2 cells.
Smad2 phosphorylation.
Cells were stably infected with lentivirus and then plated on six-well plates at a density of 100,000 cells per well. Forty-eight hours after plating, cells were washed once with HDB, starved for 1.5 to 8 h in additive-free medium, and then left untreated or treated with TGF-β1 for 30 min. Cells were rinsed in ice-cold HDB and then harvested by adding 150 μl ice-cold solubilization buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% NP-40, 0.5% deoxycholate, 0.1% sodium dodecyl sulfate) supplemented with 50 mM beta-glycerol phosphate, 20 mM NaF, and standard protease inhibitors. Fifty microliters of 4× sample buffer was than added to each sample, and the proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and blotted to nitrocellulose. Blots were treated with anti-phospho-Smad2 or anti-Smad2 antibody followed by anti-rabbit antibody conjugated to horseradish peroxidase, and bands were detected using enhanced chemiluminescence.
Cell proliferation assays and colony formation in soft agar.
MCF10A cells were stably infected with pCSC lentivirus constructs as described above. Cells were plated on 96-well plates at a density of 500 cells/well, and 48 h later cells were either treated with 100 pM TGF-β1 or left untreated. Six days after treatment, cell number was determined by using a Cyquant cell proliferation kit according to the manufacturer's instructions. To measure colony formation in soft agar, 12-well plates were prepared with 1.5-ml/well surface layers consisting of 0.6% agar resuspended in MCF10A growth medium. An additional 0.75 ml of 0.33% agar-MCF10A growth medium containing 1,000 stably infected MCF10A cells was then added to each well, and TGF-β1 (100 pM final concentration) was added before the agar hardened. Wells were refed with 20 μl of MCF10A growth medium with or without TGF-β1 (final concentration, 100 pM) twice a week for 15 days, and then colonies were visualized microscopically and counted.
RNA extraction and qPCR.
Total RNA was extracted from HeLa cells by using an RNeasy mini kit (QIAGEN, Inc., Valencia, CA) according to the manufacturer's instructions. DNase treatment was performed on-column using an RNase-free DNase kit (QIAGEN). SYBR green real-time reverse transcription-PCRs were performed by the microarray/quantitative PCR (qPCR) scientific core facility at the Salk Institute (http://cores.salk.edu/microarray). Sequences of primer pairs specific for human Cripto and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) were as follows: ATGGCCATTTCTAAAGTCTTTGAACT (Cripto-forward), GATGGACGAGCAAATTCCTGAT (Cripto-reverse), TGGAAGGACTCATGACCACAG (GAPDH-forward), and CAGTCTTCTGGGTGGCAGTGA (GAPDH-reverse).
Design of Cripto lentiviral shRNA vectors.
Target sequences within the human Cripto gene were identified and selected using the Sfold program (http://sfold.wadsworth.org/sirna.pl). The design of short hairpin RNA (shRNA) and production of lentiviral shRNA vectors were carried out essentially as previously described (44). Briefly, an 83-mer oligonucleotide containing the human Cripto shRNA sequence and a T3 oligonucleotide (5′-CTCGAAATTAACCCTCACTAAAGGG-3′) were used to PCR amplify a fragment which was then subcloned into the lentiviral vector in which shRNA expression is driven by an H1 promoter (44). The 83-mers used to generate the Cripto 1 (C1) and Cripto 2 (C2) shRNA vectors were as follows: for C1, 5′-CTGTCTAGACAAAAAGCAAGCTGACCCTGAAGTTCTCTCTTGAA GAACTTCAGGGTCAGCTTGCGGGGATCTGTGGTCTCATACA-3′, and for C2, 5′-CTGTCTAGACAAAAAGCAAGCTGACCCTGAAGTTCTCTCTTGAAGAACTTCAGGGTCAGCTTGCGGGGATCTGTGGTCTCATACA-3′.
RESULTS
Cripto binds TGF-β and reduces cross-linking of TGF-β to TβRI.
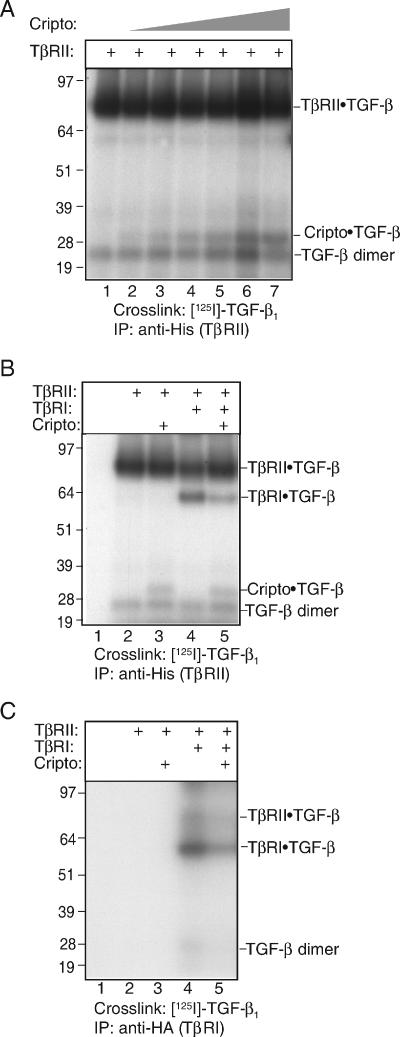
293T cells were transfected with a fixed amount of TβRII DNA together with increasing amounts of Cripto DNA and then subjected to labeling and covalent cross-linking with 125I-TGF-β1 followed by immunoprecipitation using an antibody directed against TβRII. In the absence of transfected Cripto, bands corresponding to cross-linked 125I-TGF-β1-TβRII complexes and a [125I]-TGF-β1 dimer are evident (Fig. 1A, lane 1). When Cripto DNA was included, a 125I-TGF-β1-Cripto cross-linked band of ∼32 kDa appeared and increased in intensity as the amount of Cripto DNA was increased (Fig. 1A, lanes 2 to 7). The fact that 125I-TGF-β1-Cripto cross-linked complexes were coimmunoprecipitated with an antibody directed against TβRII indicates that TβRII, TGF-β1, and Cripto form a stable ternary complex.
FIG. 1.
Cripto binds TGF-β1 in the presence of TβRII and reduces cross-linking of TGF-β1 to TβRI. 293T cells were transfected with empty vector or the indicated constructs, labeled with 125I-TGF-β1, and then subjected to covalent cross-linking and immunoprecipitation (IP) with an antibody directed against TβRII (A and B) or TβRI (C) as described in Materials and Methods. Bands corresponding to 125I-TGF-β1 cross-linked complexes are indicated. For panel A, 293T cells were transfected with the following amounts of DNA: 78 ng (lane 2), 156 ng (lane 3), 312 ng (lane 4), 625 ng (lane 5), 1.25 μg (lane 6), and 2.5 μg (lane 7). Numbers to the left indicate molecular sizes in kilodaltons.
We next tested the effect of Cripto on the ability of 125I-TGF-β1 to cross-link to its type I receptor, TβRI. 293T cells were transfected as specified (Fig. 1B and 1C), labeled with 125I-TGF-β1, and then subjected to immunoprecipitation with the indicated antibodies. Similarly to what was demonstrated in Fig. 1A, 125I-TGF-β1 formed cross-linked complexes with both TβRII and Cripto, as visualized following anti-His immunoprecipitation (Fig. 1B, lanes 2 to 5). When TβRII and TβRI were cotransfected, 125I-TGF-β1 formed cross-linked complexes with both receptor types, as expected (Fig. 1B, lane 4). However, addition of Cripto to the cotransfection resulted in reduced intensity of the TβRI band (Fig. 1B, compare lanes 4 and 5), indicating that Cripto interferes with the ability of TβRI to bind and cross-link to 125I-TGF-β1. This conclusion is supported by the observation that cross-linking of 125I-TGF-β1 to TβRI is similarly reduced, as visualized following immunoprecipitation with an anti-HA antibody targeting TβRI (Fig. 1C, compare lanes 4 and 5). In contrast to what we have observed following immunoprecipitation with anti-TβRII antibodies, we were unable to observe a labeled Cripto band following anti-TβRI immunoprecipitation (Fig. 1C, lane 5), suggesting that TβRI and Cripto are apparently not present in the same complexes. In summary, these cross-linking data point to a model in which Cripto and TβRI compete for binding to TβRII-TGF-β complexes, providing a mechanism of Cripto inhibition of TGF-β-dependent receptor assembly.
Cripto blocks TGF-β signaling in 293T cells and HepG2 cells.
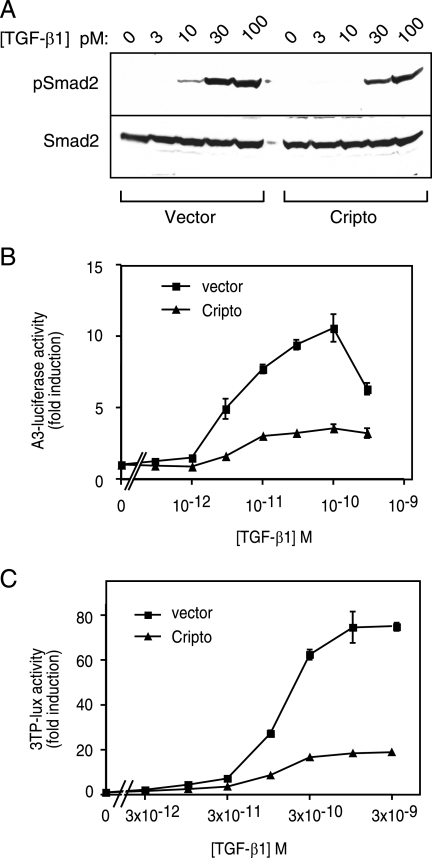
To determine whether Cripto affects TGF-β signaling, we first tested its ability to modulate TGF-β-dependent Smad2 phosphorylation. 293T cells were stably infected with lentivirus containing either Cripto or empty vector and then treated with a range of TGF-β1 doses. In control cells, phospho-Smad2 was clearly visible following treatment with 10 pM TGF-β1 and reached near-maximal levels following treatment with 30 pM TGF-β1 (Fig. 2A). By contrast, phospho-Smad2 was not detected in Cripto-infected cells treated with 10 pM TGF-β1 and phospho-Smad2 levels were clearly reduced following treatment with 30 pM and 100 pM TGF-β1 relative to corresponding phospho-Smad2 levels observed in vector-infected cells (Fig. 2A). Therefore, Cripto can mitigate TGF-β signaling as measured by phosphorylation of Smad2. Since phospho-Smad2 levels directly reflect the TβRI kinase activity resulting from TβRI recruitment into TGF-β-TβRII complexes, these results further support a role for Cripto as an inhibitor of TGF-β receptor assembly.
FIG. 2.
Cripto blocks TGF-β signaling in 293T cells and HepG2 cells. (A) 293T cells were infected with lentivirus containing empty vector or Cripto vector and then treated with a range of TGF-β1 doses as indicated. Resulting phospho-Smad2 (pSmad2) and total Smad2 levels were determined by Western blotting as described in Materials and Methods. (B) 293T cells or (C) HepG2 cells were transfected with either empty vector or Cripto DNA and either A3-luciferase/FAST-2/CMV-β-Gal (B) or 3TP-lux/CMV-β-Gal (C) as described in Materials and Methods. Cells were treated with the indicated doses of TGF-β1, and luciferase activities were measured and normalized relative to β-galactosidase activities. Data are presented as increases (n-fold) in luciferase activity of cells treated with TGF-β1 relative to activity of untreated cells, and experiments were performed in triplicate, with error bars representing standard errors of the means.
Next, we utilized TGF-β-responsive luciferase constructs to measure the effects of Cripto on TGF-β signaling. 293T cells and HepG2 cells were transiently transfected with reporter constructs together with either Cripto or empty vector and then treated with a range of TGF-β1 doses. In both 293T cells (Fig. 2B) and HepG2 cells (Fig. 2C), TGF-β1 caused a dose-dependent increase in luciferase expression that was blunted when cells were transfected with Cripto. At maximal doses of TGF-β1, Cripto blocked luciferase induction by ∼4-fold relative to cells transfected with empty vector (Fig. 2B and C). Together with the cross-linking data and phospho-Smad2 data presented above, these results provide additional evidence for a model in which Cripto interferes with receptor assembly to cause a decrease in TGF-β signaling.
Roles of the EGF-like and CFC domains of Cripto in TGF-β binding.
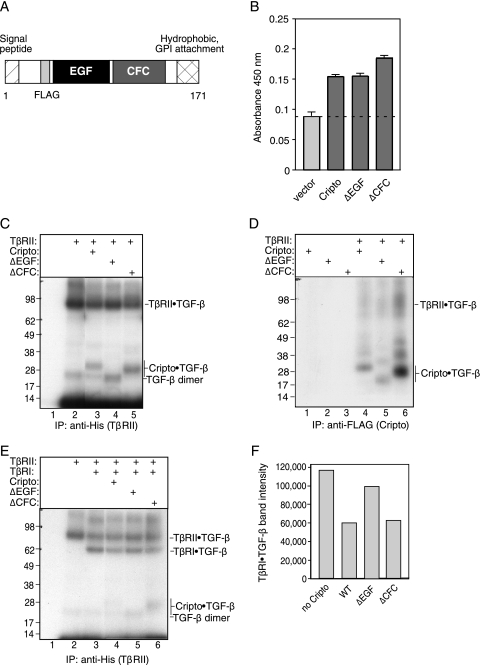
Cripto and other EGF-CFC family members share two conserved cysteine-rich domains, an N-terminal EGF-like domain and a C-terminal CFC domain (41). Each of these modular domains can have activity in the absence of the other, and both have been implicated in specific protein-protein interactions and signaling functions (4, 40, 46). The domain structure of mouse Cripto is illustrated in Fig. 3A. This diagram indicates the locations of the signal peptide, the introduced FLAG epitope, the EGF-like domain, the CFC domain, and the C-terminal hydrophobic region required for GPI anchor attachment.
FIG. 3.
Roles of the EGF-like and CFC domains of Cripto in binding and cross-linking to TGF-β. (A) Diagram of mouse Cripto, indicating the positions of the signal peptide, N-terminal FLAG epitope, EGF-like domain (EGF), CFC domain (CFC), and C-terminal site of GPI anchor attachment. (B) Empty vector or the indicated Cripto constructs were transfected in triplicate into 293T cells, and the resulting cell surface expression of these constructs in intact cells was measured using anti-FLAG antibody in an ELISA-based assay as described in Materials and Methods. (C to E) 293T cells were transfected with empty vector or the indicated constructs and labeled with 125I-TGF-β1, and extracts were subjected to immunoprecipitation (IP) with either anti-His antibody directed against TβRII (C and E) or anti-FLAG antibody targeting Cripto proteins (D) as described in Materials and Methods. Locations of the 125I-TGF-β1 dimer and 125I-TGF-β1 cross-linked bands corresponding to TβRII, TβRI, and the Cripto constructs are indicated. Numbers to the left indicate molecular sizes in kilodaltons. (F) The bands from panel E corresponding to TβRI-TGF-β complexes were quantitated using densitometry, and the resulting band intensities are presented. WT, wild type.
Three FLAG-tagged Cripto constructs were evaluated in this study: wild-type Cripto; ΔEGF, in which the EGF-like domain has been deleted; and ΔCFC, in which the CFC domain has been deleted. During posttranslational processing, N-terminal and C-terminal sequences are removed, resulting in a mature Cripto protein consisting almost entirely of the EGF-like and CFC domains (30) (Fig. 3A). Therefore, the ΔEGF mutant consists essentially of the CFC domain and vice versa. In order to confirm expression of these FLAG-tagged proteins at the cell surface, we transfected constructs into 293T cells and detected Cripto proteins with anti-FLAG antibody by using an intact cell ELISA-based assay (18). As shown in Fig. 3B, each of these Cripto constructs was expressed at the cell surface at similar levels.
We next compared the abilities of wild-type Cripto and the Cripto deletion mutants to bind and cross-link to TGF-β1 in the presence or absence of TGF-β signaling receptors. 293T cells were transfected with the indicated constructs (Fig. 3C), labeled with 125I-TGF-β1, and then subjected to covalent cross-linking followed by immunoprecipitation with an antibody directed against TβRII (anti-His). 125I-TGF-β1 cross-linked bands corresponding to TβRII as well as wild-type and mutant Cripto forms are indicated (Fig. 3C). As shown in Fig. 1, 125I-TGF-β1 formed a cross-linked complex with both TβRII and wild-type Cripto (Fig. 3C, lane 3). In addition, 125I-TGF-β1 clearly formed a cross-linked complex with the ΔCFC mutant but not the ΔEGF mutant when these constructs were coexpressed with TβRII (Fig. 3C, lanes 2, 4, and 5). This result demonstrates that the CFC domain is not required for TGF-β1 binding to Cripto and indicates, rather, that the EGF-like domain binds TGF-β.
Next, we tested whether TβRII is required for binding of TGF-β1 to wild-type and mutant forms of Cripto. 293T cells were again transfected with Cripto constructs in the absence or presence of TβRII, labeled with 125I-TGF-β1, and then subjected to covalent cross-linking followed by immunoprecipitation with an antibody directed against Cripto (anti-FLAG). As shown in Fig. 3D, cross-linking of 125I-TGF-β1 to Cripto or the Cripto mutants was not observed in the absence of cotransfected TβRII (Fig. 3D, lanes 1 to 3). However, labeled bands containing Cripto and its mutant forms were evident following cotransfection of TβRII (Fig. 3D, lanes 4 to 6). Therefore, TβRII appears to be required for TGF-β binding to Cripto and, similarly to TβRI, Cripto apparently lacks affinity for TGF-β unless TGF-β is first bound to TβRII. Consistent with the data from Fig. 3C, both wild-type Cripto and the ΔCFC mutant formed cross-linked complexes with 125I-TGF-β1 when they were coexpressed with TβRII (Fig. 3D, lanes 4 and 6). A faint band was also observed following cross-linking of 125I-TGF-β1 to the ΔEGF mutant, indicating this mutant weakly binds TGF-β (Fig. 3D, lane 5). However, the relative intensities of the bands corresponding to the ΔEGF-125I-TGF-β1 and ΔCFC-125I-TGF-β1 complexes (Fig. 3D, lanes 5 and 6) indicate that the EGF-like domain plays a predominant role in TGF-β binding.
In parallel experiments, we measured the effects of the Cripto deletion mutants on cross-linking of 125I-TGF-β1 to TβRI. As predicted, and as demonstrated in Fig. 1, 125I-TGF-β1 formed a cross-linked complex with both TβRII and TβRI when these receptor proteins were coexpressed (Fig. 3E, lane 3). Consistent with data presented in Fig. 1B, the intensity of the band corresponding to the TβRI-125I-TGF-β1 complex was reduced following cotransfection of wild-type Cripto (Fig. 3E, compare lanes 3 and 4). The relative intensity of the labeled TβRI band was similarly reduced when the ΔCFC mutant was cotransfected but was affected only minimally by cotransfection of the ΔEGF mutant (Fig. 3E, lanes 3 to 6). The intensities of the bands from Fig. 3E corresponding to TβRI-TGF-β complexes were quantitated using densitometry and are presented in Fig. 3F. In summary, these cross-linking data indicate that the EGF-like domain of Cripto binds TGF-β and enables Cripto to block cross-linking of TGF-β to TβRI. In addition, these results suggest that the CFC domain plays little if any role in these actions.
The EGF-like domain mediates inhibition of TGF-β signaling.
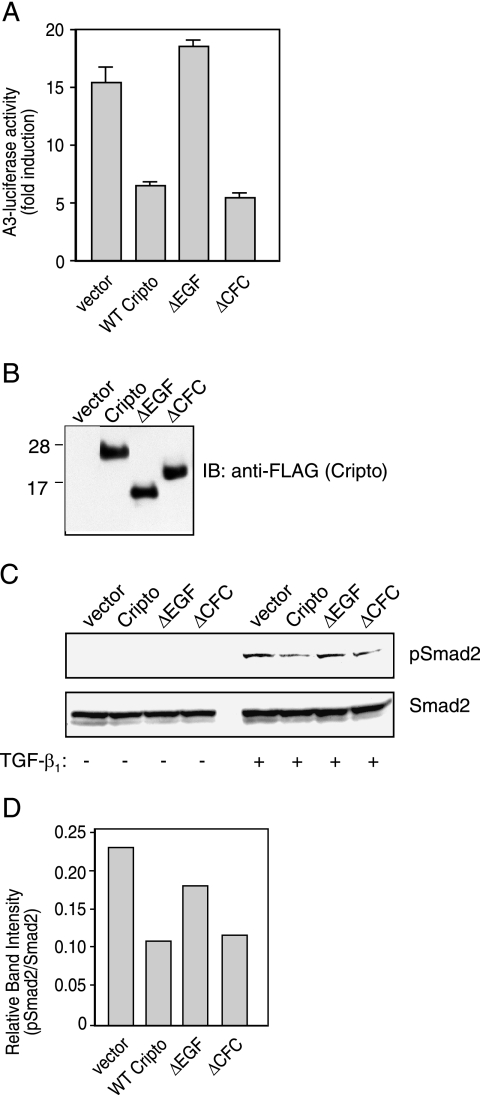
Next, we tested the effects of wild-type and mutant forms of Cripto on TGF-β signaling by transfecting Cripto constructs into 293T cells and measuring their effects on induction of a TGF-β-responsive luciferase reporter gene. Figure 4A shows that following transfection of empty vector into 293T cells, TGF-β1 treatment caused an ∼15-fold induction of luciferase relative to untreated cells. Luciferase induction in response to TGF-β1 was reduced ∼3-fold following transfection of either wild-type Cripto or the ΔCFC mutant but was not inhibited following transfection of the ΔEGF mutant (Fig. 4A). This result indicates once again that the EGF-like domain of Cripto is required for antagonism of TGF-β1 signaling and that the minimal ability of the CFC domain (ΔEGF mutant) to bind TGF-β1 is apparently insufficient to mediate TGF-β1 antagonism under these assay conditions. Together, these data indicate that the EGF-like domain of Cripto is both necessary and sufficient for the inhibition of TGF-β signaling in 293T cells.
FIG. 4.
Roles of the EGF-like and CFC domains of Cripto in antagonism of TGF-β-dependent signaling. (A) 293T cells were transfected in triplicate with vector or the indicated Cripto constructs and A3-luciferase/FAST-2/CMV-β-Gal as described in Materials and Methods. Cells were left untreated or treated with 100 pM TGF-β1, and resulting luciferase activities were normalized relative to β-galactosidase activities. Values are presented as increase (n-fold) in luciferase activities from TGF-β-treated cells relative to untreated cells. (B) MCF10A cells were infected with lentivirus containing empty vector or the indicated Cripto constructs, and expression of resulting Cripto-FLAG proteins was determined by Western blotting as described in Materials and Methods. Numbers to the left indicate molecular sizes in kilodaltons. (C) MCF10A cells were infected as described for panel B and then were left untreated or treated with 30-pM TGF-β1 doses as indicated. Resulting phospho-Smad2 (pSmad2) and total Smad2 levels were determined by Western blotting as described in Materials and Methods. (D) Phospho-Smad2 bands from panel C were quantitated using densitometry and normalized relative to corresponding Smad2 bands. WT, wild type; IB, immunoblotting.
We next tested whether Cripto could affect TGF-β signaling in MCF10A cells, nontransformed human breast epithelial cells that have undergone spontaneous immortalization and resemble normal mammary epithelial cells (45). These cells are known to be TGF-β responsive (21), and they have been utilized to define the tumor suppressor actions of TGF-β (39, 47, 48). Therefore, we selected this cell line as a suitable physiologic system in which to further explore the effects of Cripto and the two Cripto mutants on TGF-β actions. We infected MCF10A cells with lentivirus containing empty vector or the Cripto constructs and, as shown by Western blotting (Fig. 4B), stably infected cells with the three Cripto forms at similar levels. We then tested the ability of TGF-β to induce Smad2 phosphorylation in these cells. Figure 4C shows that, similarly to what we demonstrated with 293T cells (Fig. 2A), treatment of Cripto-infected MCF10A cells with TGF-β1 resulted in reduced levels of phospho-Smad2 relative to cells infected with virus containing empty vector. TGF-β1 treatment of cells infected with the ΔCFC construct also yielded phospho-Smad2 levels that were lower than levels for empty vector-infected cells (Fig. 4C). However, treatment of cells expressing ΔEGF resulted in phospho-Smad levels similar to those observed for cells infected with empty vector. The results from Fig. 4C were quantitated by measuring the intensities of the phospho-Smad2 bands and then normalizing them relative to the intensities of the corresponding Smad2 bands (Fig. 4D). These data corroborate the luciferase data shown in Fig. 4A and further indicate that the EGF-like domain of Cripto is both necessary and sufficient for Cripto antagonism of TGF-β signaling.
Cripto blocks antiproliferative effects of TGF-β in MCF10A cells.
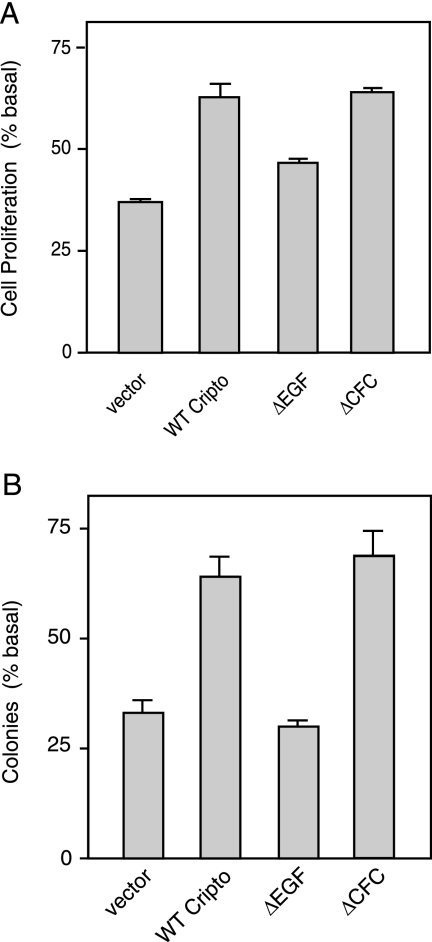
TGF-β has previously been shown to inhibit anchorage-dependent proliferation of human MCF10A cells (21), and we tested whether Cripto could block this effect. Figure 5A shows that following infection of MCF10A cells with lentivirus containing empty vector, TGF-β1 treatment reduced cellular proliferation by ∼62% relative to untreated cells. By contrast, TGF-β1 treatment of MCF10A cells expressing wild-type Cripto caused a decrease of proliferation of only ∼37% compared to untreated cells. We further tested the effects of the ΔEGF and ΔCFC mutants and found that TGF-β1 treatment caused decreases in cellular proliferation of ∼53% and ∼36%, respectively, relative to corresponding untreated cells (Fig. 5A). Therefore, as previously shown in other assays, both wild-type Cripto and the ΔCFC mutant had similar abilities to block the cytostatic effect of TGF-β on MCF10A cells. By contrast, the ΔEGF mutant had a much smaller effect on TGF-β-induced growth inhibition. Together, these data demonstrate that Cripto can attenuate the antiproliferative effects of TGF-β on MCF10A cells and provide further evidence indicating a predominant role for the EGF-like domain of Cripto in mediating TGF-β antagonism.
FIG. 5.
Cripto blocks TGF-β-dependent inhibition of cellular proliferation. MCF10A cells were infected with the indicated lentiviral vectors and then grown under (A) anchorage-dependent or (B) anchorage-independent conditions in the presence of either vehicle or 100 pM TGF-β1 as described in Materials and Methods. (A) Data are presented as the number of cells in the presence of TGF-β1 treatment divided by the number of cells in the absence of TGF-β1 treatment (% basal). (B) Data are presented as the number of colonies counted in the presence of TGF-β1 treatment divided by the number of colonies counted in the absence of TGF-β1 treatment (% basal). WT, wild type.
Although MCF10A cells are considered to be nontransformed (45), they lack expression of the cyclin-dependent kinase inhibitor p15INK4b (21) and they have been shown to form colonies in soft agar at a low frequency (36). We performed soft-agar growth assays and also found that MCF10A cells were able to form colonies following 15 days of culture. Next, we tested whether TGF-β inhibits this anchorage-independent growth of MCF10A cells and, if so, whether this inhibition is affected by Cripto. We seeded MCF10A cells infected with lentivirus containing empty vector or the Cripto constructs in the presence or absence of 100 pM TGF-β1. Figure 5B shows that TGF-β1 was able to substantially inhibit anchorage-independent growth of MCF10A cells containing empty vector, reducing the colony number by ∼67% relative to untreated cells. By contrast, TGF-β1 reduced colony formation in cells infected with Cripto virus by only ∼36% relative to untreated cells (Fig. 5B), indicating that Cripto inhibits the TGF-β-dependent antiproliferative response. This Cripto effect required the EGF-like domain since the ΔEGF mutant had essentially the same effect on TGF-β1 inhibition of colony formation as empty vector. Finally, the CFC domain was not required for this effect since the ΔCFC mutant blocked the TGF-β1 effect on colony formation to the same extent as wild-type Cripto (Fig. 5B). In summary, Cripto reduced TGF-β1-dependent growth inhibition under anchorage-dependent and anchorage-independent conditions and the EGF-like domain appeared to be both necessary and sufficient for these effects.
Reduction of endogenous Cripto expression increases TGF-β signaling.
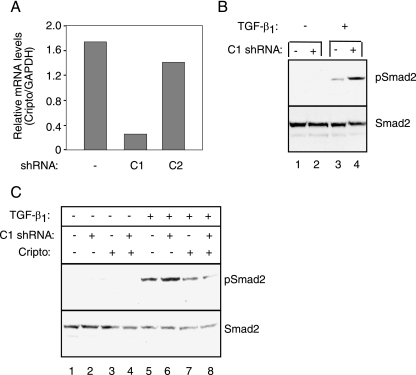
Finally, we tested whether blocking expression of endogenous Cripto could cause an enhancement of TGF-β signaling. Cripto is highly expressed in multiple cancer types, including cervical cancer (3), and here we have targeted Cripto expression in HeLa cells, a cervical carcinoma cell line. We generated lentiviral vectors designed to express shRNAs (44) and then infected HeLa cells with virus containing empty vector or one of two shRNA vectors, designated C1 and C2, each designed to target and reduce expression of human Cripto. Following infection, resulting RNA levels were measured and normalized relative to GAPDH by use of real-time qPCR. Figure 6A shows that both of the Cripto shRNA vectors caused reduction of Cripto mRNA levels relative to empty vector. However, the C1 shRNA had a much more pronounced effect than the C2 construct, reducing Cripto mRNA levels nearly fivefold relative to empty vector (Fig. 6A).
FIG. 6.
Blocking expression of endogenous Cripto by use of siRNA enhances TGF-β signaling. (A) HeLa cells were infected with lentivirus containing either empty vector or vectors (C1 or C2) encoding shRNAs designed to target human Cripto mRNA. Resulting Cripto mRNA levels (Cripto/GAPDH) were determined by qPCR as described in Materials and Methods. (B and C) HeLa cells were infected with empty vector, C1 shRNA, or Cripto lentivirus as indicated and then either left untreated or treated with 10 pM TGF-β1. Phospho-Smad2 (pSmad2) and total Smad2 levels were determined by Western blotting as described in Materials and Methods.
Having shown the ability of the C1 construct to cause reduction of endogenous Cripto expression, we next tested its ability to increase signaling by TGF-β. As shown in Fig. 6B, phospho-Smad2 was detected following treatment with 10 pM TGF-β and the level of phospho-Smad2 at this dose was substantially higher in the C1 shRNA-infected cells than in the cells infected with empty vector (Fig. 6B, compare lanes 3 and 4, top). To confirm the specificity of this effect, we tested whether it could be rescued by overexpression of a Cripto construct lacking the 5′ untranslated region sequence that is targeted by the C1 shRNA. As predicted, the C1 shRNA caused an increase in TGF-β-dependent phospho-Smad2 levels and this effect was completely abolished following the introduction of the exogenous and nontargeted Cripto construct (Fig. 6C, compare lanes 5, 6, and 8). Indeed, and consistent with data presented above (Fig. 2A and Fig. 4C), Cripto inhibited TGF-β-induced Smad2 phosphorylation and this effect appeared to be dominant to the targeting of the endogenous Cripto protein by the C1 shRNA (Fig. 6C, lanes 5, 7, and 8). Therefore, decreasing Cripto expression via siRNA enhances TGF-β signaling, as measured by Smad2 phosphorylation, and this effect is reversed by the overexpression of exogenous Cripto. Importantly, this finding supports a role for endogenous Cripto as a TGF-β inhibitor in a tumor cell line.
Model of Cripto antagonism of TGF-β signaling.
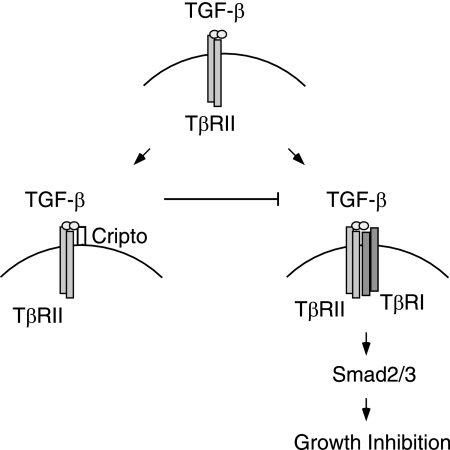
Figure 7 illustrates the mechanism we propose for Cripto antagonism of TGF-β signaling. First, TGF-β binds its type II receptor with high affinity, resulting in a TGF-β-TβRII complex. Subsequent cellular responses, such as Smad phosphorylation and growth inhibition, depend on the ability of the TGF-β-TβRII complex to recruit and activate TβRI (Fig. 7, right). The data presented here suggest a model in which Cripto binds the TGF-β-TβRII complex (Fig. 7, left) in a manner that blocks the recruitment of TβRI, thereby inhibiting TGF-β signaling.
FIG. 7.
Mechanism of Cripto antagonism of TGF-β signaling. A model illustrating the proposed mechanism by which Cripto inhibits TGF-β signaling is shown. By binding TGF-β, Cripto disrupts the ability of TGF-β to recruit its type I receptor, causing a blockade of Smad signaling and responses such as growth inhibition.
DISCUSSION
Tumorigenesis is a multistep process in which cells acquire multiple genetic and/or epigenetic changes leading to unrestricted growth and malignancy. Hanahan and Weinberg (16) enumerated six essential alterations in cell physiology that collectively dictate malignant growth: (i) self-sufficiency in growth signals, (ii) insensitivity to growth-inhibitory (antigrowth) signals, (iii) evasion of programmed cell death (apoptosis), (iv) limitless replicative potential, (v) sustained angiogenesis, and (vi) tissue invasion and metastasis. These authors proposed that most, if not all, cancers share these traits.
Several molecular actions that may contribute to these changes in cell behavior have been attributed to Cripto (46). The results presented here demonstrate the ability of Cripto to cause the second essential alteration listed above by reducing the sensitivity of cells to the cytostatic effects of TGF-β. We have shown that Cripto attenuates multiple TGF-β responses, including reporter induction, Smad phosphorylation, and inhibition of cellular proliferation, under both anchorage-dependent and anchorage-independent conditions. We have further demonstrated that targeted knockdown of Cripto expression results in increased TGF-β signaling. Therefore, endogenous Cripto can mitigate TGF-β signaling, supporting a model in which the high levels of Cripto observed in human cancers promote tumorigenesis by counteracting the growth-inhibitory effects of TGF-β.
Cripto inhibits TGF-β signaling by disrupting receptor assembly.
Cripto binds several members of the TGF-β superfamily and modulates their signaling in at least three distinct ways. First, it acts as an obligate coreceptor for nodals (38), GDF1 (8), and GDF3 (7). Second, it binds Lefty proteins and this interaction blocks Cripto coreceptor function (4). Finally, Cripto binds activins, and in contrast to its coreceptor function, this binding inhibits activin signaling (1, 15).
Here we have attempted to elucidate the molecular mechanism of Cripto antagonism of TGF-β signaling. Our results show that Cripto binds directly to TGF-β and forms complexes containing Cripto, TGF-β, and TβRII. We further demonstrate that Cripto interferes with TGF-β cross-linking to TβRI, suggesting it disrupts receptor assembly by competing with TβRI for binding to TGF-β-TβRII complexes. This is similar to our previous finding that Cripto prevents cross-linking of activin-A to ALK4 by forming inhibitory complexes containing Cripto, activin-A, and activin type II receptors (15). A key difference between these two cases, however, is that ALK4 independently binds the CFC domain of Cripto whereas TβRI does not (59, 60). In this regard, ALK4 is associated with complexes containing Cripto, activin-A, and activin type II receptors through its ability to bind directly to Cripto (15). Conversely, TβRI lacks such an ability to associate with analogous complexes containing Cripto, TGF-β, and TβRII. Indeed, and in contrast to corresponding cross-linking experiments conducted with activin-A (15), no labeled Cripto was detected in this study following anti-TβRI immunoprecipitation. Therefore, although Cripto inhibits cross-linking of activin-A and TGF-β to their respective type I receptors, it appears to form distinct inhibitory complexes that either contain ALK4 or exclude TβRI. Future studies will focus on determining the functional significance of this difference.
The EGF-like domain of Cripto binds TGF-β and inhibits TGF-β signaling.
Cripto has diverse signaling functions that have been attributed to its EGF-like and CFC domains. The EGF-like domain, for example, can act as a soluble growth factor and is sufficient to activate mitogenic signaling pathways (26). In addition, this domain independently binds TGF-β superfamily members, including nodals (30, 59, 60), GDF1 (8), and GDF3 (7), as well as Lefty proteins, which are structurally divergent TGF-β ligands that antagonize nodal/GDF1/GDF3 signaling through competition for binding to EGF-CFC proteins (4). Finally, the EGF-like domain binds activin-A and is necessary and sufficient for Cripto antagonism of activin-A signaling (15).
By contrast, fewer binding partners and functions have been attributed directly to the CFC domain. The most widely documented role of the CFC domain is its ability to bind directly to the extracellular domain of ALK4, and loss of this binding abolishes EGF-CFC coreceptor function (30, 59, 60). The importance of this interaction is supported by the fact that the nodal antagonist tomoregulin inhibits Cripto coreceptor function by binding the CFC domain and preventing it from binding ALK4 (17). The CFC domain has also been reported to bind directly to activin-B, but not activin-A, to cause antagonism of activin-B signaling (1). This finding suggests that Cripto antagonizes activin-A and activin-B via distinct mechanisms (40) and is noteworthy since it may allow Cripto to discriminate between these two ligands and modulate their activities to differing degrees. In this regard, a greater inhibitory effect of Cripto on activin-B signaling than on activin-A signaling could explain the finding that activin-B cannot fully substitute for activin-A during embryonic development (6). In addition, the binding of activin-B to the CFC domain of Cripto appears to be unique among TGF-β ligands since, as mentioned above, nodals (30, 59, 60), GDF1 (8), GDF3 (7), Leftys (4), and activin-A (15) are each thought to bind to the EGF-like domain.
As is clear from the discussion above, Cripto binds several TGF-β ligands and can play distinct and sometimes opposing roles in modulating their function. Also, both the EGF-like domain and the CFC domain have been implicated in distinct functions that are ligand specific. Therefore, the roles of these domains in modulating the function of TGF-β ligands cannot be predicted a priori. With this in mind, we have tested the roles of the EGF-like and CFC domains in TGF-β binding and effects on TGF-β signaling. We found that a Cripto mutant consisting of the EGF-like domain but not the CFC domain efficiently bound TGF-β, as visualized in cross-linking assays, and that it also inhibited TGF-β cross-linking to TβRI (Fig. 3). By contrast, the CFC domain alone bound TGF-β poorly and had little or no effect on TGF-β cross-linking to TβRI. This led us to predict that the EGF-like domain would also be sufficient to reduce TGF-β signaling. Indeed, we found that wild-type Cripto and the ΔCFC mutant inhibited TGF-β to similar extents in each of the functional assays we employed, confirming that the CFC domain was dispensable for these effects. Likewise, and consistent with our cross-linking data, the CFC domain (ΔEGF mutant) was unable or poorly able to block TGF-β signaling in the same assays, illustrating the importance of the EGF-like domain in this context. In light of these data, we propose a model in which the EGF-like domain of Cripto competes with TβRI for binding to the TGF-β-TβRII complex, resulting in reduced TGF-β signaling.
Antagonism of TGF-β signaling provides a novel mechanism of Cripto-induced tumorigenesis.
Several reports have supported a role for Cripto in promoting tumor growth. For example, overexpression of Cripto causes anchorage-independent growth and partial transformation of NOG-8 (9) and CID-9 (32) mammary epithelial cells. It has also been shown previously that targeted disruption of Cripto expression abrogates the transformed phenotype in colon carcinoma cells (10). Cripto can further promote epithelial-to-mesenchymal transition and cellular migration, suggesting it may play a role in promoting tumor metastasis (33, 51). Moreover, and in addition to these in vitro data, it has been shown that monoclonal antibodies targeting either the CFC domain (1) or the EGF-like domain (57) of Cripto can inhibit tumor growth in vivo. Finally, a role for Cripto in promoting tumor growth is supported by the fact that transgenic mice in which Cripto expression was driven by the murine mammary tumor virus promoter displayed ductal hyperplasia and mammary tumor formation (52).
TGF-β is also well established as a regulator of tumor progression and controls tissue homeostasis by inhibiting proliferation of multiple cell types, including most epithelial cells. The importance of this action is underscored by the fact that disruptions/alterations in the TGF-β signaling pathway have been observed frequently in numerous human cancers (43). However, under these circumstances complete loss of key signaling components, such as receptors and Smad proteins, is rarely observed. Rather, tumor cells are thought to escape from the cytostatic effects of TGF-β under conditions in which Smad2/3 signaling is reduced but not absent (2, 11, 37, 49). Here, we have provided evidence that Cripto can create such conditions by acting as an inhibitor of TGF-β signaling. Therefore, in conclusion, we propose that the high levels of Cripto present in human cancers may contribute to tumorigenesis by restraining the growth-inhibitory effects of TGF-β.
Acknowledgments
We thank Craig Harrison for important contributions to the early phases of this work. We also thank Sandra Guerra and Dave Dalton for providing assistance in preparing the manuscript.
This work was supported by the Foundation for Medical Research, Inc., the Robert J., Jr., and Helen C. Kleberg Foundation, NIH grant CA107420-01, and the International Human Frontier Science Program Organization.
W. Vale is a senior investigator of the Foundation for Medical Research, Inc.
Footnotes
Published ahead of print on 9 October 2006.
REFERENCES
- 1.Adkins, H. B., C. Bianco, S. G. Schiffer, P. Rayhorn, M. Zafari, A. E. Cheung, O. Orozco, D. Olson, A. De Luca, L. L. Chen, K. Miatkowski, C. Benjamin, N. Normanno, K. P. Williams, M. Jarpe, D. LePage, D. Salomon, and M. Sanicola. 2003. Antibody blockade of the Cripto CFC domain suppresses tumor cell growth in vivo. J. Clin. Investig. 112:575-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akhurst, R. J., and R. Derynck. 2001. TGF-beta signaling in cancer—a double-edged sword. Trends Cell Biol. 11:S44-S51. [DOI] [PubMed] [Google Scholar]
- 3.Bianco, C., L. Strizzi, N. Normanno, N. Khan, and D. S. Salomon. 2005. Cripto-1: an oncofetal gene with many faces. Curr. Top. Dev. Biol. 67:85-133. [DOI] [PubMed] [Google Scholar]
- 4.Branford, W. W., and H. J. Yost. 2004. Nodal signaling: CrypticLefty mechanism of antagonism decoded. Curr. Biol. 14:R341-R343. [DOI] [PubMed] [Google Scholar]
- 5.Brivanlou, A. H., F. H. Gage, R. Jaenisch, T. Jessell, D. Melton, and J. Rossant. 2003. Stem cells. Setting standards for human embryonic stem cells. Science 300:913-916. [DOI] [PubMed] [Google Scholar]
- 6.Brown, C. W., D. E. Houston-Hawkins, T. K. Woodruff, and M. M. Matzuk. 2000. Insertion of Inhbb into the Inhba locus rescues the Inhba-null phenotype and reveals new activin functions. Nat. Genet. 25:453-457. [DOI] [PubMed] [Google Scholar]
- 7.Chen, C., S. M. Ware, A. Sato, D. E. Houston-Hawkins, R. Habas, M. M. Matzuk, M. M. Shen, and C. W. Brown. 2006. The Vg1-related protein Gdf3 acts in a Nodal signaling pathway in the pre-gastrulation mouse embryo. Development 133:319-329. [DOI] [PubMed] [Google Scholar]
- 8.Cheng, S. K., F. Olale, J. T. Bennett, A. H. Brivanlou, and A. F. Schier. 2003. EGF-CFC proteins are essential coreceptors for the TGF-beta signals Vg1 and GDF1. Genes Dev. 17:31-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ciardiello, F., R. Dono, N. Kim, M. G. Persico, and D. S. Salomon. 1991. Expression of cripto, a novel gene of the epidermal growth factor gene family, leads to in vitro transformation of a normal mouse mammary epithelial cell line. Cancer Res. 51:1051-1054. [PubMed] [Google Scholar]
- 10.Ciardiello, F., G. Tortora, C. Bianco, M. P. Selvam, F. Basolo, G. Fontanini, F. Pacifico, N. Normanno, R. Brandt, M. G. Persico, et al. 1994. Inhibition of CRIPTO expression and tumorigenicity in human colon cancer cells by antisense RNA and oligodeoxynucleotides. Oncogene 9:291-298. [PubMed] [Google Scholar]
- 11.Derynck, R., R. J. Akhurst, and A. Balmain. 2001. TGF-beta signaling in tumor suppression and cancer progression. Nat. Genet. 29:117-129. [DOI] [PubMed] [Google Scholar]
- 12.Ding, J., L. Yang, Y. T. Yan, A. Chen, N. Desai, A. Wynshaw-Boris, and M. M. Shen. 1998. Cripto is required for correct orientation of the anterior-posterior axis in the mouse embryo. Nature 395:702-707. [DOI] [PubMed] [Google Scholar]
- 13.Ebert, A. D., C. Wechselberger, S. Frank, B. Wallace-Jones, M. Seno, I. Martinez-Lacaci, C. Bianco, M. De Santis, H. K. Weitzel, and D. S. Salomon. 1999. Cripto-1 induces phosphatidylinositol 3′-kinase-dependent phosphorylation of AKT and glycogen synthase kinase 3beta in human cervical carcinoma cells. Cancer Res. 59:4502-4505. [PubMed] [Google Scholar]
- 14.Gray, P. C., J. Greenwald, A. L. Blount, K. S. Kunitake, C. J. Donaldson, S. Choe, and W. Vale. 2000. Identification of a binding site on the type II activin receptor for activin and inhibin. J. Biol. Chem. 275:3206-3212. [DOI] [PubMed] [Google Scholar]
- 15.Gray, P. C., C. A. Harrison, and W. Vale. 2003. Cripto forms a complex with activin and type II activin receptors and can block activin signaling. Proc. Natl. Acad. Sci. USA 100:5193-5198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanahan, D., and R. A. Weinberg. 2000. The hallmarks of cancer. Cell 100:57-70. [DOI] [PubMed] [Google Scholar]
- 17.Harms, P. W., and C. Chang. 2003. Tomoregulin-1 (TMEFF1) inhibits nodal signaling through direct binding to the nodal coreceptor Cripto. Genes Dev. 17:2624-2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harrison, C. A., P. C. Gray, S. C. Koerber, W. Fischer, and W. Vale. 2003. Identification of a functional binding site for activin on the type I receptor ALK4. J. Biol. Chem. 278:21129-21135. [DOI] [PubMed] [Google Scholar]
- 19.Harrison, C. A., P. C. Gray, W. W. Vale, and D. M. Robertson. 2005. Antagonists of activin signaling: mechanisms and potential biological applications. Trends Endocrinol. Metab. 16:73-78. [DOI] [PubMed] [Google Scholar]
- 20.Hogan, B. L. 1996. Bone morphogenetic proteins: multifunctional regulators of vertebrate development. Genes Dev. 10:1580-1594. [DOI] [PubMed] [Google Scholar]
- 21.Iavarone, A., and J. Massague. 1997. Repression of the CDK activator Cdc25A and cell-cycle arrest by cytokine TGF-beta in cells lacking the CDK inhibitor p15. Nature 387:417-422. [DOI] [PubMed] [Google Scholar]
- 22.Kannan, S., M. De Santis, M. Lohmeyer, D. J. Riese II, G. H. Smith, N. Hynes, M. Seno, R. Brandt, C. Bianco, G. Persico, N. Kenney, N. Normanno, I. Martinez-Lacaci, F. Ciardiello, D. F. Stern, W. J. Gullick, and D. S. Salomon. 1997. Cripto enhances the tyrosine phosphorylation of Shc and activates mitogen-activated protein kinase (MAPK) in mammary epithelial cells. J. Biol. Chem. 272:3330-3335. [DOI] [PubMed] [Google Scholar]
- 23.Kirkbride, K. C., B. N. Ray, and G. C. Blobe. 2005. Cell-surface co-receptors: emerging roles in signaling and human disease. Trends Biochem. Sci. 30:611-621. [DOI] [PubMed] [Google Scholar]
- 24.Lebrin, F., M. J. Goumans, L. Jonker, R. L. Carvalho, G. Valdimarsdottir, M. Thorikay, C. Mummery, H. M. Arthur, and P. ten Dijke. 2004. Endoglin promotes endothelial cell proliferation and TGF-beta/ALK1 signal transduction. EMBO J. 23:4018-4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lewis, K. A., P. C. Gray, A. L. Blount, L. A. MacConell, E. Wiater, L. M. Bilezikjian, and W. Vale. 2000. Betaglycan binds inhibin and can mediate functional antagonism of activin signalling. Nature 404:411-414. [DOI] [PubMed] [Google Scholar]
- 26.Lohmeyer, M., P. M. Harrison, S. Kannan, M. DeSantis, N. J. O'Reilly, M. J. Sternberg, D. S. Salomon, and W. J. Gullick. 1997. Chemical synthesis, structural modeling, and biological activity of the epidermal growth factor-like domain of human cripto. Biochemistry 36:3837-3845. [DOI] [PubMed] [Google Scholar]
- 27.López-Casillas, F., S. Cheifetz, J. Doody, J. L. Andres, W. S. Lane, and J. Massagué. 1991. Structure and expression of the membrane proteoglycan betaglycan, a component of the TGF-β receptor system. Cell 67:785-795. [DOI] [PubMed] [Google Scholar]
- 28.López-Casillas, F., J. L. Wrana, and J. Massague. 1993. Betaglycan presents ligand to the TGF beta signaling receptor. Cell 73:1435-1444. [DOI] [PubMed] [Google Scholar]
- 29.Massague, J. 1998. TGF-beta signal transduction. Annu. Rev. Biochem. 67:753-791. [DOI] [PubMed] [Google Scholar]
- 30.Minchiotti, G., G. Manco, S. Parisi, C. T. Lago, F. Rosa, and M. G. Persico. 2001. Structure-function analysis of the EGF-CFC family member Cripto identifies residues essential for nodal signalling. Development 128:4501-4510. [DOI] [PubMed] [Google Scholar]
- 31.Miyoshi, H., U. Blomer, M. Takahashi, F. H. Gage, and I. M. Verma. 1998. Development of a self-inactivating lentivirus vector. J. Virol. 72:8150-8157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Niemeyer, C. C., M. G. Persico, and E. D. Adamson. 1998. Cripto: roles in mammary cell growth, survival, differentiation and transformation. Cell Death Differ. 5:440-449. [DOI] [PubMed] [Google Scholar]
- 33.Normanno, N., A. De Luca, C. Bianco, M. R. Maiello, M. V. Carriero, A. Rehman, C. Wechselberger, C. Arra, L. Strizzi, M. Sanicola, and D. S. Salomon. 2004. Cripto-1 overexpression leads to enhanced invasiveness and resistance to anoikis in human MCF-7 breast cancer cells. J. Cell. Physiol. 198:31-39. [DOI] [PubMed] [Google Scholar]
- 34.Onichtchouk, D., Y. G. Chen, R. Dosch, V. Gawantka, H. Delius, J. Massague, and C. Niehrs. 1999. Silencing of TGF-beta signalling by the pseudoreceptor BAMBI. Nature 401:480-485. [DOI] [PubMed] [Google Scholar]
- 35.Pece-Barbara, N., S. Vera, K. Kathirkamathamby, S. Liebner, G. M. Di Guglielmo, E. Dejana, J. L. Wrana, and M. Letarte. 2005. Endoglin null endothelial cells proliferate faster and are more responsive to transforming growth factor beta 1 with higher affinity receptors and an activated Alk1 pathway. J. Biol. Chem. 280:27800-27808. [DOI] [PubMed] [Google Scholar]
- 36.Rasmussen, S. B., E. Kordon, R. Callahan, and G. H. Smith. 2001. Evidence for the transforming activity of a truncated Int6 gene, in vitro. Oncogene 20:5291-5301. [DOI] [PubMed] [Google Scholar]
- 37.Roberts, A. B., and L. M. Wakefield. 2003. The two faces of transforming growth factor beta in carcinogenesis. Proc. Natl. Acad. Sci. USA 100:8621-8623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schier, A. F. 2003. Nodal signaling in vertebrate development. Annu. Rev. Cell Dev. Biol. 19:589-621. [DOI] [PubMed] [Google Scholar]
- 39.Seton-Rogers, S. E., and J. S. Brugge. 2004. ErbB2 and TGF-beta: a cooperative role in mammary tumor progression? Cell Cycle 3:597-600. [PubMed] [Google Scholar]
- 40.Shen, M. M. 2003. Decrypting the role of Cripto in tumorigenesis. J. Clin. Investig. 112:500-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shen, M. M., and A. F. Schier. 2000. The EGF-CFC gene family in vertebrate development. Trends Genet. 16:303-309. [DOI] [PubMed] [Google Scholar]
- 42.Shi, Y., and J. Massague. 2003. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell 113:685-700. [DOI] [PubMed] [Google Scholar]
- 43.Siegel, P. M., and J. Massague. 2003. Cytostatic and apoptotic actions of TGF-beta in homeostasis and cancer. Nat. Rev. Cancer 3:807-821. [DOI] [PubMed] [Google Scholar]
- 44.Singer, O., R. A. Marr, E. Rockenstein, L. Crews, N. G. Coufal, F. H. Gage, I. M. Verma, and E. Masliah. 2005. Targeting BACE1 with siRNAs ameliorates Alzheimer disease neuropathology in a transgenic model. Nat. Neurosci. 8:1343-1349. [DOI] [PubMed] [Google Scholar]
- 45.Soule, H. D., T. M. Maloney, S. R. Wolman, W. D. Peterson, Jr., R. Brenz, C. M. McGrath, J. Russo, R. J. Pauley, R. F. Jones, and S. C. Brooks. 1990. Isolation and characterization of a spontaneously immortalized human breast epithelial cell line, MCF-10. Cancer Res. 50:6075-6086. [PubMed] [Google Scholar]
- 46.Strizzi, L., C. Bianco, N. Normanno, and D. Salomon. 2005. Cripto-1: a multifunctional modulator during embryogenesis and oncogenesis. Oncogene 24:5731-5741. [DOI] [PubMed] [Google Scholar]
- 47.Tian, F., S. D. Byfield, W. T. Parks, C. H. Stuelten, D. Nemani, Y. E. Zhang, and A. B. Roberts. 2004. Smad-binding defective mutant of transforming growth factor beta type I receptor enhances tumorigenesis but suppresses metastasis of breast cancer cell lines. Cancer Res. 64:4523-4530. [DOI] [PubMed] [Google Scholar]
- 48.Tian, F., S. DaCosta Byfield, W. T. Parks, S. Yoo, A. Felici, B. Tang, E. Piek, L. M. Wakefield, and A. B. Roberts. 2003. Reduction in Smad2/3 signaling enhances tumorigenesis but suppresses metastasis of breast cancer cell lines. Cancer Res. 63:8284-8292. [PubMed] [Google Scholar]
- 49.Wakefield, L. M., and A. B. Roberts. 2002. TGF-beta signaling: positive and negative effects on tumorigenesis. Curr. Opin. Genet. Dev. 12:22-29. [DOI] [PubMed] [Google Scholar]
- 50.Wang, X. F., H. Y. Lin, E. E. Ng, J. Downward, H. F. Lodish, and R. A. Weinberg. 1991. Expression cloning and characterization of the TGF-beta type III receptor. Cell 67:797-805. [DOI] [PubMed] [Google Scholar]
- 51.Wechselberger, C., A. D. Ebert, C. Bianco, N. I. Khan, Y. Sun, B. Wallace-Jones, R. Montesano, and D. S. Salomon. 2001. Cripto-1 enhances migration and branching morphogenesis of mouse mammary epithelial cells. Exp. Cell Res. 266:95-105. [DOI] [PubMed] [Google Scholar]
- 52.Wechselberger, C., L. Strizzi, N. Kenney, M. Hirota, Y. Sun, A. Ebert, O. Orozco, C. Bianco, N. I. Khan, B. Wallace-Jones, N. Normanno, H. Adkins, M. Sanicola, and D. S. Salomon. 2005. Human Cripto-1 overexpression in the mouse mammary gland results in the development of hyperplasia and adenocarcinoma. Oncogene 24:4094-4105. [DOI] [PubMed] [Google Scholar]
- 53.Welt, C., Y. Sidis, H. Keutmann, and A. Schneyer. 2002. Activins, inhibins, and follistatins: from endocrinology to signaling. A paradigm for the new millennium. Exp. Biol. Med. (Maywood) 227:724-752. [DOI] [PubMed] [Google Scholar]
- 54.Wiater, E., and W. Vale. 2003. Inhibin is an antagonist of bone morphogenetic protein signaling. J. Biol. Chem. 278:7934-7941. [DOI] [PubMed] [Google Scholar]
- 55.Wrana, J. L., L. Attisano, J. Carcamo, A. Zentella, J. Doody, M. Laiho, X. F. Wang, and J. Massague. 1992. TGF beta signals through a heteromeric protein kinase receptor complex. Cell 71:1003-1014. [DOI] [PubMed] [Google Scholar]
- 56.Wrana, J. L., L. Attisano, R. Wieser, F. Ventura, and J. Massague. 1994. Mechanism of activation of the TGF-beta receptor. Nature 370:341-347. [DOI] [PubMed] [Google Scholar]
- 57.Xing, P. X., X. F. Hu, G. A. Pietersz, H. L. Hosick, and I. F. McKenzie. 2004. Cripto: a novel target for antibody-based cancer immunotherapy. Cancer Res. 64:4018-4023. [DOI] [PubMed] [Google Scholar]
- 58.Xu, C., G. Liguori, M. G. Persico, and E. D. Adamson. 1999. Abrogation of the Cripto gene in mouse leads to failure of postgastrulation morphogenesis and lack of differentiation of cardiomyocytes. Development 126:483-494. [DOI] [PubMed] [Google Scholar]
- 59.Yan, Y.-T., J.-J. Liu, Y. Luo, E. Chaosu, R. S. Haltiwanger, C. Abate-Shen, and M. M. Shen. 2002. Dual roles of Cripto as a ligand and coreceptor in the nodal signaling pathway. Mol. Cell. Biol. 22:4439-4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yeo, C., and M. Whitman. 2001. Nodal signals to Smads through Cripto-dependent and Cripto-independent mechanisms. Mol. Cell 7:949-957. [DOI] [PubMed] [Google Scholar]