Abstract
The histone acetyltransferase (HAT) Gcn5 plays a role in chromatin structure and gene expression regulation as a catalytic component of multiprotein complexes, some of which also contain Ada2-type transcriptional coactivators. Data obtained mostly from studies on yeast (Saccharomyces cerevisiae) suggest that Ada2 potentiates Gcn5 activity and substrate recognition. dAda2b, one of two related Ada2 proteins of Drosophila melanogaster, was recently found to play a role in complexes acetylating histone 3 (H3). Evidence of an in vivo functional link between the related coactivator dAda2a and dGcn5, however, is lacking. Here we present data on the genetic interaction of dGcn5 and dAda2a. The loss of either dGcn5 or dAda2a function results in similar chromosome structural and developmental defects. In dAda2a mutants, the nucleosomal H4 acetylation at lysines 12 and 5 is significantly reduced, while the acetylation established by dAda2b-containing Gcn5 complexes at H3 lysines 9 and 14 is unaffected. The data presented here, together with our earlier data on the function of dAda2b, provide evidence that related Ada2 proteins of Drosophila, together with Gcn5 HAT, are involved in the acetylation of specific lysine residues in the N-terminal tails of nucleosomal H3 and H4. Our data suggest dAda2a involvement in both uniformly distributed H4 acetylation and gene-specific transcription regulation.
Histone acetyltransferases (HATs) are present in most eukaryotes, playing important roles in controlling the diverse functions of the genome. Almost invariably, nucleosomal HATs are constituents of multiprotein complexes, in which accessory factors potentiate their activity, modify their substrate specificity, and play roles in targeting the complexes to specific chromosomal sites by providing surfaces for versatile protein-protein interactions (5, 13, 14, 33). Relatives of Gcn5 (general control nonrepressed protein 5), the first HAT enzyme linked to transcription activation (10), have been identified in numerous complexes ranging from yeasts (Saccharomyces cerevisiae) to humans. Most of the Gcn5-containing HAT complexes also share Ada2- and Ada3-type adaptor proteins. In fact, in a genetic screen conducted with Saccharomyces cerevisiae for coactivators required to mediate the transcriptional activation effect of strong acidic activators, Gcn5, together with a number of other genes, was identified as having an alteration/deficiency in activation (ADA) function (6). Subsequently, at least three Gcn5-containing complexes of yeast, SAGA (SPT3-TAF-GCN5 acetylase), ADA2, and A2, were separated and shown to have distinct properties (16, 30, 31).
Recently, we and others found that, in contrast with the single Ada2 gene present in Saccharomyces cerevisiae, the Drosophila melanogaster genome contains two genes, referred to as dAda2a and dAda2b, encoding related Ada2 proteins (21, 24). In several other metazoan organisms, including mouse, human, and Arabidopsis thaliana, there are also two Ada2-type coactivators (4, 11, 34).
Biochemical separation of Ada2-containing Drosophila complexes indicated that dAda2a is present in a smaller (0.8 MDa) complex and dAda2b is present in a larger (2 MDa) complex, most probably corresponding to Drosophila ADA (dADA) and Drosophila SAGA (dSAGA), respectively (21, 24, 25). Recently, Guelman et al. reported the biochemical separation of a further dAda2a-dGcn5-containing complex, ATAC (18). Other variants of Gcn5-containing HAT complexes have also been identified in both yeast and mammalian cells. They include SALSA (SAGA altered and Spt8 absent) (32), SLIK (SAGA-like) (28), and STAGA (SPT3-TAF-GCN5 acetylase) (23). In all of these, the Gcn5 and Ada2 proteins are common constituents (for a review, see reference 13), which raises the question of how the functional divergence of these complexes is determined.
In a number of independent studies, a direct physical interaction between Ada2 and Gcn5 has been demonstrated both in vitro and in vivo, and the SANT domain of Ada2 has been implicated in the interaction (8, 22, 34). Ada2 also associates physically with TATA binding protein and a number of acidic activators (3). Numerous studies on the Ada2-Gcn5 interaction thus suggest that Ada2 is required for the assembly of Gcn5-containing complexes, within which it is involved in activator and TATA binding protein recruitment and, by virtue of its SANT domain, also effects Gcn5 activity (1-3, 7, 22, 34, 35). Accordingly, the deletion of the Ada2 SANT domain has the same negative effect as that of Gcn5 deletion on the activation of the yeast PHO promoter (2).
Recombinant yeast Gcn5 (yGcn5) acetylates free histone 3 (H3), but exhibits little activity towards histones assembled into nucleosome particles. Together with Ada2 and Ada3 proteins, yGcn5 acetylates an expanded set of lysines and exhibits a preference for the lysines of nucleosomal H3 substrate. The results of in vitro acetylation assays performed using H3 peptides and nucleosomal H3 as a substrate led to the conclusion that the Ada2 and Ada3 proteins play roles in enhancing Gcn5 HAT activity and determining the enzyme substrate specificity (17, 21). yGcn5 mutations were also found to reduce acetylation at each of the four acetylation sites of the H4 N terminus (36).
Mutant alleles of Drosophila Ada2a, Ada2b, and Gcn5 have recently been reported (12, 25, 29). Immunostaining of polytene chromosomes with antibodies specific for differently acetylated forms of H3 and H4 revealed that, with the lack of dAda2b, the acetylation of H3 K9 and K14 is greatly reduced (25, 29). However, dAda2b mutations did not affect the acetylation level of H4. Carre et al. found that dGcn5 mutations abolished the K9 and K14 acetylation of H3 but had no effect on H4 K8 acetylation (12). While these studies significantly extended the knowledge on the function of Gcn5-containing Drosophila HAT complexes, they also left several questions unanswered. Most importantly, is there indeed a functional link between dGcn5 and dAda2a? To address this question, we studied the dAda2a-dGcn5 genetic interaction. After establishing a functional link between the two factors in vivo, we extended our previous studies on the acetylation state of nucleosomal histones in dAda2 mutants to reveal that dAda2a is involved in the acetylation of lysines 5 and 12 of H4. The data presented here, combined with recent results reported by ourselves and others, demonstrate that related adaptor proteins present in several metazoan organisms can provide HAT complexes with functional diversity by targeting them to different histone residues.
MATERIALS AND METHODS
Recombinant DNA and quantitative real-time PCR (Q-RT-PCR).
The transgene constructs pCaSpeR4-DtlAda2aRpb4 (P[DtlAdaRpb4]), pCaSpeR4-DtlRpb4 (P[DtlRpb4]), and pCaSpeR4-DtlAda2a (P[DtlAda2a]) were described earlier (20, 25). P[DtlAdaRpb4] carries the entire Ada2a/Rpb4/Dtl locus, P[DtlRpb4] has a nonsense mutation in the dAda2a coding region, and from P[DtlAda2a], the Rpb4 region is deleted. The upstream activator sequence promoter-driven dGcn5 transgene was generated by the insertion of a cDNA fragment from clone LD17356 (generated in the Berkeley EST sequencing project), encompassing the dGcn5 coding region, into the P element insertion vector pUAST with the help of PCR. The structure of the plasmid thus obtained was verified by nucleotide sequencing.
Plasmid constructs for yeast two-hybrid experiments were generated using pBTM116 and pGAD424 vectors and PCR-generated cDNA fragments of the desired genes. Detailed descriptions of the constructs used are available on request. Two-hybrid experiments were performed as described in the Clontech manual.
For the quantitative determination of transcripts of early-response ecdysone genes, larvae were staged at the second- to third-instar molt, and 42 h later (which corresponds to the time of spiracle eversion in wild types), total RNA was isolated with the QIAGEN RNeasy kit according to the manufacturer's instructions. First-strand cDNA was synthetized from 1 μg RNA by using TaqMan reverse transcription reagent (ABI). The relative abundances of Broad-Complex-, E74A-, and E75A-specific mRNAs were quantified by Q-RT-PCR (ABI Prism 7300) using 18S rRNA as control. The primers for ecdysone early-response transcripts were as follows: BR-C L, GCCCTGGTGGAGTTCATCTA; BR-C R, CAGATGGCTGTGTGTGTCCT; Eip74 AL, GTTGCCGGAACATTATGGAT; Eip74 AR, ATCAGCCGAATTGTCAATCA; Eip75 AL, GCGGTCCAGAATCAGCAG; and Eip75 AR, GAGGATGTGGAGGAGGATGA. 18S rRNA-specific primers 18Fw and 18Rev have been described previously (25). Cycle threshold values were set against a calibration curve ranging over 2 orders of magnitude.
Drosophila stocks, genetic crosses and phenotype analysis.
Fly stocks were raised, and crosses were performed at 25°C on standard medium containing nipagin. Deletion 189, encompassing the Ada2a/Rpb4/Dtl locus, has been described previously (20, 25). dGcn5-null and hypomorph alleles dGcn5E333st and dGcn5C137T were kindly provided by C. Antoniewski (12). The dGcn55 allele was generated by remobilizing a P element located in line CB-0434-3R at approximately 5 kb downstream to the dGcn5 coding region within the best gene. Improper jump-outs were first selected by genetic crosses and then characterized by PCR amplification of fragments corresponding to the dGcn5 coding region. Finally, a fragment containing the endpoints of the deletion was sequenced to determine that dGcn55 is a null allele in which a deletion removed two-thirds of the dGcn5 coding region and part of the adjacent best gene.
dAda2a and dGcn5 alleles were kept as heterozygotes with TM6C, Tb Sb, or T(2;3)TSTL, Cy; Tb Hu balancer chromosomes and mutants were selected on the basis of the Tb+ phenotype.
To produce dAda2a dGcn5 double mutants, dGcn5E333st and d189 or, alternatively, dGcn5C137T and d189 alleles were recombined into the same chromosome.
dAda2a-null dGcn5 hypomorph mutants (P[DtlRpb4]/+; dGcn5E333std189/dGcn5C137Td189) were obtained from crosses P[DtlRpb4]; dGcn5E333std189/TM6C × dGcn5C137Td189/TM6C. The overexpression of Gcn5 in an Ada2a mutant background was achieved by crossing P[act-GAL4]; P[DtlRpb4]d189/T(2;3)TSTL to P[UAS-Gcn5]; d189/TM6C. The genotypes P[DtlRpb4]/+; P[DtlAda2a]79/1d189/d189 and +/+; P[DtlAda2a]79/1d189/P[DtlRpb4]d189 represent dAda2a hypomorphs and for simplicity are labeled dAda2ahyp1 and dAda2ahyp2, respectively. dAda2ahyp1 in a wild-type or heterozygous dGcn5 background was obtained from the crosses P[DtlRpb4]; P[DtlAda2a]79/1 d189/TM6C × d189/TM6C and P[DtlRpb4]; P[DtlAda2a]79/1 d189/TM6C × dGcn5E333std189/TM6C, respectively. In order to obtain dAda2ahyp2 in combination with normal or overexpressed dGcn5 levels, the following crosses were performed: P[DtlAda2a]79/1d189/TM6C × P[act-GAL4]; P[DtlRpb4]d189/T(2;3)TSTL and P[UAS-Gcn5]; P[DtlAda2a]79/1d189/TM6C × P[act-GAL4]; P[DtlRpb4]d189/T(2;3)TSTL, respectively. Transgene carrier lines were generated by injection of embryos and selection for the mini white marker in the offspring as described earlier (20, 25).
Immunohistochemistry.
H3 and H4 acetylation in dAda2a, dAda2b, and dGcn5 mutants and wild-type animals was compared by the immunostaining of polytene chromosomes obtained from the salivary glands of wandering larvae (or dAda2a larvae corresponding to this stage, according to their age). H3-AcK14-, H3-AcK9-, and H4-AcK8-specific antibodies were from Upstate; H4-AcK5 and H4-AcK12 were from ABCAM and from SEROTEC. Ecdysone receptor antibody (EcRB) was from the Developmental Studies Hybridoma Bank, University of Iowa. Mouse anti-polymerase II (Pol II) (7G5) antibodies were raised against specific peptides as reported previously (15). The secondary antibodies were Alexa Fluor 488-conjugated goat anti-mouse immunoglobulin and Alexa Fluor 555-conjugated goat anti-rabbit immunoglobulin (Molecular Probes). Primary antibodies were used at 100- to 500-fold and secondary antibodies at 500-fold dilutions. Polytene chromosome preparation and immunostaining were performed as described in reference 27.
In vitro and in vivo ecdysone treatments.
For in vitro ecdysone treatment, larvae were synchronized at the second- to third-instar molt and collected 24 h later at mid-L3 stage. The salivary glands were removed and placed into Robb medium. Each gland was divided into two parts; one part was ecdysone treated, and the other was mock treated. For ecdysone treatment, 20 μM 20-OH-ecdysone (Sigma) was added to the medium and the lobes of the glands were incubated at 25°C for 2 h. Following incubation, ecdysone and mock-treated control salivary glands were used to prepare polytene chromosome squashes. Chromosome preparations were visualized under a phase-contrast microscope and photographed, and the widths of the puffs were determined by comparison with a nearby band as reference. Data were analyzed by averaging the widths of puffs observed in ecdysone-treated and mock-treated lobes of the same glands.
For in vivo ecdysone treatment, L3 larvae were placed on autoclaved yeast containing 1 mM 20-OH-ecdysone.
RESULTS
Genetic interaction between dAda2a and dGcn5.
We isolated a small deletion (d189) in the 90F cytological region of the Drosophila melanogaster chromosome, which removes the regulatory regions of the adjacent dAda2a/Rpb4 and Dtl genes and results in an early larva lethal phenotype (Fig. 1) (20, 25). With the use of transgenes corresponding to specific functions affected by the deletion, we determined that the loss of Rpb4 and Dtl function results in L1 and L2 lethality, respectively (Fig. 1) (20, 25, 26). When a transgene (P[DtlRpb4]) encompassing both transcription units with a stop codon in the dAda2a coding region is introduced into the d189 background, the Rpb4 and Dtl functions are restored and the transgene carriers survive until late L3. A further transgene with an intact dAda2a region results in a complete phenotypic rescue, providing evidence that the L3 lethality is a result of the absence of the dAda2a function. P[DtlRpb4] transgene carrier d189 homozygotes therefore represent clean dAda2a loss-of-function mutants (these will be referred to as dAda2a189).
FIG. 1.
(A) Schematic view of the Drosophila 90F chromosomal region. The position of deletion 189 and the genes affected by it as well as the regions present in the used transgene constructs are shown. The rescue ability of particular transgenes and their combinations is indicated.
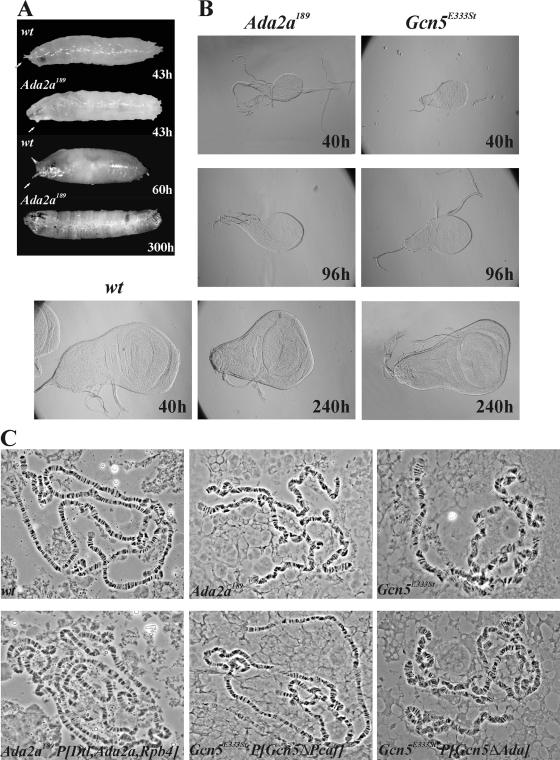
A characteristic feature of dAda2a189 homozygotes is that they survive for an extended period (up to 2 weeks) in the L3 stage but fail to pupariate or form only malformed structures covered with brownish cuticle (Fig. 2A). During their extended L3 stage, dAda2a larvae feed and reach a size similar to that of their wild-type siblings but never start wandering. The imaginal discs, central nervous system, and gonads of dAda2a189 larvae are significantly smaller than those of the wild-type controls (Fig. 2B and data not shown). The polytene chromosomes of dAda2a189 animals are fragile, displaying a distorted banding pattern (Fig. 2C). dGcn5-null mutants generated in this and other laboratories have similar phenotypes regarding lethality, underdeveloped discs, and polytene chromosome structural defects (Fig. 2B and C) (12). A transgene corresponding to the wild-type dGcn5 allele restores the normal polytene chromosome phenotype of dGcn5 mutants; however, a transgene with a deletion in the HAT domain or in the region involved in ADA2 interaction fails to restore the normal chromosome structure of dGcn5 mutants (Fig. 2C).
FIG. 2.
dAda2a and dGcn5 mutants display phenotypes indicating developmental defects. (A) dAda2a189 mutants fail to pupariate or form malformed pupae. Third-instar dAda2a189 larvae reach sizes similar to those of their wild-type (wt) siblings but do not start wandering and fail to evert spiracles (arrows), and only after 8 to 10 days in L3 do some of them form primitive pupa-like structures covered with brownish cuticle. (B) Imaginal discs of dAda2a189 and dGcn5E333st third-instar larvae are underdeveloped compared with similarly aged wild-type controls. During their extended L3 stage, the imaginal discs of dAda2a and dGcn5 mutants grow and reach the size of the fully developed wild-type discs after 8 to 10 days of L2/L3 molting, but they are abnormal in regards to both their shape and their structure. (C) Polytene chromosomes of dAda2a189 and dGcn5E333st mutants display similarly abnormal structures with respect to reduced condensation, short arms, and distorted banding pattern. Wild-type (wt) dAda2a and dGcn5 transgenes or a Gcn5 transgene lacking the PCAF region restores the normal chromosome structure of the corresponding mutants, while dGcn5 transgenes with a deletion in the ADA-interacting region or in the HAT domain fail in the rescue.
dGcn5 dAda2a double-null mutants or a combination of dAda2a189 and a hypomorph dGcn5 allele (dGcn5C137T [12]) results in a phenotype stronger than that of either of the two mutations alone; dAda2a189 dGcn5C137T animals are L2 and early L3 lethal. The coexpression of an upstream activator sequence promoter-driven dGcn5 transgene and an act-GAL4 driver in a dAda2a189 background results in a partial phenotypic rescue: dAda2a189 P[UAS-Gcn5] P[act-GAL4] L3 larvae have polytene chromosomes that are indistinguishable from the wild type in morphological features, and they form pupae, though they do not hatch (data not shown). These observations indicate a genetic interaction between dAda2a and dGcn5.
Yeast two-hybrid experiments also indicate dGcn5 dAda2a interaction: while we detected only weak interaction between dAda2a and dAda3, a strong interaction between dAda2a and dGcn5 was repeatedly observed (Fig. 3A).
FIG. 3.
Functional interaction of dGcn5 and dAda2a. (A) Yeast two-hybrid assay indicates strong interaction between dAda2a and dGcn5. Constructs tested for interaction contained fusions (DNA-binding domain in pBTM and acidic activation domain in pGAD424) as follows: 1, dAda2a-none (negative control); 2, dRpb4-dRpb7 (positive control); 3, dAda2a-dAda3; 4, dAda2b-dAda3; 5, dAda2a-dGcn5; and 6, dADA2b-dGcn5. (B) The null dGcn5E333st mutation has a negative effect on the development of dAda2ahyp1. The phenotype change is shown as the fraction of animals reaching the adult (A), pharat adult (PhA), and early pupa (EP) stages in the wild-type (light gray bars) and dGcn5 heterozygous background (white bars). (C) The overexpression of dGcn5 from a transgene (dark gray bars) has a positive effect on the development of dAda2ahyp2 (labels are as described in panel B). The inserts show the fractions of adults displaying the outstretched wing (os, white) and wild-type (wt, black) phenotypes. Resulting from the different chromosomal background (see Materials and Methods), the fractions of dAda2ahyp1 and dAda2ahyp2 adults in the two experiments (columns in panel B and C) are different. Error bars indicate deviations.
Results obtained from an analysis of the phenotypes of dAda2a transgene carriers provide further support for the genetic interaction of dAda2a and dGcn5. In several independent dAda2a transgene carrier lines that we established, the insertion of a genomic fragment encompassing the Ada2a locus with its regulatory region fully rescues the Ada2a189 phenotype. In line P[DtlAda2a]79/1, however, the transgene expression results in only a partial phenotypic rescue, most probably because of the low level of expression determined by the site of integration. The dAda2a189 P[DtlAda2a]79/1 genotype can be therefore considered a hypomorph dAda2a (dAda2ahyp) allele (see Materials and Methods). Nearly 85% of dAda2ahyp1 animals with a wild-type dGcn5 background hatch, but 30% of them display an outstretched wing (os) phenotype (Fig. 3B). In a +/dGcn5E333st heterozygous background, however, two-thirds of the dAda2ahyp1 animals perish as pupae or pharat adults, and 70% of those which emerge as adults have an os phenotype (Fig. 3B, insert). In contrast, an excess amount of dGcn5 partially suppresses the phenotypic defects of the hypomorph dAda2a mutation; dAda2ahyp2 P[UAS-Gcn5] P[act-GAL4] adults emerge in higher numbers than do their control siblings without the dGcn5 transgene (Fig. 3C), and only a small proportion of them display the os phenotype (Fig. 3C, insert). Together with earlier biochemical data, these results strongly suggest a functional interaction between dAda2a and dGcn5.
Failure in ecdysone response in dAda2a mutants.
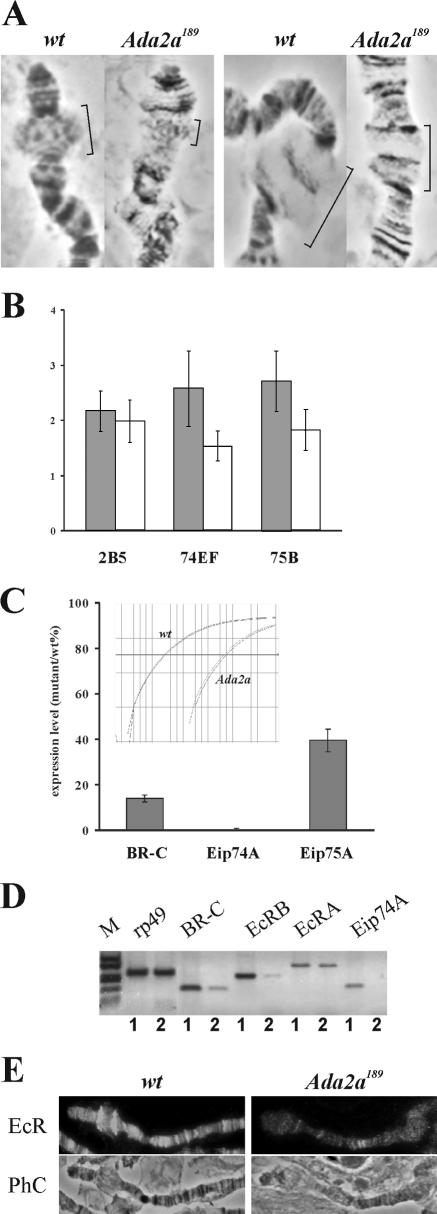
The phenotypic features of dAda2a mutants indicate a developmental block at the time of the larva-pupa transition. Since major developmental transitions during the onset of metamorphosis are triggered by the steroid hormone ecdysone, we wondered whether the induction of ecdysone-responsive genes in dAda2a mutants was affected. Late larval and prepupal pulses of ecdysone trigger a sequential induction of puffs in the giant polytene chromosomes of the larval and prepupal salivary glands. The puffs correspond to a loose chromatin structure where genes are actively transcribed. Among the early responding puffs in the salivary glands are those in the cytological regions 2B5, 74EF, and 75B. Importantly, during normal development, puffs corresponding to the above regions are visible in the last stage of the third instar, from the beginning of the wandering phase, when the larvae stop feeding and prepare for puparium formation. To establish whether the dAda2a mutation has an effect on ecdysone-induced puff formation, we staged wild-type and dAda2a189 larvae at the second- to third-instar molt and then sampled at regular intervals. Polytene chromosome squashes were prepared from salivary glands dissected from wandering wild-type animals and dAda2a189 animals of similar age, and the presence and size of puffs in the cytological regions 2B5, 74EF, and 75B were determined. Figure 4A illustrates salivary gland squashes for control wild-type and dAda2a larvae. The puffs present in the regions 2B5, 74EF, and 75B in the wild-type animals are significantly larger than those in the dAda2a mutants. The reduced size of early ecdysone-responsive puffs was also observed in dGcn5 mutants (12). Since the ecdysone levels change dynamically in this stage, and the response to the hormone is rapid, there is inevitably some degree of heterogeneity in the developmental age of the late third instars, which makes the timing of the comparison critical. This is of particular concern when the puffs of wild-type and dAda2a mutants are compared, since the development of the latter in the L3 stage is slowed down considerably compared with that of their control siblings. With this in mind, we also tested whether dAda2a mutants retained their abilities to form puffs in response to in vitro ecdysone treatment. For ectopic ecdysone treatment, salivary glands were dissected from mid-third-instar larvae and cultured at 25°C for 2 h either in the absence or in the presence of ecdysone. Figure 4B reveals that ecdysone treatment induces puff formation at 2B5, 74EF, and 75B in the salivary glands from the wild-type larvae. In the dAda2a mutants, the puffs are consistently smaller. Thus, the reduced abilities of these loci to be induced by ecdysone in late-third-instar dAda2a salivary glands can be overcome only partially by ectopic ecdysone treatment. In accord with this observation, we found that an in vivo increase in the ecdysone level of dAda2a mutant L3 larvae results in a partial phenotypic rescue: animals whose control siblings remain in the L3 stage for more than 10 days mostly form deformed pupa-like structures when placed on ecdysone-containing medium for 1 day (data not shown). The failure of ecdysone-induced transcription induction in dAda2a mutants can also be observed by comparing the levels of hormone-induced mRNA in mutant and wild-type animals. Early puffs at 2B5, 74EF, and 75B contain complex transcription units (Broad-Complex, Eip74, and Eip75, respectively), each encoding a family of transcription factors. Q-RT-PCRs indicated that the mRNA level corresponding to a representative transcript from each locus is markedly decreased in the dAda2a samples compared with that of the control late-L3 wild-type larvae (Fig. 4C). The difference in the levels of the Eip74A message of the wild-type and dAda2a mutants was consistently the highest upon repetition, reaching more than 2 log differences. These results indicate that the ecdysone-triggered transcription activation that directs developmental responses is severely attenuated in the dAda2a mutants.
FIG. 4.
(A) The formation of ecdysone-induced early-response puffs is reduced in dAda2a mutants. (A) Phase-contrast images of polytene chromosome region of wild-type control (wt) and dAda2a189 third-instar larvae, depicting the cytological regions 2B5 (left) and 74EF-75B (right). Brackets show the regions corresponding to early-response puffs. (B) The average size of early puffs (expressed as the average width of puffs in arbitrary units) following ectopic ecdysone treatment is significantly smaller in the dAda2a189 mutants than in the wild-type controls. The size of the puffs was determined as described in Materials and Methods. Each column represents the average of measurements on at least 10 dissected salivary glands. Error bars indicate deviations. (C) The level of representative mRNAs transcribed in early puffs is decreased in the dAda2a189 mutants. mRNA levels in L3 larvae were determined by quantitative RT-PCR in four independent experiments and are presented as the ratios between mutants and the wild type (wt). The insert illustrates the cycle threshold plot of a Q-PCR performed for the comparison of Eip74A mRNA levels in wild-type and in dAda2a189 mutants. Two parallels were run and are shown for both samples. One division of the chart represents a twofold difference in the amount of PCR product. (D) RT-PCR detection indicates reduced levels of ecdysone receptor subunit and ecdysone-induced messages. 1, wild type; 2, dAda2a189. Note the reduced level of E74A and BR-C mRNA similar to that seen in panel C, while no change in the level of rp49 message is detectable. (E) dADA2a chromosomes show reduced EcR-specific antibody staining. Phase-contrast (PhC) and EcR-specific antibody-stained image of the end of polytene X chromosomes of wild-type and dAda2a L3 larvae are shown.
A possible underlying reason of the reduced expression of ecdysone-induced genes could be that the lack of Ada2a interferes with the expression of the ecdysone receptor (EcR). By RT-PCR detection of specific messages, we observed that the level of EcRB and, to a lesser extent, the level of EcRA are indeed decreased in Ada2a mutants (Fig. 4D). In accord with that observation, the immunostaining of polytene chromosomes with EcR-specific antibody revealed decreased staining in dAda2a mutants compared to the staining intensities observed in wild-type animals (Fig. 4E). Altogether, these data clearly indicate that the loss of the dAda2a function interferes with hormonal induction of a set of genes required for the progress of the Drosophila developmental program.
The decreased level of gene expression, however, is not the result of a general failure of transcriptional activation in dAda2a mutants. On the one hand, different ecdysone-induced genes are affected to different extents (Fig. 4D) and, on the other hand, a number of genes are not affected or, on the contrary, show increased expression in dAda2a mutants. The expression of rp49 (Fig. 4D), Dp53, rosy, and the microtubule-associated protein coding gene MAP205, for example, is not affected by the loss of dAda2a function, while the levels of rosy and MAP205 are decreased significantly in dAda2b mutants (25) (data not shown). On the contrary, the level of FROST mRNA is increased to severalfold in dAda2a mutants and to a lesser extent in dAda2b mutants compared to that of the wild type (data not shown).
The lack of dAda2a function results in decreased H4 acetylation.
Gcn5 is the catalytic component of several complexes which acetylate nucleosomal histones. Both we and others recently determined that H3 acetylation at lysines 9 and 14 is significantly reduced in Ada2b mutants of Drosophila. dAda2a mutations, however, do not affect the acetylation of these lysine residues. Furthermore, the loss of either of the dAda2 functions does not change the acetylation of H4 at lysine 8 (25, 29). To extend these studies, we compared the acetylation of further lysine residues of H4 in dAda2a, dAda2b, and dGcn5 mutants. We assessed the H4 acetylation level via the immunostaining of polytene chromosomes with antibodies specific for H4 acetylated at K5 (H4-AcK5), and K12 (H4-AcK12). As a control, we used RNA Pol II antibody. Fluorescent microscopic images of wild-type, dAda2a, dAda2b, and dGcn5 polytene chromosomes stained with antibodies recognizing Pol II and H4 acetylated at different lysine residues are presented in Fig. 5A. All staining and data recording procedures were performed under identical conditions. The comparison of the staining intensities indicates that the levels of H4-AcK12 and H4-AcK5 are significantly less in the dAda2a mutants than in the wild type. The detection of K12 and K5 acetylated H4 in extracts of dAda2a larvae by Western blotting also indicates significant reductions in the levels of H4-AcK12 and H4-AcK5 (Fig. 5D). Similarly, dGcn5 mutation results in reduced H4 K12 and also H4 K5 acetylation (Fig. 5A and D). In contrast, neither the loss of dAda2a nor the loss of dGcn5 changed the level of H4-AcK8 to an extent detectable by immunostaining (data not shown) (see references 12 and 25). Chromosomes of dAda2b mutants do not reveal decreased acetylation of either of the two lysines of H4 tested in these experiments (Fig. 5A and D). We also did not observe a change in Ada2a or Gcn5 mutants in the intensity of H4-AcK16 staining, detectable on the X chromosomes as part of the dosage compensation (Fig. 6).
FIG. 5.
The effects of dAda2b, dAda2a, and dGcn5 mutations on the H4 K12 and H4 K5 acetylation of polytene chromosomes. (A) Chromosomes immunostained with polyclonal antibodies specific for individual acetylated lysine residues of H4, as indicated on the left, and a Pol II-specific monoclonal antibody (Pol II 7G5) are shown. Genotypes are indicated at the top. The images demonstrating immunostaining in different mutants were obtained with identical data-recording settings. wt, wild type. (B) Merged images of (left) phase-contrast and acetylated H4-stained (red) and (right) Pol II- (green) and acetylated H4-stained region of wild-type 3R chromosomes reveal similar distributions of H4-AcK12 and AcK5 staining mostly in the condensed chromosomal regions. It may be noted that some of the stronger bands seen in the phase-contrast images do not display acetylated H4 staining (stars) and that acetylated H4 and Pol II colocalization is observable in a few regions (open arrowheads), while in other regions, strong polymerase-specific, but no acetylated H4, signal is detected (closed arrowheads). (C) Identical regions of wild-type and dAda2a chromosomes costained for K12 acetylated H4 (red) and Pol II (green). For better comparison, the H4-AcK12 signal on the dAda2a chromosome is enhanced. Despite the distorted banding pattern of the dAda2a chromosome, similar distributions of K12 acetylated H4 are observable on wild-type and mutant chromosomes, though for specific bands, the H4-AcK12 signal on the dAda2a chromosome is absent or greatly reduced (arrows). (D) Western blot of protein extracts of wild-type (1), dAda2a (2), dGcn5 (3), and dAda2b (4) L3 larvae developed with antibodies as indicated on the left. For the detection of H4-AcK5 and H3, the same blot was developed consecutively with the two specific antibodies.
FIG. 6.
dAda2a mutation does not affect H4 K16, H3 K14, and K9 and K18 acetylation. Polytene chromosomes of wild-type (wt), dAda2a, and dGcn5 L3 larvae stained with DAPI (4′,6′-diamidino-2-phenylindole) and specific antibodies as indicated on the left. On the top, DAPI-stained images are shown of the same chromosomes which were used for acetylated H4 K16 detection.
The acetylation of H3 at K9 and K14 has been shown to depend on Ada2b-containing GCN5 HAT complexes. Neither the acetylation of these sites nor the acetylation of H3K18 is affected by Ada2a.
A full-length Gcn5 transgene restores the level of K12 acetylation to the level of the wild type in Gcn5 mutants, while transgenes with deletions in the ADA interaction or in the HAT domain do not (Fig. 7). This indicates that a normal level of H4 acetylation at K12 requires Gcn5 Ada2a interaction.
FIG. 7.
H4 K12 acetylation in dGcn5 mutants is restored by a dGcn5 transgene lacking the PCAF region but not by transgenes with mutations in the ADA interaction regions or HAT domains. For controls, images obtained with Pol II-specific antibody staining are shown. wt, wild type.
dAda2a and dGcn5 chromosomes stained with H4-AcK12-specific antibodies exhibit reduced staining intensities along the entire length of the chromosomes. A comparison of the staining patterns obtained with Pol II-specific and H4-AcK5- or H4-AcK12-specific antibodies on wild-type chromosomes reveals K5- and K12-acetylated H4 localized mostly in bands staining less intensively for Pol II, although the acetylated H4 and Pol II stainings overlap at several positions. Notably, intensive staining of H4-AcK5 or H4-AcK12 is not detected in puffs or interband regions (Fig. 5B). The structure of dAda2a chromosomes makes it difficult to obtain a detailed staining pattern; nonetheless, a comparison of the H4-AcK12 and Pol II-specific antibody-stained wild-type and dAda2a chromosome regions clearly reveals that some of the bands staining intensely for H4-AcK12 in the wild type are missing in the dAda2a mutant (Fig. 5C).
DISCUSSION
The colocalization of Gcn5 and Ada2 in HAT complexes has been demonstrated in several independent studies. First, yeast complexes containing both Gcn5 and Ada2 were separated biochemically and their HAT activities were demonstrated (13, 16, 17). Recently, two Gcn5-containing complexes, presumed to be dSAGA and dADA, one containing dAda2a and the other dAda2b, were partially separated from Drosophila cell extracts (21, 24). More recently ATAC, a further complex containing both dGcn5 and dAda2a, has been identified (18). Nonetheless, direct proof of the functional interaction of Drosophila Gcn5 with both dAda2 proteins has not been reported so far.
Our previous genetic analysis clearly demonstrated that the two dAda2s are functionally distinct and both we and, independently from us, Qi et al. have shown that one of them, Ada2b, is involved in histone acetylation (25, 29). Those studies, however, did not indicate dAda2a involvement in histone modification. Moreover, Carre et al. recently reported that a deletion variant of dGcn5 (Gcn5ΔAda), lacking the domain believed to be involved in Gcn5-Ada2 interaction, appeared normally distributed on polytene chromosomes and restored H3 acetylation in dGcn5 mutants. However, dGcn5 mutants were not rescued by the Gcn5ΔAda transgene and were arrested at puparium formation (12). The observations detailed above prompted us to search for direct evidence of the dGcn5-dAda2a interaction in vivo. For this, we studied the genetic interaction of dGcn5 and dAda2a. Our observation that the characteristic phenotypic features of the dAda2 mutants are suppressed or enhanced, depending on the dGcn5 genetic background, indicates an in vivo functional interplay between the two proteins. The finding that the level of dGcn5 alters the manifestation of a hypomorph dAda2a allele (Fig. 3) and vice versa (data not shown) is in accord with the proposed Ada2a-Gcn5 functional link, the former playing a role in facilitating the HAT activity of the latter. As expected from this scenario, the effect resulting from a change in the level of dGcn5 is compensated for by a change in the opposite direction in the dAda2a level or, vice versa, changes in dAda2a level are compensated by changes in the level of dGcn5. The similar phenotypes of dGcn5 and dAda2a mutants, with characteristic developmental defects at the larva-pupa transition (Fig. 2) (for details, see reference 12), indicate that the two proteins play essential roles in the regulatory hierarchy controlled by the steroid hormone ecdysone at the end of the third-larval instar. The observation that puff formation at chromosomal regions containing early ecdysone response genes is reduced in both dAda2a and dGcn5 mutants (Fig. 4) supports this conclusion and indicates a lack of appropriate transcriptional activation upon the appearance of the regulatory signal. The reduced level of puff formation correlates with the failure in hormone-induced gene activation, as indicated by the drastically reduced level of mRNAs corresponding to the early ecdysone-induced genes (Fig. 4C and D). The lack of transcription induction is not simply a result of a decreased hormone level, since dAda2a mutants are defective in puff formation even if the hormone is ectopically provided (Fig. 4B). Furthermore, the expression of EcR mRNA and localization of EcR to polytene chromosomes is also reduced in dAda2a mutants. Overall, we conclude from these observations that the lack of the dAda2a (and similarly, the lack of the dGcn5) function reduces transcription activation at specific loci. The effect is gene specific since the lack of Ada2a did not affect the expression of several genes we tested, and in other cases, an increase in the mRNA level in the absence of dAda2a can be observed.
The specificity of recombinant yGcn5 and partially purified Gcn5-containing HAT complexes for particular lysine residues of the core histones has been analyzed on both free and nucleosomal histone substrates (9, 17). These studies indicated that yGcn5 has a preference for the lysines of H3, and that the components of the Gcn5-containing complexes influence both HAT activity and specificity. From these studies, it was concluded that the major targets of yGcn5 HAT are the lysines of nucleosomal H3. However, other studies also suggested the involvement of yGcn5-containing complexes in the acetylation of histones other than H3. By analyzing the in vivo effects of changes in the specific lysines in the N termini of the histones in the absence and in the presence of yGcn5, Zhang et al. found that the loss of Gcn5 caused a diminution of acetylation at each of the four lysines in the N terminus of H4 (36). Recently, we and Qi et al. observed that the lack of dAda2b greatly reduced the K9 and K14 acetylation of H3, while not affecting H4 acetylation (25, 29). In concert with these data, Carre et al. reported that mutations of dGcn5 reduced H3 K9 and K14 acetylation but had no effect on the acetylation of H4 K8 (12).
The acetylation state of other lysine residues of H4 in dGcn5 mutants, however, was not tested rigorously. The immunostainings and Western blots presented here show that, in dAda2a mutants, the extent of H4 K12 and K5 acetylation is significantly reduced (Fig. 5). On the other hand, polytene dAda2a chromosomes do not display a significant change in K9- and K14-acetylated H3 staining and neither of the dAda2s has a strong effect on H3 K18 or H4 K8 and K16 acetylation (Fig. 6 and data not shown) (12, 25, 29).
Polytene chromosomes of dGcn5-null mutants display reduced H4-AcK5 and H4-AcK12 staining, and transgenes with a deletion in the ADA-interacting region or in the HAT domain fail in rescuing the acetylation (Fig. 7).
The staining of wild-type chromosomes with anti-H4-AcK5 and anti-H4-AcK12 antibodies reveals a banded pattern, indicating that regions with high DNA content are enriched in these types of modifications (Fig. 5B). Strong staining is also visible in the centromeric heterochromatic region for both H4-AcK5 and H4-AcK12.
The colocalization of Pol II- and K5- or K12-acetylated H4 staining occurs in some regions along the chromosome, but, as a general rule, intensely transcribed loci are not enriched in the acetylation deposited by dAda2a-containing complexes (Fig. 5B). A similar distribution of dAda2b-dependent acetylation of H3 (K14 and K9) and its loss in dAda2b mutants were observed earlier (25, 29). Taken together, these data indicate that dAda2a and dAda2b are functionally similar in playing roles in histone acetylation distributed along the chromosomes in rough proportion with the DNA content. However, dAda2a and dAda2b participate in dGcn5-containing complexes that acetylate specific residues of H4 and H3, respectively.
dAda2a and dAda2b also differ in that the complexes containing them participate in the transcriptional activation of distinct sets of genes: the hormone induction of early-response genes requires the dAda2a function (Fig. 4), while dAda2b is involved in p53-mediated processes among others (25). Accordingly, we propose dual functions for Ada2-Gcn5-containing complexes: the deposition of uniformly distributed H3 and H4 acetylation along the chromosomes and the targeted acetylation of histones at specific loci. A decrease in the former might contribute to the observed structural change in dAda2a chromosomes, while a loss of the latter function results in an altered transcription activity at specific loci. Our data clearly indicate that, regarding the first function, dAda2a- and dAda2b-containing complexes have distinct preferences for H4 and H3, respectively. Whether the same specificity of dAda2a and dAda2b-containing complexes exists in promoter-specific modifications remains to be determined.
Guelman et al. recently reported the identification of a novel Ada2a-containing HAT complex, ATAC, in Drosophila (18). In an in vitro acetylation assay, ATAC was found to display strong nucleosomal H4 activity. The specificity of ATAC for in vivo histone acetylation is at present unknown. Therefore, it is an intriguing possibility that in vivo the Ada2b-containing SAGA complexes acetylate H3 lysines, while the Ada2a-containing ATAC complexes target lysines at the N terminus of H4. It should be noted, however, that in the ATAC complex, the presence of an additional protein with a putative HAT domain was also detected (18). Alternatively, our data do not exclude the possibility that dAda2a, and perhaps also dAda2b, participates in gene-specific transcription regulation independently of dGcn5. Indeed, several independent observations indicate the presence of dAda2a independently of dGcn5, which might reflect a dGcn5-independent role for dAda2a. On glycerol gradient sedimentation of Drosophila embryo extract, a significant fraction of dAda2a did not cosediment with dGcn5 but was present in fractions corresponding to smaller Mw complexes (24). Immunocolocalization studies of polytene chromosomes revealed that dAda2a was enriched at many sites independently of dGcn5, and only a little overlap was observed between the two proteins (21). Furthermore, the amount of dAda2a and dGcn5 does not seem to be tightly linked at different stages of Drosophila development (21). Our observation that dGcn5 dAda2a double mutants cause a stronger phenotype than that of either null mutant alone can also be interpreted as an indication of separate functions of the two proteins.
The observations that dAda2 and dGcn5 (and also dAda2b) mutants survive until the late-larva stage suggest that the uniformly deposited acetylation by the complexes containing these factors either is not essential or can be accomplished without these factors or can be compensated for by other acetyl transferases. On the other hand, the gene-specific effects of dGcn5- and dAda2a-containing complexes, which lead to the onset of the metamorphosis program or those carried out by Ada2b-containing complexes in regulating p53-mediated processes, cannot be compensated for. It is interesting that the partial phenotypic rescue resulting from the increased ecdysone level in hormone-fed larvae indicates further complexity in the gene regulatory circuits: while these animals form cuticle readily in response to the hormone pulse, no response in the development in their imaginal discs is apparent and they perish as malformed pupae.
A dual role for yGcn5 in promoter-targeted acetylation and in maintaining low levels of uniform acetylation in surrounding regions has also been proposed by Howe et al. (19). Our findings on the role of Drosophila Ada2-Gcn5-containing complexes are in full accord with their suggestion and also with observations that yeast cells can tolerate large decreases in histone acetylation without affecting the cell viability (19).
Acknowledgments
We thank Katalin Ökrös for her expert technical help. We are grateful to Cristophe Antoniewsky and László Tora for kindly providing dGcn5C137T, dGcn5E333st stocks and Gcn5 ΔPcaf ΔAda ΔHat transgenes and Pol II antibody, respectively, and for critical comments and discussion of the results prior to publication. We thank IstvánTombácz and Norbert Pardi for their contribution to the selection of dGcn5 mutants and Zsuzsanna Újfaludi for her help with Q-PCR. SelenMuratoglu and Zsolt Tóth participated in the early phase of these studies. We are grateful for their contribution. We are very grateful to L. Lebedeva for her help with polytene chromosome immunostaining.
This work was supported by grants from the Hungarian Science Fund (OTKA T046414) and EU FP-6 (LSHG-CT-2004-502950). A.C. is a European Community RTN Marie Curie research fellow, supported by grant HPRN-CT-2004-504228.
Footnotes
Published ahead of print on 9 October 2006.
REFERENCES
- 1.Balasubramanian, R., M. G. Pray-Grant, W. Selleck, P. A. Grant, and S. Tan. 2002. Role of the Ada2 and Ada3 transcriptional coactivators in histone acetylation. J. Biol. Chem. 277:7989-7995. [DOI] [PubMed] [Google Scholar]
- 2.Barbaric, S., H. Reinke, and W. Horz. 2003. Multiple mechanistically distinct functions of SAGA at the PHO5 promoter. Mol. Cell. Biol. 23:3468-3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barlev, N. A., R. Candau, L. Wang, P. Darpino, N. Silverman, and S. L. Berger. 1995. Characterization of physical interactions of the putative transcriptional adaptor, ADA2, with acidic activation domains and TATA-binding protein. J. Biol. Chem. 270:19337-19344. [DOI] [PubMed] [Google Scholar]
- 4.Barlev, N. A., A. V. Emelyanov, P. Castagnino, P. Zegerman, A. J. Bannister, M. A. Sepulveda, F. Robert, L. Tora, T. Kouzarides, B. K. Birshtein, and S. L. Berger. 2003. A novel human Ada2 homologue functions with Gcn5 or Brg1 to coactivate transcription. Mol. Cell. Biol. 23:6944-6957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belotserkovskaya, R., and S. L. Berger. 1999. Interplay between chromatin modifying and remodeling complexes in transcriptional regulation. Crit. Rev. Eukaryot. Gene Expr. 9:221-230. [DOI] [PubMed] [Google Scholar]
- 6.Berger, S. L., B. Pina, N. Silverman, G. A. Marcus, J. Agapite, J. L. Regier, S. J. Triezenberg, and L. Guarente. 1992. Genetic isolation of ADA2: a potential transcriptional adaptor required for function of certain acidic activation domains. Cell 70:251-265. [DOI] [PubMed] [Google Scholar]
- 7.Bhaumik, S. R., and M. R. Green. 2002. Differential requirement of SAGA components for recruitment of TATA-box-binding protein to promoters in vivo. Mol. Cell. Biol. 22:7365-7371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boyer, L. A., M. R. Langer, K. A. Crowley, S. Tan, J. M. Denu, and C. L. Peterson. 2002. Essential role for the SANT domain in the functioning of multiple chromatin remodeling enzymes. Mol. Cell 10:935-942. [DOI] [PubMed] [Google Scholar]
- 9.Brown, C. E., T. Lechner, L. Howe, and J. L. Workman. 2000. The many HATs of transcription coactivators. Trends Biochem. Sci. 25:15-19. [DOI] [PubMed] [Google Scholar]
- 10.Brownell, J. E., J. Zhou, T. Ranalli, R. Kobayashi, D. G. Edmondson, S. Y. Roth, and C. D. Allis. 1996. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell 84:843-851. [DOI] [PubMed] [Google Scholar]
- 11.Candau, R., P. A. Moore, L. Wang, N. Barlev, C. Y. Ying, C. A. Rosen, and S. L. Berger. 1996. Identification of human proteins functionally conserved with the yeast putative adaptors ADA2 and GCN5. Mol. Cell. Biol. 16:593-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carre, C., D. Szymczak, J. Pidoux, and C. Antoniewski. 2005. The histone H3 acetylase dGcn5 is a key player in Drosophila melanogaster metamorphosis. Mol. Cell. Biol. 25:8228-8238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carrozza, M. J., R. T. Utley, J. L. Workman, and J. Cote. 2003. The diverse functions of histone acetyltransferase complexes. Trends Genet. 19:321-329. [DOI] [PubMed] [Google Scholar]
- 14.Fischle, W., Y. Wang, and C. D. Allis. 2003. Histone and chromatin cross-talk. Curr. Opin. Cell Biol. 15:172-183. [DOI] [PubMed] [Google Scholar]
- 15.Georgieva, S., D. B. Kirschner, T. Jagla, E. Nabirochkina, S. Hanke, H. Schenkel, C. de Lorenzo, P. Sinha, K. Jagla, B. Mechler, and L. Tora. 2000. Two novel Drosophila TAFIIs have homology with human TAFII30 and are differentially regulated during development. Mol. Cell. Biol. 20:1639-1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grant, P. A., L. Duggan, J. Cote, S. M. Roberts, J. E. Brownell, R. Candau, R. Ohba, T. Owen-Hughes, C. D. Allis, F. Winston, S. L. Berger, and J. L. Workman. 1997. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 11:1640-1650. [DOI] [PubMed] [Google Scholar]
- 17.Grant, P. A., A. Eberharter, S. John, R. G. Cook, B. M. Turner, and J. L. Workman. 1999. Expanded lysine acetylation specificity of Gcn5 in native complexes. J. Biol. Chem. 274:5895-5900. [DOI] [PubMed] [Google Scholar]
- 18.Guelman, S., T. Suganuma, L. Florens, S. K. Swanson, C. L. Kiesecker, T. Kusch, S. Anderson, J. R. Yates III, M. P. Washburn, S. M. Abmayr, and J. L. Workman. 2006. Host cell factor and an uncharacterized SANT domain protein are stable components of ATAC, a novel dAda2A/dGcn5-containing histone acetyltransferase complex in Drosophila. Mol. Cell. Biol. 26:871-882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Howe, L., D. Auston, P. Grant, S. John, R. G. Cook, J. L. Workman, and L. Pillus. 2001. Histone H3 specific acetyltransferases are essential for cell cycle progression. Genes Dev. 15:3144-3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Komonyi, O., G. Papai, I. Enunlu, S. Muratoglu, T. Pankotai, D. Kopitova, P. Maroy, A. Udvardy, and I. Boros. 2005. DTL, the Drosophila homolog of PIMT/Tgs1 nuclear receptor coactivator-interacting protein/RNA methyltransferase, has an essential role in development. J. Biol. Chem. 280:12397-12404. [DOI] [PubMed] [Google Scholar]
- 21.Kusch, T., S. Guelman, S. M. Abmayr, and J. L. Workman. 2003. Two Drosophila Ada2 homologues function in different multiprotein complexes. Mol. Cell. Biol. 23:3305-3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marcus, G. A., N. Silverman, S. L. Berger, J. Horiuchi, and L. Guarente. 1994. Functional similarity and physical association between GCN5 and ADA2: putative transcriptional adaptors. EMBO J. 13:4807-4815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martinez, E., T. K. Kundu, J. Fu, and R. G. Roeder. 1998. A human SPT3-TAFII31-GCN5-L acetylase complex distinct from transcription factor IID. J. Biol. Chem. 273:23781-23785. [DOI] [PubMed] [Google Scholar]
- 24.Muratoglu, S., S. Georgieva, G. Papai, E. Scheer, I. Enunlu, O. Komonyi, I. Cserpan, L. Lebedeva, E. Nabirochkina, A. Udvardy, L. Tora, and I. Boros. 2003. Two different Drosophila ADA2 homologues are present in distinct GCN5 histone acetyltransferase-containing complexes. Mol. Cell. Biol. 23:306-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pankotai, T., O. Komonyi, L. Bodai, Z. Ujfaludi, S. Muratoglu, A. Ciurciu, L. Tora, J. Szabad, and I. Boros. 2005. The homologous Drosophila transcriptional adaptors ADA2a and ADA2b are both required for normal development but have different functions. Mol. Cell. Biol. 25:8215-8227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Papai, G., O. Komonyi, Z. Toth, T. Pankotai, S. Muratoglu, A. Udvardy, and I. Boros. 2005. Intimate relationship between the genes of two transcriptional coactivators, ADA2a and PIMT, of Drosophila. Gene 348:13-23. [DOI] [PubMed] [Google Scholar]
- 27.Pile, L. A., and D. A. Wassarman. 2002. Localizing transcription factors on chromatin by immunofluorescence. Methods 26:3-9. [DOI] [PubMed] [Google Scholar]
- 28.Pray-Grant, M. G., D. Schieltz, S. J. McMahon, J. M. Wood, E. L. Kennedy, R. G. Cook, J. L. Workman, J. R. Yates III, and P. A. Grant. 2002. The novel SLIK histone acetyltransferase complex functions in the yeast retrograde response pathway. Mol. Cell. Biol. 22:8774-8786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qi, D., J. Larsson, and M. Mannervik. 2004. Drosophila Ada2b is required for viability and normal histone H3 acetylation. Mol. Cell. Biol. 24:8080-8089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saleh, A., V. Lang, R. Cook, and C. J. Brandl. 1997. Identification of native complexes containing the yeast coactivator/repressor proteins NGG1/ADA3 and ADA2. J. Biol. Chem. 272:5571-5578. [DOI] [PubMed] [Google Scholar]
- 31.Sendra, R., C. Tse, and J. C. Hansen. 2000. The yeast histone acetyltransferase A2 complex, but not free Gcn5p, binds stably to nucleosomal arrays. J. Biol. Chem. 275:24928-24934. [DOI] [PubMed] [Google Scholar]
- 32.Sterner, D. E., R. Belotserkovskaya, and S. L. Berger. 2002. SALSA, a variant of yeast SAGA, contains truncated Spt7, which correlates with activated transcription. Proc. Natl. Acad. Sci. USA 99:11622-11627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sterner, D. E., and S. L. Berger. 2000. Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev. 64:435-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stockinger, E. J., Y. Mao, M. K. Regier, S. J. Triezenberg, and M. F. Thomashow. 2001. Transcriptional adaptor and histone acetyltransferase proteins in Arabidopsis and their interactions with CBF1, a transcriptional activator involved in cold-regulated gene expression. Nucleic Acids Res. 29:1524-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Syntichaki, P., and G. Thireos. 1998. The Gcn5.Ada complex potentiates the histone acetyltransferase activity of Gcn5. J. Biol. Chem. 273:24414-24419. [DOI] [PubMed] [Google Scholar]
- 36.Zhang, W., J. R. Bone, D. G. Edmondson, B. M. Turner, and S. Y. Roth. 1998. Essential and redundant functions of histone acetylation revealed by mutation of target lysines and loss of the Gcn5p acetyltransferase. EMBO J. 17:3155-3167. [DOI] [PMC free article] [PubMed] [Google Scholar]