Abstract
The sarcoplasmic reticulum (SR) plays a critical role in excitation-contraction coupling by regulating the cytoplasmic calcium concentration of striated muscle. The histidine-rich calcium-binding protein (HRCBP) is expressed in the junctional SR, the site of calcium release from the SR. HRCBP is expressed exclusively in muscle tissues and binds calcium with low affinity and high capacity. In addition, HRCBP interacts with triadin, a protein associated with the ryanodine receptor and thought to be involved in calcium release. Its calcium binding properties, localization to the SR, and interaction with triadin suggest that HRCBP is involved in calcium handling by the SR. To determine the function of HRCBP in vivo, we inactivated HRC, the gene encoding HRCBP, in mice. HRC knockout mice exhibited impaired weight gain beginning at 11 months of age, which was marked by reduced skeletal muscle and fat mass, and triadin protein expression was upregulated in the heart of HRC knockout mice. In addition, HRC null mice displayed a significantly exaggerated response to the induction of cardiac hypertrophy by isoproterenol compared to their wild-type littermates. The exaggerated response of HRC knockout mice to the induction of cardiac hypertrophy is consistent with a regulatory role for HRCBP in calcium handling in vivo and suggests that mutations in HRC, in combination with other genetic or environmental factors, might contribute to pathological hypertrophy and heart failure.
Excitation-contraction (E-C) coupling is the pathway by which depolarization of the muscle cell membrane is coupled to contraction. The sarcoplasmic reticulum (SR) plays a critical role in E-C coupling by regulating the concentration and distribution of intracellular calcium. Depolarization of the cardiomyocyte plasma membrane activates L-type calcium channels known as dihydropyridine receptors, which permit the influx of extracellular calcium. The resulting increase in the local calcium concentration activates calcium release channels, known as ryanodine receptors, in the SR. Activated ryanodine receptors rapidly release large calcium stores from the SR to the cytoplasm, increasing the intracellular calcium concentration. This increase in intracellular calcium results in calcium binding to troponin C and the subsequent uncovering of myosin binding sites on actin filaments. This enables myosin to walk along actin, producing contraction. Relaxation occurs when the cytoplasmic calcium concentration is restored to the basal level via reuptake of calcium into the SR by the sarcoplasmic-endoplasmic reticulum ATPase (SERCA) and via calcium extrusion from the cell through the Na+-Ca2+ exchanger (6, 7, 43).
The SR is a specialized network of endoplasmic reticulum that surrounds the myofibrils in striated muscle (7, 43). The SR serves as the major intracellular storage compartment of exchangeable calcium, and the calcium handling properties of the SR mediate contraction. The SR can be loosely divided into two regions based on protein content, morphology, and sedimentation properties (7, 8, 10, 19, 32, 43). The majority of SR consists of a tubular network of light or longitudinal SR, while the heavy or junctional SR consists of terminal cisternae budding from the longitudinal SR. The longitudinal SR membrane is enriched with SERCA and serves as the primary site for calcium reuptake into the SR following contraction. The junctional SR is the site of calcium release from the SR via the ryanodine receptor. A multiprotein release channel complex associated with the ryanodine receptor localizes to the junctional SR and includes calsequestrin, junctin, triadin, and probably multiple additional proteins (8, 24, 67). Calsequestrin, the most abundant calcium binding protein in the SR, possesses a high capacity for calcium, but binds calcium with only moderate affinity via long acidic amino acid repeats (6, 47, 65). Calsequestrin effectively decreases the amount of free calcium in the SR lumen and concentrates calcium at the site of release from the SR. Consistent with this model for the role of calsequestrin in the SR, cardiomyocytes that overexpress calsequestrin have increased calcium in the SR (33, 59, 62), whereas cardiomyocytes expressing reduced levels or a mutant form of calsequestrin have reduced SR calcium stores (64). In addition to serving as the major calcium buffering protein in the SR, calsequestrin may also play a role in calcium release (33, 59, 62). Junctin and triadin may also modulate calcium release from the junctional SR, although the precise roles of these proteins in E-C coupling remain unclear (25, 35-39, 50, 61).
The histidine-rich calcium-binding protein (HRCBP) is another SR protein that appears to be involved in calcium handling. HRCBP was originally identified as a low-density lipoprotein binding protein (29). However, HRCBP localizes to the SR of striated muscle and to putative calciosomes within arterial smooth muscle, suggesting that the biological function of the protein does not involve low-density lipoprotein binding (29, 51, 55). Rather, HRCBP may play a role in SR calcium handling since the protein binds calcium with high capacity and moderate affinity in vitro (52). Calcium binding to HRCBP appears to occur via several long repeats of acidic amino acids in a manner analogous to calcium binding to calsequestrin (30, 52). However, unlike calsequestrin, HRCBP also contains a C-terminal laminin-type epidermal growth factor-like domain that may bind zinc and serve as a domain for interaction with other proteins (30, 52, 55). Subcellular fractionation studies indicate that HRCBP is localized to the junctional SR (15). Consistent with these observations, HRCBP colocalizes and interacts with triadin, a member of the release channel complex localized primarily in the junctional SR (16, 44, 56, 57). Its calcium binding properties and localization to the SR support a likely role for HRCBP in calcium handling by the SR.
Consistent with a role in calcium handling, overexpression of HRCBP in cultured neonatal rat cardiomyocytes results in increases in SR calcium storage capacity and in depolarization-induced calcium release (34). Similarly, adult rat cardiomyocytes overexpressing HRCBP also display an increase in calcium storage capacity and exhibit impaired calcium release and contractility (18). These studies suggest that a primary role for HRCBP is in calcium storage within the SR. This notion is consistent with the high-capacity, moderate-affinity calcium binding properties of HRCBP. However, while calsequestrin is the major calcium storage protein in the junctional SR, previous studies have indicated that HRCBP only comprises about 1% of the total protein content of the SR in skeletal muscle (6, 17). Therefore, although overexpression of HRCBP clearly results in an increase in calcium storage capacity in cultured rat cardiomyocytes, the low abundance of HRCBP suggests that it may be more likely to play a regulatory role in calcium handling in vivo. Consistent with the notion that HRCBP plays a regulatory role in calcium handling, overexpression of HRCBP in the heart results in impaired recovery of the calcium transient without any change in the amount of releasable SR calcium stores (23).
To begin to define the function of HRCBP in vivo, we inactivated HRC, the gene encoding HRCBP, in mice. Beginning at 1 year of age, HRC null mice display a significant decrease in body weight compared to wild-type controls, and this decrease in body weight is marked by reduced muscle and fat pad mass compared to wild-type littermates. In addition, HRC knockout mice exhibit a significantly exaggerated hypertrophic response to prolonged β-adrenergic receptor stimulation with isoproterenol compared to wild-type littermate controls. This increased hypertrophic response under conditions of cardiac stress is consistent with a regulatory role for HRCBP in SR calcium handling in vivo.
MATERIALS AND METHODS
Generation of an antibody directed against mouse HRCBP.
A fragment of the mouse HRC cDNA encoding the C-terminal 418 nucleotides was subcloned into the pMal-c2x plasmid (New England Biolabs) to generate an expression construct with the C terminus of HRCBP fused to maltose binding protein (MBP). Sequence analysis confirmed that the HRCBP C terminus was in frame with MBP. This construct (pMal-c2x-HRCBP) was used to generate and purify 1.5 mg of fusion protein (MBP-HRCBP) following the protocol for the pMal protein fusion and purification system (New England Biolabs). The protein was further purified from sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels lightly stained with an aqueous solution containing 0.05% Coomassie brilliant blue. The band corresponding to MBP-HRCBP was excised and used for antibody production in rabbits following standard procedures, performed by Animal Pharm Services, Inc. (Healdsburg, CA). To affinity purify antibody from immune serum, the same HRC 3′ cDNA fragment described above was subcloned into pGex-2T (GE Healthcare) to generate a glutathione S-transferase (GST) fusion construct. The GST-HRCBP fusion protein was purified following the GST Gene Fusion System protocol (GE Healthcare) and covalently linked to Affigel 15 resin (Bio-Rad) to generate a column for affinity purification of HRCBP-specific antibodies from the serum. Antibody was then affinity purified from the serum following the acid elution protocol described in the Affigel 15 instructions (Bio-Rad), and specificity was confirmed by Western blot analysis. Nucleotides 1834 to 2251 from mouse HRC cDNA (GenBank accession no. NM_010473) were used to generate the fusion constructs described above.
Protein immunoblots and immunohistochemistry.
To prepare tissue homogenates for Western analyses, adult tissues were quickly removed and placed in 3 volumes of ice-cold MMB, which contains 20 mM sodium-PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)], pH 6.8, 10% sucrose, 2 μM leupeptin, 100 μM phenylmethylsulfonyl fluoride, and 500 μM benzamidine. Tissues were homogenized for 30 s using the Powergen 35 tissue homogenizer (Fisher) at low to medium speed. Homogenized tissues were then centrifuged at 8,000 × g for 20 min, and the supernatant was mixed with SDS-PAGE loading buffer to obtain a solution with a final concentration of 50 mM Tris, pH 6.8, 2% SDS, 10% glycerol, 30 mM dithiothreitol, and 0.01% bromophenol blue. Homogenates were boiled for 10 min and frozen. Proteins were separated by SDS-PAGE and transferred overnight to Imobilon polyvinylidene difluoride membranes (Millipore). Membranes were blocked with TBST (13 mM Tris, pH 7.4, 150 mM sodium chloride, 0.05% Tween 20) plus 10% nonfat dry milk for 1 h at room temperature, incubated with primary antibody for 1 h at room temperature, washed with TBST plus 1% milk, incubated with peroxidase-conjugated secondary antibody for 1 h at room temperature, and washed again. Signal was detected using the ECL Plus enhanced chemiluminescence detection kit and Hyperfilm-ECL (Amersham Biosciences). Mouse monoclonal C3-33 (anti-ryanodine receptor 2; MA3-916 from Affinity Bioreagents) was used at 2 μg/ml. Rabbit anti-HRCBP was used at a 1:100 dilution. Rabbit anticalsequestrin (PA1-913 from Affinity Bioreagents) was used at a 1:2,500 dilution. Mouse monoclonal GE 4.90 (antitriadin; MA3-927 from Affinity Bioreagents) and mouse monoclonal 12G10 (anti-α-tubulin, obtained from the Developmental Studies Hybridoma Bank at the University of Iowa) were both used at a 1:1,000 dilution. Mouse anti-phospholamban monoclonal antibody from clone A1 (Upstate Cell Signaling Solutions) was used at a 1:400 dilution. Peroxidase-conjugated goat anti-mouse immunoglobulin G (IgG) and goat anti-rabbit IgG were both obtained from Sigma and used at a 1:25,000 dilution.
For immunohistochemical staining, hearts were dissected from adult mice, incubated in phosphate-buffered saline (PBS), pH 7.4, at 37°C for 10 min, and fixed overnight in 10% neutral buffered formalin. Hearts were then dehydrated, cleared with xylenes, and embedded for frontal sections in paraffin. Sections cut at a thickness of 7 μm were then rehydrated and subjected to antigen retrieval (Antigen Retrieval Citra; Biogenex). After treatment of slides with 3% hydrogen peroxide to deplete endogenous peroxidase activity, the tissue sections were blocked with PBS containing 3% normal goat serum and 0.1% Triton X-100. Sections were then incubated overnight with or without rabbit anti-HRCBP (1:50) in PBS plus 0.1% Triton X-100, washed with PBS, incubated with secondary antibody (1:250 peroxidase-conjugated goat anti-rabbit IgG), and washed again. Peroxidase activity was detected by treatment with nickel-DAB stain (DAB peroxidase substrate kit; Vector Laboratories).
Generation of HRC null mice and genotyping.
To generate an HRC targeting vector, we initially isolated a genomic clone encompassing the HRC locus by screening a mouse Sv129 genomic library (Lambda FixII; Stratagene) with the mouse HRC cDNA. A 2.6-kb XhoI-SacI fragment of the HRC locus, which encompasses exons 2 through 6, was used as the 3′ homology arm. A 1.2-kb BamHI-Psp1406 fragment that encompasses the majority of the HRC promoter and enhancer but does not include the transcriptional start site (2) was used as the 5′ homology arm. Homology arms were cloned into plasmid NeoTKXho (kind gift of R. Behringer), which contains the PGK-Neo and MC1-TK cassettes for positive and negative selection, respectively (13).
The HRC targeting vector was linearized with NotI, gel purified, and introduced into KG-1 embryonic stem (ES) cells, which were derived from the 129SvEv mouse strain, using standard procedures for ES cell electroporation (31). Electroporated cells were then plated on Sto-1 feeder cells, which were previously treated with mitomycin C (Sigma) to block DNA replication in the feeder cells (31). ES cells were simultaneously subjected to positive selection in 180 μg/ml G418 (Gibco-BRL) and negative selection in 2 μM ganciclovir (kind gift of Syntex Chemicals, Boulder, CO). Following selection for 10 days, 384 colonies were picked and grown to confluence on Sto-1 feeder cells in flat-bottom 96-well plates in the presence of 180 μg/ml G418. Expanded ES cell clones were then frozen in 96-well plates and were also further expanded in the absence of feeder cells for DNA preparation and Southern analysis according to standard procedures (31). Correct gene targeting resulted in the replacement of the transcriptional and translation start sites and the entire first exon of HRC, which accounts for the majority of the HRCBP protein. Correct targeting also resulted in the introduction of an additional EcoRI site into the HRC locus, which was detected using probes that are external to the targeting vector at both the 5′ and 3′ ends. The 3′ probe, an EcoRI-SacI fragment that resides outside the region of the HRC locus used for the homology arms, detects a 7.9-kb EcoRI fragment from the wild-type allele and a 3.8-kb band from the targeted allele. The 5′ probe, a SacI-BamHI fragment that also resides outside the region of the HRC locus that was used for the homology arms, detects a 7.9-kb EcoRI fragment by Southern analysis from the wild-type allele and a 4.7-kb band from the targeted allele. ES cell clones were screened independently with both the 3′ and 5′ probes. Sixteen clones were properly targeted based on Southern analyses with both probes, and 2 of these were expanded and used to generate high-percentage chimeric mice by injection into the blastocysts of C57BL/6 mice.
Male chimeric mice were bred to wild-type C57BL/6 mice and screened for germ line transmission of the mutant allele by Southern analyses using the 3′ probe described above. Following germ line transmission of the allele, HRC heterozygous mice were backcrossed to wild-type C57BL/6 mice for seven generations. Seven generations of backcrossing results in heterozygous mice that carry the allele of interest, in this case the HRC mutant allele, in the context of a recipient genome that is 99.2% C57BL/6 (46). Age- and sex-matched mice of each genotype were generated as littermates for use in experiments in which different genotypes were compared. All experiments using animals complied with federal and institutional guidelines and were reviewed and approved by the UCSF Institutional Animal Care and Use Committee.
Analysis of SR calcium handling in cardiac microsomes.
Hearts from five age- and sex-matched adult mice per group were pooled, and homogenates were prepared following the initial steps of a cardiac SR microsome preparation protocol described previously (45). Briefly, hearts were minced with a razor blade in 5 volumes of MMB. Tissue was then homogenized for 30 s using a Powergen 35 tissue homogenizer (Fisher) at low to medium speed. Samples were further homogenized using a PFTE mortar and pestle. For this step, the pestle was spun using a Sears Craftsman drill (9101121) set to 60% power with a Staaco variable autotransformer. The homogenized tissues were then centrifuged at 8,000 × g for 20 min. All steps were performed on ice or at 4°C. Supernatants were filtered through three layers of gauze and frozen in liquid nitrogen. Homogenates were normalized for protein content by Bradford assay, and protein levels were confirmed by Western analysis of α-tubulin levels.
For analysis of calcium uptake, uptake buffer (20 mM imidazole, pH 7.0, 100 mM KCl, 5 mM MgCl2, 5 mM potassium oxalate, and 10 mM NaN3) containing cardiac microsome homogenate at a concentration of 750 μg/ml and 2 μM Fura-2 (Molecular Probes) was incubated with continuous stirring in a quartz cuvette at 37°C in a Photon Technology International fluorimeter. To assess extramicrosomal calcium levels, the ratio of light emitted by Fura-2 at 510 nm when excited at 340 nm to light emitted when excited at 380 nm was measured. Calcium uptake was stimulated by the addition of 2.5 mM K+-ATP. After calcium levels stabilized, 1 μM CaCl2 was added, and uptake was allowed to proceed for 8 min, when an additional 1 μM CaCl2 was added. This process was continued until calcium uptake was no longer observed, at which time 10 mM EGTA was added to chelate the remaining extramicrosomal calcium. To assess the activity of the ryanodine receptor, [3H]ryanodine binding to cardiac microsome preps was measured using a modification of a method described previously (68). Briefly, 250 μg of cardiac microsome homogenate was added to ryanodine binding buffer (25 mM imidazole, pH 7.4, 1 M KCl, 3 mM CaCl2, 950 μM EGTA), and 5 nM [3H]ryanodine (0.25 μCi/ml) in a total reaction volume of 250 μl, and the samples were incubated at 37°C for 90 min. Reactions were then filtered through 0.45-μm HA filters (Millipore), which retained the microsome membranes. The filters were dissolved in 4 ml ethylene glycol monoethyl ether (Sigma). The samples were then treated with 4 ml Scintiverse (Sigma), and radioactive ryanodine bound to the receptor was quantified using an LS6500 multipurpose scintillation counter (Beckman Coulter). Calcium loading capacity was assessed in a similar manner. Cardiac homogenates (100 μg/ml) in the uptake buffer described above were incubated with 50 μM 45CaCl2 (5 μCi/ml) and 3 mM K+-ATP for 20 min at 37°C. Samples were then filtered through 0.45-μm HA filters, and the filters were processed as described above for the ryanodine binding assays.
Voluntary exercise assay.
Voluntary exercise assays were conducted essentially as described previously (1). Briefly, either 3-month-old or 11-month-old wild-type and HRC knockout mice were housed individually in oversized microisolator rodent cages (25 cm by 48 cm by 25 cm) containing standard metal rodent wheels (11.5 cm in diameter) suspended from the wire cage top. A bicycle computer (Sigma Sport model BC 800) was mounted on each wheel such that the magnet passed the sensor with each revolution of the wheel. The distance run on the wheels was recorded from 6 p.m. to 10 a.m. each night for five consecutive nights. The exercise assays were performed under standard light/dark conditions, and mice were provided unlimited access to food and water throughout the course of the experiment. To assess exercise-induced cardiac hypertrophy, age- and sex-matched male littermates were allowed to run as described above for 28 consecutive nights since previous studies indicated that 4 weeks of running was sufficient to induce mild physiologic hypertrophy (1). Eleven-month-old wild-type and HRC null mice were housed in a cage containing either a mouse wheel or no wheel for the duration of the experiment. After 28 days, the mice were weighed and sacrificed, the hearts were removed and weighed, and the length of the tibia was measured with calipers.
Induction of cardiac hypertrophy with isoproterenol.
Cardiac hypertrophy was induced in 6-month-old wild-type and HRC null male mice in a 99.2% C57BL/6 background with isoproterenol (9, 22). Isoproterenol (60 mg/kg body weight/day) in PBS or PBS alone was administered for 10 days using subcutaneously implanted miniosmotic pumps (Alzet model 2002). After 10 days of isoproterenol treatment, mice were weighed and euthanized. Hearts were then removed, incubated in PBS at 37°C for 10 min, weighed, and fixed overnight in either 4% paraformaldehyde or 10% neutral buffered formalin. In each case, the left tibia was also removed and cleaned and the length was measured with calipers. In some cases, hearts were dehydrated, cleared with xylenes, and embedded for frontal sections in paraffin. Five-micrometer sections from representative hearts for each group were stained with hematoxylin and eosin.
RESULTS
Murine HRCBP localizes to striated muscle.
To examine the distribution and localization of HRCBP in mice, we generated a polyclonal antibody to the highly conserved epidermal growth factor-like C-terminal domain of HRCBP (55). Western analyses of tissue and organ homogenates with anti-HRCBP confirmed that murine HRCBP is found specifically in striated muscle (Fig. 1). Anti-HRCBP detected a single 160-kDa band, corresponding to HRCBP, in skeletal and cardiac muscle (Fig. 1, lanes 1 and 2). This band was not present in stomach, intestine, bladder, brain, or kidney (Fig. 1, lanes 3 to 7), which is consistent with the protein distribution previously reported for HRCBP in rabbits (29, 30) and with the muscle-specific expression pattern directed by the human HRC promoter in transgenic mice (2). The human promoter is also active in developing arterial smooth muscle cells in mouse embryos (2). In contrast, we did not detect HRCBP in the aorta, suggesting that the protein is absent or only weakly expressed in the aorta of the adult mouse (data not shown). However, this observation is consistent with the expression of HRCBP in the adult rabbit, where the protein and mRNA were present in small arteries and arterioles but were not detected in the aorta (29, 30, 51).
FIG. 1.
HRCBP localizes specifically to striated muscle in mice. Homogenates of skeletal muscle (lane 1), heart (lane 2), stomach (lane 3), intestine (lane 4), bladder (lane 5), brain (lane 6), and kidney (lane 7) were analyzed by Western blotting with anti-HRCBP (top) or anti-α-tubulin (bottom). Similar levels of α-tubulin were detected in all samples. The 160-kDa band corresponding to mouse HRCBP was only detected in skeletal muscle and heart (lanes 1 and 2). Bars on the right depict the position of Full Range Rainbow molecular weight markers (Amersham).
Generation of HRC null mice.
The tissue distribution, localization to the sarcoplasmic reticulum, and calcium binding properties of HRCBP are consistent with a role in calcium handling by the SR. In addition, ex vivo analyses of HRCBP overexpression in isolated rat cardiomyocytes showed an increase in SR calcium storage, which further suggests that HRCBP may be important for calcium handling (18, 34). To determine the function of HRCBP in vivo, we inactivated the HRC gene in mice.
To generate HRC null mice, we replaced the first exon and the transcriptional and translational start sites of the HRC gene with a neomycin resistance cassette by homologous recombination (Fig. 2A). Recombination also introduced an additional EcoRI site 5′ to exon 2, which allowed us to distinguish between the wild-type and null alleles by Southern analyses using probes located outside the targeting vector at both the 5′ and 3′ ends (Fig. 2A). Both probes were used independently to identify gene targeting in ES cells (data not shown), and the 3′ probe was used to discriminate the wild-type and targeted alleles in mice (Fig. 2B). Using this Southern blot strategy, we confirmed the genotypes of wild-type mice and mice heterozygous and homozygous for the HRC null allele (Fig. 2B). In addition, we used the HRCBP antibody to examine HRCBP protein levels in striated muscle from age- and sex-matched littermates (Fig. 2C). When equivalent amounts of skeletal muscle and heart homogenates (as determined by equivalent α-tubulin expression) were analyzed by Western blotting, the 160-kDa band corresponding to HRCBP was detected in striated muscle tissue from wild-type and heterozygous mice but not in tissue from homozygous null mice, indicating that targeting resulted in a null allele (Fig. 2C).
FIG. 2.
Generation of HRC null mice. (A) Schematic representation of the targeting strategy for generating the HRC null allele. The transcriptional and translational start sites and exon 1 were replaced by a neomycin resistance cassette by recombination of homologous flanking sequences. Blue boxes represent HRC exons. Yellow and green boxes represent neomycin (NEO) and thymidine kinase (TK) selection cassettes (arrows indicate orientation), respectively. The locations of the 3′ and 5′ probes, which reside outside the homology arms, are shown. E, EcoRI; B, BamHI; P, Psp1406I; S, SalI; X, XhoI. (B) Southern blot analysis of tail DNA from wild type (+/+), heterozygous (+/−), and homozygous HRC null (−/−) mice. Arrows indicate the position of the wild-type (wt) and HRC null (mut) alleles in EcoRI-digested genomic DNA detected with the 3′ probe indicated in panel A. (C) Immunoblotting confirms the absence of HRCBP in striated muscle from HRC null mice. Skeletal muscle and heart homogenates from age- and sex-matched wild type (+/+), heterozygous (+/−), and homozygous HRC null (−/−) mice were probed with anti-HRCBP and anti-α-tubulin. Similar levels of α-tubulin were observed in every sample. HRCBP was only present in skeletal muscle and heart from wild-type and heterozygous mice and was absent in muscle from HRC null mice. Bars on the right depict the position of Full Range Rainbow molecular weight markers. BSA, bovine serum albumin. (D) HRCBP is absent from hearts of HRC knockout mice. The dark staining shows the localization of HRCBP in the heart as detected by immunohistochemistry with anti-HRCBP (α-HRCBP). The panel on the left shows that HRCBP was present in all four chambers of the heart in wild-type mice (wt), while the center panel shows that HRCBP expression was absent from HRC null hearts (ko). The panel on the right shows the absence of HRCBP in a wild-type heart section that received no primary antibody (No Ab control). Arrowheads denote selected sites of strong expression in the wild-type antibody-treated section and the corresponding regions in the knockout and control sections. LA, left atrium; LV, left ventricle; RA, right atrium; RV, right ventricle.
We also examined the expression of HRCBP within the adult myocardium in wild-type and HRC knockout mice (Fig. 2D). Sections through hearts of adult wild-type mice treated with anti-HRCBP showed robust staining in both the left and right ventricles as well as the left and right atria (Fig. 2D, left panel). Furthermore, HRCBP was widely distributed throughout each chamber and did not localize to a specific region within the myocardium. In the ventricles, HRCBP was detected in the myocardium of the compact and trabecular layers and in the myocardial cushions (Fig. 2D, left panel). Previous studies showed that the human HRC promoter directs strong expression to the ventricles in transgenic mice but only weak or no expression to the atria (2). It is possible that differences in transcriptional activation, reflected by the promoter studies, do not directly correlate with steady-state protein expression, which is indicated by the immunohistochemical studies shown here (Fig. 2D). Alternatively, the observed discrepancy in atrial expression may reflect differences between human and mouse HRC regulation, or it might indicate that additional cis-regulatory elements outside the proximal promoter region are required for atrial expression of HRC. In contrast, no signal was detected in HRC null hearts using the anti-HRCBP antibody, providing additional evidence that HRCBP protein was absent in HRC null mice (Fig. 2D, center panel). A control stained wild-type heart, in which no anti-HRCBP antibody was added, also showed no detectable staining, demonstrating the specificity of the antibody for immunohistochemistry (Fig. 2D, right panel).
HRC null mice exhibit impaired weight gain.
Homozygous HRC null mice were present at Mendelian frequency at weaning (Table 1), were viable into adulthood, and were fertile. Interestingly, however, HRC null mice weighed significantly less than age- and sex-matched wild-type controls beginning at 11 months of age (Fig. 3A), and HRC null mice exhibited significant weight loss from 32.3 g at 11 months to 28.3 g at 13 months of age (P = 0.0002). The weight difference between HRC knockout mice and their wild-type littermates became more profound by 13 months of age, when wild-type male mice had an average body weight of 35.6 g compared to 28.3 g for HRC knockout mice (P < 0.0001). To determine if this weight difference impacted all organs and tissues equally, we measured heart, kidney, skeletal muscle, and abdominal fat pad weights from 13-month-old wild-type and HRC null mice (Fig. 3B). HRC null mice displayed a significant reduction in skeletal muscle and fat pad weights, while the heart and kidney weights remained unchanged (Fig. 3B). Consistent with our observations, heart and skeletal muscle tissue from 6-month-old HRC null mice displayed no obvious evidence of abnormalities when analyzed histologically (data not shown). At 13 months of age, skeletal muscle fibers from the quadriceps, soleus, and plantaris muscles appeared to be slightly smaller in cross-sectional area, which is consistent with the observed reduction in muscle mass, although no other abnormalities in muscle histology, fibrosis, or prevalence of centrally located nuclei were observed (data not shown).
TABLE 1.
HRC null mice are present at Mendelian frequencies at weaninga
| HRC genotype | No. of mice with genotype:
|
|
|---|---|---|
| Observed | Expected | |
| +/+ | 43 | 36 |
| +/− | 74 | 72 |
| −/− | 27 | 36 |
A total of 154 mice generated by crosses of mice heterozygous for HRC were genotyped at weaning. The numbers of mice of each genotype are presented here (observed) and were not significantly different from the numbers predicted by standard Mendelian inheritance (expected), as determined by chi-square analysis (chi-square value = 3.67, 2 degrees of freedom). +/+, wild-type mice; +/−, HRC heterozygous mice; −/−, HRC null mice.
FIG. 3.
HRC null mice have impaired weight gain after 11 months of age. (A) Wild-type (wt) and HRC null (ko) male mice had similar body weights at 5, 6, and 8 months of age. In contrast, HRC null mice displayed a significant reduction in body weight compared to wild-type controls at 11 months of age (32.3 ± 0.76 g, n = 10 for HRC null; 35.8 ± 1.4 g, n = 10 for wild type [P = 0.037]) and 13 months of age (28.3 ± 0.56 g, n = 16 for HRC null; 35.6 ± 0.94 g, n = 16 for wild type [P < 0.0001]). (B) HRC null mice had decreased skeletal muscle and fat pad weight at 13 months of age. While hearts and kidneys from wild-type and HRC null mice had similar weights, skeletal muscles of the crural group and abdominal fat pads from HRC null mice weighed significantly less than those from wild-type mice (89% ± 2% of wild type, n = 3, P = 0.0354 for skeletal muscle; 75% ± 6% of wild type for fat pad, n = 8, P = 0.0194). Organ and tissue weights were normalized to tibia length for each animal and are expressed as percentage of wild-type weight. P values were calculated using unpaired, two-tailed t test analyses. Error bars represent the standard error of the mean for each group.
HRC null hearts exhibit normal calcium handling.
Because HRCBP has been shown to interact with triadin (44, 56, 57), which is associated with the ryanodine receptor (24, 25, 67), we considered the possibility that HRC null mice might have defects in calcium binding in the heart. Therefore, we tested whether HRCBP might be involved in calcium release from the SR. We compared the [3H]ryanodine binding activity of microsome homogenate preparations from the hearts of wild-type and HRC null mice since binding of ryanodine to low-affinity sites on the ryanodine receptor is dependent on the activation state of the receptor and provides an indirect measure of calcium release (14). We observed no significant difference in [3H]ryanodine binding to homogenates from wild-type and HRC null hearts in three independent experiments (Fig. 4A). The SR calcium loading capacities of homogenates from wild-type mice were also similar (Fig. 4B), demonstrating that loss of HRCBP has no effect on SR calcium storage capacity in this assay and suggesting that HRCBP is not involved in SR calcium storage. This is in contrast to previous overexpression studies in tissue culture that have implicated a role for HRCBP in calcium storage but is consistent with a more recent report demonstrating that the overexpression of HRCBP in the heart in vivo does not alter the SR calcium load (18, 23, 34). Finally, we observed no difference in the rates of calcium uptake by homogenates derived from wild-type and HRC null hearts using a fluorimetric assay (Fig. 4C). In contrast to mice that overexpress HRC and show depressed calcium uptake rates in the heart, these data suggest that calcium uptake into the cardiac SR of HRC null mice is normal, at least in vitro (23).
FIG. 4.
Normal calcium handling in cardiac muscle from HRC null mice. (A) Ryanodine binding as an indirect measure of SR calcium release channel activity in wild-type (wt) and HRC knockout (ko) mice. Ryanodine binding to wild type (105.5 ± 14.5 fmol ryanodine/mg) and knockout (84.8 ± 20 fmol ryanodine/mg) homogenates was not statistically different. Data are expressed as the mean values plus standard error from experiments performed in duplicate on three independently prepared sets of homogenates. (B) Calcium loading capacity of the SR from wild-type and HRC null mice. The SR calcium loads of homogenates from wild-type (47.9 ± 7.7 nmol Ca2+/mg) and HRC knockout (65.4 ± 12.1 nmol Ca2+/mg) mice were not significantly different. Data expressed are the mean values plus standard error of the means from four experiments performed on two independently prepared sets of homogenates. (C) Fluorimetric assay for calcium uptake by the SR of wild-type and HRC null mice. Calcium uptake was initiated by the addition of ATP, and CaCl2 was added in 1-μM increments (arrowheads). No difference in the shape of the calcium uptake curve was observed between homogenates from wild-type and HRC knockout mice. The traces shown are representative of three independent experiments performed on two independent sets of homogenate preparations.
Increased cardiac expression of triadin in HRC null mice.
We investigated the possibility that the expression of other SR-associated proteins might be altered as a compensatory response to the lack of HRCBP in the heart (Fig. 5). The expression levels of calsequestrin, ryanodine receptor, and phospholamban, a SERCA-associated regulatory protein (7), appeared to be identical in wild-type and HRC null mouse hearts (Fig. 5A to C). α-Tubulin, a cytoplasmic protein, also was not differentially expressed between wild-type and HRC null mouse hearts (Fig. 5D). In contrast, triadin expression was significantly increased in the absence of HRCBP (Fig. 5E and F). Interestingly, triadin and HRCBP have been shown to interact, and triadin is associated with the calcium release channel complex, which supports the possibility that increased triadin expression may potentially compensate, at least in part, for the loss of HRCBP.
FIG. 5.
Hearts from HRC null mice have increased levels of triadin expression. Cardiac homogenates from age- and sex-matched wild-type (wt) and HRC null (ko) mice were analyzed by Western blotting with antibodies to calsequestrin (CSQ), the cardiac ryanodine receptor (RyR), phospholamban (PLB), α-tubulin, and triadin. Calsequestrin (A), ryanodine receptor (B), the pentameric form of phospholamban (C), and α-tubulin (D) levels were nearly identical in wt and ko homogenates. The similar α-tubulin levels in wild-type and HRC null homogenates confirmed that approximately equal of amounts of protein were analyzed in the two samples. In contrast to calsequestrin, ryanodine receptor, and phospholamban, triadin expression was increased in HRC knockout mouse hearts (E). Bars on the left in panels A to E depict the position of Full Range Rainbow molecular weight markers. (F) Triadin expression is significantly increased in HRC null compared to wild-type cardiac tissue (1.52 ± 0.23-fold over wild type, P = 0.0027). Triadin levels from three independent pools of cardiac extract from age- and sex-matched knockout and wild-type mice were analyzed in independent experiments, and the expression of triadin was normalized to the α-tubulin level for each independent sample. Data are expressed relative to the wild-type triadin protein level, and the P values were calculated using a two-tailed, paired t test. Error bars represent the standard error of the mean for each group.
HRC null mice display increased susceptibility to isoproterenol-induced cardiac hypertrophy.
Although the hearts of HRC null mice are morphologically indistinguishable from those of their wild-type littermates, we considered the possibility that loss of HRCBP may result in defects that are not readily apparent under normal laboratory conditions. To address this possibility, we employed a voluntary exercise assay to determine whether HRC knockout mice had a reduced capacity for exercise due to deficits in either cardiac or skeletal muscle function at 3 months and 11 months of age (Fig. 6). At 3 months of age, which was prior to the onset of any changes in overall body weight or skeletal muscle weight in HRC knockout mice, no statistical difference in running ability was observed (Fig. 6A). Similarly, at 11 months of age, when significant weight differences were already apparent, no significant difference in voluntary running ability was observed between wild-type and knockout mice (Fig. 6B). These data demonstrate that HRCBP function is not required for normal exercise ability in a voluntary running assay over a 5-day period.
FIG. 6.
HRC null mice show no defects in exercise capacity or exaggeration of exercise-induced cardiac hypertrophy compared to wild-type littermates in a voluntary wheel running assay. Three-month-old (A) or 11-month-old (B) male mice were placed in a cage with a rodent wheel, and the distance each mouse ran in a 16-h period each night for five consecutive nights was recorded. Wild-type (wt) and HRC null (ko) mice ran 3.5 ± 0.5 km/night and 4.5 ± 1.2 km/night at 3 months, respectively, and 2.3 ± 0.3 km/night and 2.2 ± 0.3 km/night at 11 months, respectively. The mean nightly distance run plus the standard error of the mean for three mice of each genotype at 3 months and five mice at 11 months is presented. (C) Exercise-induced cardiac hypertrophy was measured in 11-month-old wild-type and HRC knockout mice after 28 days of voluntary wheel running. This exercise regimen resulted in a statistically significant increase in heart weight/tibia length in both wild-type (114% ± 5% of wild-type no-run control, n = 5, P = 0.047) and HRC knockout mice (119% ± 6% of wild-type no-run control, n = 5, P = 0.015). The difference in the normalized heart weight/tibia length between wild-type (114%) and HRC knockout mice (119%) following the voluntary exercise regimen was not statistically different. P values were calculated using unpaired, two-tailed t test analyses. Data are presented as the percentage of the wild-type no-run control. Error bars represent the standard error of the mean for each group.
Although HRC null mice exhibited no difference in voluntary exercise performance compared to their wild-type littermates, we assessed whether voluntary exercise induced similar levels of cardiac hypertrophy in wild-type and knockout mice (Fig. 6C). We subjected 11-month-old wild-type and HRC null mice to voluntary exercise for 28 consecutive nights, which has been shown previously to induce mild physiological hypertrophy in mice (1). No significant difference was observed in the distances run by the two groups of mice over the 28-day period in spite of the fact that knockout mice were significantly smaller at that age (Fig. 3A). Wild-type and HRC knockout mice both displayed a significant increase in heart weight/tibia length induced by exercise (Fig. 6C). Hearts from the exercised wild-type mice were 13.6% larger than hearts from sedentary wild-type controls (P = 0.047), while the hearts of the exercised HRC knockout mice were 19.2% larger than the sedentary wild-type control hearts (P = 0.015). Although the trend suggested a slight exaggeration in hypertrophy in knockout compared to wild-type mice subjected to voluntary exercise, the difference in the heart weight/tibia length ratio between wild-type and knockout mice was not statistically significant, indicating that HRC knockout mice do not display an exaggerated response to exercise-induced physiologic cardiac hypertrophy.
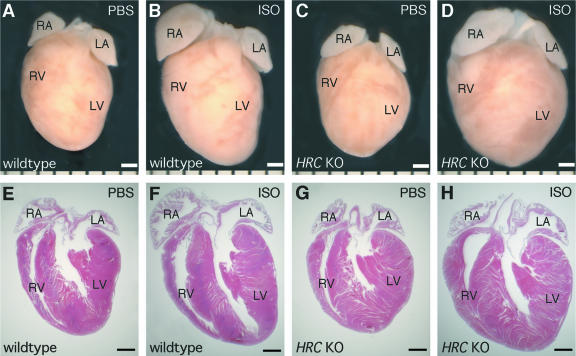
To determine the consequences of loss of HRCBP in the heart in the presence of a pathological stimulus, we examined the responses of sex-matched wild-type and HRC null mice to cardiac hypertrophy induced by chronic β-adrenergic receptor stimulation at 6 months of age, which was prior to any obvious differences in body weight. To do this, we surgically implanted HRC null and wild-type male mice with miniosmotic pumps containing either isoproterenol in PBS or PBS alone. Prolonged subcutaneous administration of isoproterenol, a β-adrenergic receptor agonist, has been shown previously to induce cardiac hypertrophy (22). After 10 days of treatment, we removed the hearts and examined them for evidence of cardiac hypertrophy (Fig. 7). Hearts from wild-type (Fig. 7A and E) and HRC null (Fig. 7C and G) mice treated with PBS alone were similar in size. Treatment with isoproterenol resulted in a dramatic increase in the size of hearts from wild-type mice (Fig. 7, compare panels A and E to panels B and F). Likewise, treatment of HRC null mice with isoproterenol resulted in a strong hypertrophic response (Fig. 7, compare panels C and G to panels D and H). The hypertrophic response to isoproterenol treatment was substantially greater in HRC null mice than in wild-type mice (Fig. 7, compare panels B and F to panels D and H), indicating an exaggerated hypertrophic response in mice lacking HRCBP.
FIG. 7.
Exaggerated hypertrophic response to isoproterenol in HRC null mice. Isoproterenol (ISO) treatment resulted in a dramatic increase in heart size in wild type (B and F) and HRC null (D and H) mice when compared to mice treated with PBS alone, where wild type (A and E) and HRC null (C and G) mouse hearts were similar in size. Hearts from HRC null (D and H) mice treated with isoproterenol were larger than the hearts of isoproterenol-treated wild-type (B and F) mice. Photographs are of whole-mount (A to D) and hematoxylin-and-eosin-stained frontal sections (E to H). The bar is equal to 1 mm. KO, knockout; LA, left atrium; LV, left ventricle; RA, right atrium; RV, right ventricle.
We quantified the extent of hypertrophy induced by isoproterenol in HRC knockout and heterozygous mice by determining the heart weight/tibia length ratios for each group and normalizing them to PBS-treated wild-type mice (Fig. 8). Consistent with the results shown in Fig. 7, there was no difference in the sizes of hearts from wild-type and HRC null mice treated with PBS alone. However, while isoproterenol treatment resulted in a significant increase in the size of hearts from wild-type mice (28%), the hypertrophic response was significantly exaggerated in hearts from HRC null mice (46%), and this increase in heart size in HRC null mice treated with isoproterenol was significantly greater than that of wild-type mice treated with isoproterenol (Fig. 8). It appears that this increase in heart size in HRC null mice may be due to additional concentric hypertrophy and not to further progression to dilated cardiomyopathy, since we observed no signs of chamber dilation (Fig. 7). Interestingly, heterozygous mice treated with isoproterenol showed an intermediate increase in heart size, suggesting that mice with only a single copy of the HRC gene may have an intermediate phenotype. Our finding that HRC null hearts exhibited a greater hypertrophic response to isoproterenol treatment than wild-type hearts suggests that HRC null hearts may have defects in cardiac function and are unable to compensate to preserve cardiac function under pathological conditions without an exaggerated hypertrophic response.
FIG. 8.
HRC null mice display an exaggerated response to cardiac hypertrophy induced by isoproterenol. Isoproterenol treatment resulted in a statistically significant increase in heart weight/tibia length ratio in mice of each genotype at 6 months of age (**, P < 0.0001 in every case). No significant differences in heart weight/tibia length ratio were observed among the three genotypes when mice were treated with PBS alone. However, HRC null mice treated with isoproterenol showed a significantly greater increase in heart weight/tibia length ratio than isoproterenol-treated wild-type mice (146% ± 5% versus 128% ± 3% of PBS-treated +/+ mice; P = 0.0132). Heterozygous mice treated with isoproterenol exhibited an intermediate heart weight/tibia length ratio ratio (137% ± 4% of PBS-treated +/+ mice). P values were determined by two-tailed, unpaired t test analyses. Data are presented as the mean percentage of the heart weight/tibia length ratio ratio of the PBS-treated wild-type (+/+) mice, and error bars represent the standard error of the mean for each group (n = 19 for each group).
DISCUSSION
Cardiac hypertrophy is a compensatory response to increased load or defects in myocyte contractility that results in the activation of a fetal gene transcriptional program, elevated protein synthesis, increased myocyte size, and enlargement of the heart (21, 48, 49). The studies presented here demonstrate that loss of HRCBP function in vivo results in an increased response to a pathological β-adrenergic hypertrophic stimulus (Fig. 7 and 8). Several recent reports have described transgenic or knockout mice with normal hearts that also show an exaggerated response to a hypertrophic stimulus. In mouse models of heart disease, cardiac hypertrophy is commonly induced by aortic constriction or treatment with isoproterenol. Most of the reports describing normal hearts that display an exaggerated response to hypertrophic induction involved genetic perturbations of proteins known or suspected to play a role in regulating the hypertrophic response. Knockouts of histone deacetylase 5, histone deacetylase 9, calsarcin 1, and FHL2, a cardiac specific LIM domain transcription factor, all provide examples in which mice had normal hearts, but showed an increased hypertrophic response to isoproterenol or aortic constriction (12, 20, 40, 66). Similarly, when α-myosin heavy chain was replaced by β-myosin heavy chain, transgenic hearts had moderately reduced contractility and a dramatically greater hypertrophic response to isoproterenol, even though those hearts appeared to be morphologically normal in the absence of isoproterenol treatment (41). It is likely that in each of these models, mice have morphologically normal hearts because they have already undergone compensatory changes to preserve cardiac function under normal conditions. As a result, these hearts have a reduced capacity to compensate in response to a pathological stimulus other than to undergo additional hypertrophy (41).
It is interesting that we did not observe a significant difference in exercise-induced hypertrophy between wild-type and HRC knockout mice (Fig. 6). This may indicate a difference in the requirement for HRCBP function in physiological versus pathological hypertrophy. Alternatively, voluntary exercise may not have induced a robust enough hypertrophic response overall to reveal the underlying defect in HRC null mice. Consistent with this notion, exercise-induced hypertrophy has been shown previously to cause only relatively mild hypertrophy in wild-type mice (1). While exercise did induce significant hypertrophy in both wild-type and HRC knockout mice (Fig. 6C), it was less exaggerated than isoproterenol-induced hypertrophy (Fig. 8), and it may not have been sufficient to exhaust the compensatory capacity in the knockout animals.
Previous overexpression studies in cultured rat cardiomyocytes suggested that HRCBP may play a role in calcium storage by the SR (18, 34). However, HRCBP only accounts for approximately 1% of junctional SR protein, while calsequestrin is far more abundant (17). These observations suggest that calsequestrin probably accounts for the bulk of calcium buffering in the SR (17). Furthermore, the releasable calcium content of the cardiac SR was not altered in transgenic mice overexpressing HRCBP in the heart (23). Consistent with the notion that HRCBP does not play a major role in calcium buffering, we observed no differences in calcium storage capacity in isolated SR preparations from wild-type and HRC null hearts (Fig. 4).
Previous studies have demonstrated that HRCBP interacts with triadin, another SR protein (44, 56). Triadin may be involved in sensing luminal calcium levels and mediating ryanodine receptor activity in response to changes in luminal calcium (25). The interaction between the two proteins suggests that HRCBP may also modulate the ryanodine receptor in response to luminal calcium levels (44, 56, 57). Although we observed no differences in ryanodine receptor activity in cardiac homogenates from wild-type and HRC null mice (Fig. 4), it is possible that HRCBP modulates calcium release from the ryanodine receptor in a manner that cannot be detected in an in vitro ryanodine binding assay. Alternatively, the increase in triadin expression (Fig. 5) and other compensatory changes may partially offset a calcium handling defect in HRC null hearts under normal conditions, but this compensation may be unable to preserve normal contractility under pathological conditions, such as chronic β-adrenergic receptor stimulation (Fig. 7 and 8). Interestingly, overexpression of HRCBP in the heart also resulted in the upregulation of triadin expression (18, 23). However, the predominantly skeletal muscle isoform of triadin was increased in those studies, while we observed an increase in the levels of the cardiac isoform of triadin (18, 23). Furthermore, the skeletal muscle isoform of triadin may inhibit the activity of the ryanodine receptor while the cardiac isoform appears to stabilize the open conformation of the release channel (25, 50, 54, 61). Because these two isoforms of triadin appear to play different roles in regulating ryanodine receptor activity, it is possible that the skeletal muscle isoform may be upregulated to compensate for the gain of HRCBP function while the cardiac isoform may be upregulated to compensate for the loss of HRCBP function.
Changes in metabolism also accompany cardiac hypertrophy (4, 58). The hypertrophied heart is marked by an increase in glycolysis and decreased phosphocreatine/ATP ratios, suggesting that muscle metabolism may be altered by contractile defects (4, 11, 58, 60, 63). Since HRC null mice display exaggerated cardiac hypertrophy induced by isoproterenol and suffer from an age-dependent impairment in weight gain compared to wild-type controls, it will be important for future studies to determine if alterations in myocardial metabolism are present in HRC null mice and if these changes contribute to the exaggerated response to β-adrenergic receptor stimulation. It is also possible that changes in cardiac or skeletal muscle function in HRC null mice may result in overall changes in energy usage, resulting in secondary loss of fat and muscle weight, but this hypothesis remains to be tested directly.
Our observation that HRC null mice display an increased hypertrophic response to an external stimulus may have important implications for pathological cardiac hypertrophy in humans. Changes in calcium handling and in the expression of SR proteins have been reported in cardiac hypertrophy and heart failure previously (3, 5, 28). Mutations in the ryanodine receptor and calsequestrin genes are associated with catecholaminergic polymorphic ventricular tachycardia, and mutation of the phospholamban gene is associated with lethal dilated cardiomyopathy, providing additional evidence that normal calcium handling is required for proper cardiac function (26, 27, 42, 53). The exaggerated response of HRC null mice to a pathological hypertrophic stimulus suggests that mutations in HRC, in combination with other genetic and environmental factors, may also contribute to pathological cardiac hypertrophy and heart failure in humans.
Acknowledgments
We thank Yong Ji, Ben Wilkins, and Jeff Molkentin for invaluable assistance in implementing techniques required for these studies. We are grateful to Ken Chien, Anthony Baker, Andrew Lokuta, Leslie Leinwand, and Brooke Harrison for helpful discussions. We appreciate the critical comments on the manuscript provided by David Gardner. We also thank Preston Ford, Marina Haugland, Morgan Royce-Tolland, Jean Regard, Miao-Hsueh Chen, Yajun Li, Andrew Tauscher, and Alex Fay for assistance with these studies.
E.J.J. was supported in part by a predoctoral fellowship from the American Heart Association, Western States Affiliate, and by a graduate fellowship from the Gladstone Institutes of Cardiovascular Disease. A.B.H. was supported in part by a predoctoral fellowship from the Howard Hughes Medical Institute. This work was supported by grants HL64658 and AR52130 from the NIH to B.L.B.
Footnotes
Published ahead of print on 9 October 2006.
REFERENCES
- 1.Allen, D. L., B. C. Harrison, A. Maass, M. L. Bell, W. C. Byrnes, and L. A. Leinwand. 2001. Cardiac and skeletal muscle adaptations to voluntary wheel running in the mouse. J Appl. Physiol. 90:1900-1908. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, J. P., E. Dodou, A. B. Heidt, S. J. De Val, E. J. Jaehnig, S. B. Greene, E. N. Olson, and B. L. Black. 2004. HRC is a direct transcriptional target of MEF2 during cardiac, skeletal, and arterial smooth muscle development in vivo. Mol. Cell. Biol. 24:3757-3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arai, M., H. Matsui, and M. Periasamy. 1994. Sarcoplasmic reticulum gene expression in cardiac hypertrophy and heart failure. Circ. Res. 74:555-564. [DOI] [PubMed] [Google Scholar]
- 4.Ashrafian, H., C. Redwood, E. Blair, and H. Watkins. 2003. Hypertrophic cardiomyopathy: a paradigm for myocardial energy depletion. Trends Genet. 19:263-268. [DOI] [PubMed] [Google Scholar]
- 5.Balke, C. W., and S. R. Shorofsky. 1998. Alterations in calcium handling in cardiac hypertrophy and heart failure. Cardiovasc. Res. 37:290-299. [DOI] [PubMed] [Google Scholar]
- 6.Beard, N. A., D. R. Laver, and A. F. Dulhunty. 2004. Calsequestrin and the calcium release channel of skeletal and cardiac muscle. Prog. Biophys. Mol. Biol. 85:33-69. [DOI] [PubMed] [Google Scholar]
- 7.Bers, D. M. 2002. Cardiac excitation-contraction coupling. Nature 415:198-205. [DOI] [PubMed] [Google Scholar]
- 8.Bers, D. M. 2004. Macromolecular complexes regulating cardiac ryanodine receptor function. J. Mol. Cell Cardiol. 37:417-429. [DOI] [PubMed] [Google Scholar]
- 9.Bueno, O. F., B. J. Wilkins, K. M. Tymitz, B. J. Glascock, T. F. Kimball, J. N. Lorenz, and J. D. Molkentin. 2002. Impaired cardiac hypertrophic response in calcineurin Abeta-deficient mice. Proc. Natl. Acad. Sci. USA 99:4586-4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campbell, K. P., and A. E. Shamoo. 1980. Chloride-induced release of actively loaded calcium from light and heavy sarcoplasmic reticulum vesicles. J. Membr. Biol. 54:73-80. [DOI] [PubMed] [Google Scholar]
- 11.Carvajal, K., and R. Moreno-Sanchez. 2003. Heart metabolic disturbances in cardiovascular diseases. Arch. Med. Res. 34:89-99. [DOI] [PubMed] [Google Scholar]
- 12.Chang, S., T. A. McKinsey, C. L. Zhang, J. A. Richardson, J. A. Hill, and E. N. Olson. 2004. Histone deacetylases 5 and 9 govern responsiveness of the heart to a subset of stress signals and play redundant roles in heart development. Mol. Cell. Biol. 24:8467-8476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheah, S. S., and R. R. Behringer. 2000. Gene-targeting strategies. Methods Mol. Biol. 136:455-463. [DOI] [PubMed] [Google Scholar]
- 14.Coronado, R., J. Morrissette, M. Sukhareva, and D. M. Vaughan. 1994. Structure and function of ryanodine receptors. Am. J. Physiol. 266:C1485-C1504. [DOI] [PubMed] [Google Scholar]
- 15.Damiani, E., and A. Margreth. 1991. Subcellular fractionation to junctional sarcoplasmic reticulum and biochemical characterization of 170 kDa Ca(2+)- and low-density-lipoprotein-binding protein in rabbit skeletal muscle. Biochem. J. 277:825-832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Damiani, E., E. Picello, L. Saggin, and A. Margreth. 1995. Identification of triadin and of histidine-rich Ca(2+)-binding protein as substrates of 60 kDa calmodulin-dependent protein kinase in junctional terminal cisternae of sarcoplasmic reticulum of rabbit fast muscle. Biochem. Biophys. Res. Commun. 209:457-465. [DOI] [PubMed] [Google Scholar]
- 17.Damiani, E., G. Tobaldin, E. Bortoloso, and A. Margreth. 1997. Functional behaviour of the ryanodine receptor/Ca(2+)-release channel in vesiculated derivatives of the junctional membrane of terminal cisternae of rabbit fast muscle sarcoplasmic reticulum. Cell Calcium 22:129-150. [DOI] [PubMed] [Google Scholar]
- 18.Fan, G. C., K. N. Gregory, W. Zhao, W. J. Park, and E. G. Kranias. 2004. Regulation of myocardial function by histidine-rich, calcium-binding protein. Am. J. Physiol. Heart Circ. Physiol. 287:H1705-H1711. [DOI] [PubMed] [Google Scholar]
- 19.Forbes, M. S., L. A. Hawkey, S. K. Jirge, and N. Sperelakis. 1985. The sarcoplasmic reticulum of mouse heart: its divisions, configurations, and distribution. J. Ultrastruct. Res. 93:1-16. [DOI] [PubMed] [Google Scholar]
- 20.Frey, N., T. Barrientos, J. M. Shelton, D. Frank, H. Rutten, D. Gehring, C. Kuhn, M. Lutz, B. Rothermel, R. Bassel-Duby, J. A. Richardson, H. A. Katus, J. A. Hill, and E. N. Olson. 2004. Mice lacking calsarcin-1 are sensitized to calcineurin signaling and show accelerated cardiomyopathy in response to pathological biomechanical stress. Nat. Med. 10:1336-1343. [DOI] [PubMed] [Google Scholar]
- 21.Frey, N., and E. N. Olson. 2003. Cardiac hypertrophy: the good, the bad, and the ugly. Annu. Rev. Physiol. 65:45-79. [DOI] [PubMed] [Google Scholar]
- 22.Friddle, C. J., T. Koga, E. M. Rubin, and J. Bristow. 2000. Expression profiling reveals distinct sets of genes altered during induction and regression of cardiac hypertrophy. Proc. Natl. Acad. Sci. USA 97:6745-6750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gregory, K. N., K. S. Ginsburg, I. Bodi, H. Hahn, Y. M. Marreez, Q. Song, P. A. Padmanabhan, B. A. Mitton, J. R. Waggoner, F. Del Monte, W. J. Park, G. W. Ii, D. M. Bers, and E. G. Kranias. 2006. Histidine-rich Ca binding protein: a regulator of sarcoplasmic reticulum calcium sequestration and cardiac function. J. Mol. Cell Cardiol. 40:653-665. [DOI] [PubMed] [Google Scholar]
- 24.Guo, W., and K. P. Campbell. 1995. Association of triadin with the ryanodine receptor and calsequestrin in the lumen of the sarcoplasmic reticulum. J. Biol. Chem. 270:9027-9030. [DOI] [PubMed] [Google Scholar]
- 25.Gyorke, I., N. Hester, L. R. Jones, and S. Gyorke. 2004. The role of calsequestrin, triadin, and junctin in conferring cardiac ryanodine receptor responsiveness to luminal calcium. Biophys. J. 86:2121-2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haghighi, K., F. Kolokathis, A. O. Gramolini, J. R. Waggoner, L. Pater, R. A. Lynch, G. C. Fan, D. Tsiapras, R. R. Parekh, G. W. Dorn II, D. H. Maclennan, D. T. Kremastinos, and E. G. Kranias. 2006. A mutation in the human phospholamban gene, deleting arginine 14, results in lethal, hereditary cardiomyopathy. Proc. Natl. Acad. Sci. USA 103:1388-1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haghighi, K., F. Kolokathis, L. Pater, R. A. Lynch, M. Asahi, A. O. Gramolini, G. C. Fan, D. Tsiapras, H. S. Hahn, S. Adamopoulos, S. B. Liggett, G. W. Dorn II, D. H. MacLennan, D. T. Kremastinos, and E. G. Kranias. 2003. Human phospholamban null results in lethal dilated cardiomyopathy revealing a critical difference between mouse and human. J. Clin. Investig. 111:869-876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hasenfuss, G. 1998. Alterations of calcium-regulatory proteins in heart failure. Cardiovasc. Res. 37:279-289. [DOI] [PubMed] [Google Scholar]
- 29.Hofmann, S. L., M. S. Brown, E. Lee, R. K. Pathak, R. G. Anderson, and J. L. Goldstein. 1989. Purification of a sarcoplasmic reticulum protein that binds Ca2+ and plasma lipoproteins. J. Biol. Chem. 264:8260-8270. [PubMed] [Google Scholar]
- 30.Hofmann, S. L., J. L. Goldstein, K. Orth, C. R. Moomaw, C. A. Slaughter, and M. S. Brown. 1989. Molecular cloning of a histidine-rich Ca2+-binding protein of sarcoplasmic reticulum that contains highly conserved repeated elements. J. Biol. Chem. 264:18083-18090. [PubMed] [Google Scholar]
- 31.Hogan, B., R. Beddington, F. Costantini, and E. Lacy. 1994. Manipulating the mouse embryo, 2nd ed. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 32.Jones, L. R., and S. E. Cala. 1981. Biochemical evidence for functional heterogeneity of cardiac sarcoplasmic reticulum vesicles. J. Biol. Chem. 256:11809-11818. [PubMed] [Google Scholar]
- 33.Jones, L. R., Y. J. Suzuki, W. Wang, Y. M. Kobayashi, V. Ramesh, C. Franzini-Armstrong, L. Cleemann, and M. Morad. 1998. Regulation of Ca2+ signaling in transgenic mouse cardiac myocytes overexpressing calsequestrin. J. Clin. Investig. 101:1385-1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim, E., D. W. Shin, C. S. Hong, D. Jeong, D. H. Kim, and W. J. Park. 2003. Increased Ca2+ storage capacity in the sarcoplasmic reticulum by overexpression of HRC (histidine-rich Ca2+ binding protein). Biochem. Biophys. Res. Commun. 300:192-196. [DOI] [PubMed] [Google Scholar]
- 35.Kirchhefer, U., H. A. Baba, G. Hanske, L. R. Jones, P. Kirchhof, W. Schmitz, and J. Neumann. 2004. Age-dependent biochemical and contractile properties in atrium of transgenic mice overexpressing junctin. Am. J. Physiol. Heart Circ. Physiol. 287:H2216-H2225. [DOI] [PubMed] [Google Scholar]
- 36.Kirchhefer, U., H. A. Baba, Y. M. Kobayashi, L. R. Jones, W. Schmitz, and J. Neumann. 2002. Altered function in atrium of transgenic mice overexpressing triadin 1. Am. J. Physiol. Heart Circ. Physiol. 283:H1334-H1343. [DOI] [PubMed] [Google Scholar]
- 37.Kirchhefer, U., L. R. Jones, F. Begrow, P. Boknik, L. Hein, M. J. Lohse, B. Riemann, W. Schmitz, J. Stypmann, and J. Neumann. 2004. Transgenic triadin 1 overexpression alters SR Ca2+ handling and leads to a blunted contractile response to beta-adrenergic agonists. Cardiovasc. Res. 62:122-134. [DOI] [PubMed] [Google Scholar]
- 38.Kirchhefer, U., J. Neumann, H. A. Baba, F. Begrow, Y. M. Kobayashi, U. Reinke, W. Schmitz, and L. R. Jones. 2001. Cardiac hypertrophy and impaired relaxation in transgenic mice overexpressing triadin 1. J. Biol. Chem. 276:4142-4149. [DOI] [PubMed] [Google Scholar]
- 39.Kirchhefer, U., J. Neumann, D. M. Bers, I. B. Buchwalow, L. Fabritz, G. Hanske, I. Justus, B. Riemann, W. Schmitz, and L. R. Jones. 2003. Impaired relaxation in transgenic mice overexpressing junctin. Cardiovasc. Res. 59:369-379. [DOI] [PubMed] [Google Scholar]
- 40.Kong, Y., J. M. Shelton, B. Rothermel, X. Li, J. A. Richardson, R. Bassel-Duby, and R. S. Williams. 2001. Cardiac-specific LIM protein FHL2 modifies the hypertrophic response to beta-adrenergic stimulation. Circulation 103:2731-2738. [DOI] [PubMed] [Google Scholar]
- 41.Krenz, M., and J. Robbins. 2004. Impact of beta-myosin heavy chain expression on cardiac function during stress. J. Am. Coll. Cardiol. 44:2390-2397. [DOI] [PubMed] [Google Scholar]
- 42.Lahat, H., E. Pras, and M. Eldar. 2003. RYR2 and CASQ2 mutations in patients suffering from catecholaminergic polymorphic ventricular tachycardia. Circulation 107:e29. [Online.] [DOI] [PubMed] [Google Scholar]
- 43.Lamb, G. D. 2000. Excitation-contraction coupling in skeletal muscle: comparisons with cardiac muscle. Clin. Exp. Pharmacol. Physiol. 27:216-224. [DOI] [PubMed] [Google Scholar]
- 44.Lee, H. G., H. Kang, D. H. Kim, and W. J. Park. 2001. Interaction of HRC (histidine-rich Ca(2+)-binding protein) and triadin in the lumen of sarcoplasmic reticulum. J. Biol. Chem. 276:39533-39538. [DOI] [PubMed] [Google Scholar]
- 45.Lokuta, A. J., T. B. Rogers, W. J. Lederer, and H. H. Valdivia. 1995. Modulation of cardiac ryanodine receptors of swine and rabbit by a phosphorylation-dephosphorylation mechanism. J. Physiol. 487:609-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Markel, P., P. Shu, C. Ebeling, G. A. Carlson, D. L. Nagle, J. S. Smutko, and K. J. Moore. 1997. Theoretical and empirical issues for marker-assisted breeding of congenic mouse strains. Nat. Genet. 17:280-284. [DOI] [PubMed] [Google Scholar]
- 47.Milner, R. E., K. S. Famulski, and M. Michalak. 1992. Calcium binding proteins in the sarcoplasmic/endoplasmic reticulum of muscle and nonmuscle cells. Mol. Cell. Biochem. 112:1-13. [DOI] [PubMed] [Google Scholar]
- 48.Molkentin, J. D. 2000. Calcineurin and beyond: cardiac hypertrophic signaling. Circ. Res. 87:731-738. [DOI] [PubMed] [Google Scholar]
- 49.Morkin, E. 2000. Control of cardiac myosin heavy chain gene expression. Microsc. Res. Tech. 50:522-531. [DOI] [PubMed] [Google Scholar]
- 50.Ohkura, M., K. Furukawa, H. Fujimori, A. Kuruma, S. Kawano, M. Hiraoka, A. Kuniyasu, H. Nakayama, and Y. Ohizumi. 1998. Dual regulation of the skeletal muscle ryanodine receptor by triadin and calsequestrin. Biochemistry 37:12987-12993. [DOI] [PubMed] [Google Scholar]
- 51.Pathak, R. K., R. G. Anderson, and S. L. Hofmann. 1992. Histidine-rich calcium binding protein, a sarcoplasmic reticulum protein of striated muscle, is also abundant in arteriolar smooth muscle cells. J. Muscle Res. Cell Motil. 13:366-376. [DOI] [PubMed] [Google Scholar]
- 52.Picello, E., E. Damiani, and A. Margreth. 1992. Low-affinity Ca(2+)-binding sites versus Zn(2+)-binding sites in histidine-rich Ca(2+)-binding protein of skeletal muscle sarcoplasmic reticulum. Biochem. Biophys. Res. Commun. 186:659-667. [DOI] [PubMed] [Google Scholar]
- 53.Priori, S. G., C. Napolitano, N. Tiso, M. Memmi, G. Vignati, R. Bloise, V. Sorrentino, and G. A. Danieli. 2001. Mutations in the cardiac ryanodine receptor gene (hRyR2) underlie catecholaminergic polymorphic ventricular tachycardia. Circulation 103:196-200. [DOI] [PubMed] [Google Scholar]
- 54.Rezgui, S. S., S. Vassilopoulos, J. Brocard, J. C. Platel, A. Bouron, C. Arnoult, S. Oddoux, L. Garcia, M. De Waard, and I. Marty. 2005. Triadin (Trisk 95) overexpression blocks excitation-contraction coupling in rat skeletal myotubes. J. Biol. Chem. 280:39302-39308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ridgeway, A. G., H. Petropoulos, A. Siu, J. K. Ball, and I. S. Skerjanc. 1999. Cloning, tissue distribution, subcellular localization and overexpression of murine histidine-rich Ca2+ binding protein. FEBS Lett. 456:399-402. [DOI] [PubMed] [Google Scholar]
- 56.Sacchetto, R., E. Damiani, F. Turcato, A. Nori, and A. Margreth. 2001. Ca(2+)-dependent interaction of triadin with histidine-rich Ca(2+)-binding protein carboxyl-terminal region. Biochem. Biophys. Res. Commun. 289:1125-1134. [DOI] [PubMed] [Google Scholar]
- 57.Sacchetto, R., F. Turcato, E. Damiani, and A. Margreth. 1999. Interaction of triadin with histidine-rich Ca(2+)-binding protein at the triadic junction in skeletal muscle fibers. J. Muscle Res. Cell Motil. 20:403-415. [DOI] [PubMed] [Google Scholar]
- 58.Sambandam, N., G. D. Lopaschuk, R. W. Brownsey, and M. F. Allard. 2002. Energy metabolism in the hypertrophied heart. Heart Fail. Rev. 7:161-173. [DOI] [PubMed] [Google Scholar]
- 59.Sato, Y., D. G. Ferguson, H. Sako, G. W. Dorn II, V. J. Kadambi, A. Yatani, B. D. Hoit, R. A. Walsh, and E. G. Kranias. 1998. Cardiac-specific overexpression of mouse cardiac calsequestrin is associated with depressed cardiovascular function and hypertrophy in transgenic mice. J. Biol. Chem. 273:28470-28477. [DOI] [PubMed] [Google Scholar]
- 60.Spindler, M., K. W. Saupe, M. E. Christe, H. L. Sweeney, C. E. Seidman, J. G. Seidman, and J. S. Ingwall. 1998. Diastolic dysfunction and altered energetics in the alphaMHC403/+ mouse model of familial hypertrophic cardiomyopathy. J. Clin. Investig. 101:1775-1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Terentyev, D., S. E. Cala, T. D. Houle, S. Viatchenko-Karpinski, I. Gyorke, R. Terentyeva, S. C. Williams, and S. Gyorke. 2005. Triadin overexpression stimulates excitation-contraction coupling and increases predisposition to cellular arrhythmia in cardiac myocytes. Circ. Res. 96:651-658. [DOI] [PubMed] [Google Scholar]
- 62.Terentyev, D., S. Viatchenko-Karpinski, I. Gyorke, P. Volpe, S. C. Williams, and S. Gyorke. 2003. Calsequestrin determines the functional size and stability of cardiac intracellular calcium stores: mechanism for hereditary arrhythmia. Proc. Natl. Acad. Sci. USA 100:11759-11764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tian, R., N. Musi, J. D'Agostino, M. F. Hirshman, and L. J. Goodyear. 2001. Increased adenosine monophosphate-activated protein kinase activity in rat hearts with pressure-overload hypertrophy. Circulation 104:1664-1669. [DOI] [PubMed] [Google Scholar]
- 64.Viatchenko-Karpinski, S., D. Terentyev, I. Gyorke, R. Terentyeva, P. Volpe, S. G. Priori, C. Napolitano, A. Nori, S. C. Williams, and S. Gyorke. 2004. Abnormal calcium signaling and sudden cardiac death associated with mutation of calsequestrin. Circ. Res. 94:471-477. [DOI] [PubMed] [Google Scholar]
- 65.Yano, K., and A. Zarain-Herzberg. 1994. Sarcoplasmic reticulum calsequestrins: structural and functional properties. Mol. Cell. Biochem. 135:61-70. [DOI] [PubMed] [Google Scholar]
- 66.Zhang, C. L., T. A. McKinsey, S. Chang, C. L. Antos, J. A. Hill, and E. N. Olson. 2002. Class II histone deacetylases act as signal-responsive repressors of cardiac hypertrophy. Cell 110:479-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang, L., J. Kelley, G. Schmeisser, Y. M. Kobayashi, and L. R. Jones. 1997. Complex formation between junctin, triadin, calsequestrin, and the ryanodine receptor. Proteins of the cardiac junctional sarcoplasmic reticulum membrane. J. Biol. Chem. 272:23389-23397. [DOI] [PubMed] [Google Scholar]
- 68.Zucchi, R., R. J. Cerniway, S. Ronca-Testoni, R. R. Morrison, G. Ronca, and G. P. Matherne. 2002. Effect of cardiac A(1) adenosine receptor overexpression on sarcoplasmic reticulum function. Cardiovasc. Res. 53:326-333. [DOI] [PubMed] [Google Scholar]