Abstract
BRF1 posttranscriptionally regulates mRNA levels by targeting ARE-bearing transcripts to the decay machinery. We previously showed that protein kinase B (PKB) phosphorylates BRF1 at Ser92, resulting in binding to 14-3-3 and impairment of mRNA decay activity. Here we identify an additional regulatory site at Ser203 that cooperates in vivo with Ser92. In vitro kinase labeling and wortmannin sensitivity indicate that Ser203 phosphorylation is also performed by PKB. Mutation of both serines to alanine uncouples BRF1 from PKB regulation, leading to constitutive mRNA decay even in the presence of stabilizing signals. BRF1 protein is labile because of proteasomal degradation (half-life, <3 h) but becomes stabilized upon phosphorylation and is less stable in PKBα−/− cells. Surprisingly, phosphorylation-dependent protein stability is also regulated by Ser92 and Ser203, with parallel phosphorylation required at these sites. Phosphorylation-dependent binding to 14-3-3 is abolished only when both sites are mutated. Cell compartment fractionation experiments support a model in which binding to 14-3-3 sequesters BRF1 through relocalization and prevents it from executing its mRNA decay activity, as well as from proteasomal degradation, thereby maintaining high BRF1 protein levels that are required to reinstate decay upon dissipation of the stabilizing signal.
Posttranscriptional regulation of mRNA levels is an important mechanism for control of gene expression. Regulation of mRNA turnover rates enables adjustment of the steady-state levels of transcripts and consequently the optimum level of proteins adapted to current physiological needs. Many transcripts under posttranscriptional control are inherently unstable with short half-lives that can, however, be increased in response to appropriate stimuli. Short-lived transcripts bear cis elements that target them to the mRNA decay machinery; of these, the most prevalent is the AU-rich element (ARE) that is located in the 3′ untranslated region (UTR) and is present in up to 8% of all transcripts (4). Unstable ARE-bearing transcripts have been described from a diverse group of genes such as those for cytokines, proto-oncogenes, growth factors, and cell cycle regulators. The ARE generally serves as a binding site for destabilizing AU-binding proteins (AUBPs) that sets in motion a chain of events initiated by deadenylation and decapping and culminate in the destruction of the transcript. AUBPs that have been identified with known mRNA decay-promoting properties are the CCCH tandem Zn finger proteins of the ZFP36/Tis11 family tristetraprolin (TTP), BRF1 (synonyms, ZFP36L1 and Tis11b), BRF2, and ZFP36L3 (6, 10, 24, 41) and the KH domain RNA binding protein KSRP (16). Conversely, HuR is an example of a stabilizing AUBP (14, 29), and in the case of AUF1, different isoforms can exert either a stabilizing or a destabilizing effect (37, 38).
The salient features of posttranscriptional regulation are rapidity and reversibility. The default state of most ARE-bearing transcripts is instability; stabilization prompted by exogenous signals leads to rapid mRNA accumulation and amplification of gene expression with a consequent increase in their protein levels. When the signal dissipates, excess ARE mRNA that has accumulated must be rapidly degraded in order to reinstate the previous physiological state of the cell. Well-described physiological examples of ARE mRNA stabilization are TNF-α and CDX2 production from macrophages stimulated with interleukin-1 (IL-1) or bacterial lipopolysaccharide during infection (10, 20), IL-3 production in mast cells in response to immunoglobulin E (IgE)-linked allergens (47), and IL-2 production from T cells following immune stimulation (28). In addition, stress stimuli such as UV exposure (17), heat shock and ubiquitinylation (25, 26), hypoxia (35), and oncogenesis (32) have been reported to lead to ARE mRNA stabilization.
AUBPs are obvious distal targets for signaling pathways in linking membrane-derived stimuli to the mRNA decay machinery. KSRP (7) and AUF1 (46) activities are negatively regulated by phosphorylation. Stabilizing effects have been reported for the c-Jun kinase in regulating IL-2 (12) and IL-3 (31) mRNAs. The p38-MK2 pathway regulates tumor necrosis factor alpha (TNF-α) mRNA and biosynthesis in an ARE-dependent fashion (11, 19, 33). The target of p38-MK2 in macrophages is TTP, which becomes hyperphosphorylated at multiple sites upon lipopolysaccharide induction with several different putative kinases proposed (9). A critical phosphorylation at Ser178 by MK2 leads to 14-3-3 binding and inhibition of TTP activity (13, 42).
Another mode of stabilization is via the phosphatidylinositol 3-kinase (PI3-K)-protein kinase B (PKB) pathway, which exerts a repressive role on BRF1 activity. Our previous work identified Ser92 as an important phosphoregulatory site targeted by PKB (39). With an in vitro assay, we showed that BRF1 phosphorylation by PKB led to 14-3-3 binding and loss of ARE mRNA decay-promoting activity. Replacement of Ser92 with alanine rendered BRF1 refractory to PKB inhibition. In the present work, we identify Ser203 as a second PKB regulatory site and report that Ser92 and Ser203 cooperate to regulate the in vivo decay activity of this AUBP.
BRF1 knockout mice are embryonic lethal, suggesting a vital role in development (44). In addition, the BRF1 gene is circadianly expressed in peripheral organs such as the heart and liver (34, 43) and thus may be responsible for imposing circadian expression on its target transcripts. These observations support the view that posttranscriptional regulation by BRF1 may play a complex biological role. As it is under circadian expression, the BRF1 protein is expected to be short-lived in order to reflect this mode of expression. Here, we show that BRF1 protein is indeed rapidly degraded according to prediction; however, the protein is stabilized by phosphorylation. Unexpectedly, phosphorylation-dependent protein stabilization is conferred by the same phosphoregulatory sites that also control its mRNA decay activity. We propose a model whereby phosphorylation-dependent binding of BRF1 to 14-3-3 leads to inhibition of both its activity and degradation, thereby uniting these seemingly unconnected phenomena, and that this coordinated regulation ensures that sufficient BRF1 protein is available to rapidly reinstall mRNA decay.
MATERIALS AND METHODS
Plasmids.
Plasmid pTet-off was purchased from Clontech. Plasmids bsd-HisBRF1wt and bsd-HisBRF1S90/92A have been described previously (39). bsd-HisBRF1S203A and the bsd-HisBRF1S90/92/203A triple mutant were constructed by site-directed mutagenesis with TV420 (5′-TTGCCTTTGCTGGGTTTCCCAGTGC-3′) as an upstream primer, TV419 (5′-AGGAATGCTGGAGGCGGGGACGG-3′) as a downstream primer, and bsd-HisBRF1wt and bsd-HisBRF1S90/92A as templates. pTRE-BRF1wt, pTRE-BRF1S203A, and pTRE-BRF1S90/92/203A were constructed by insertion of a XbaI-KpnI(blunt) fragment from bsd-HisBRF1wt, bsd-HisBRF1S203A, and bsdHisBRF1S90/92/203A into an XbaI-SalI(blunt) site of pTRE-myc-GFP (43). Construction of plasmids SP6-ARE (39), (m/p)-HA-PKBα (2), T7-ARE−, and pQE-30-BRF1wt (41) has been described previously. The pSRL vector (Tet-β-globin-IL-3 3′ UTR reporter) was generated from pTet-BBB-AREGMCSF (48) by exchanging the blunted BamHI-BstXI fragment for the blunted BamHI-SphI fragment of pMXh-β-IL-3(UTR)wt (31). pQE-30-BRF1S203A and pQE-30-BRF1wt were generated by insertion of a BamHI(blunt)-XhoI fragment from bsd-HisBRF1wt or bsd-HisBRF1S203A, respectively, into the SmaI-SalI sites of pQE-30 (QIAGEN). The pQE-30-BRF1wt143-233 and BRF1S203A143-233 plasmids were constructed by insertion of the 240-bp SacI/BtrI fragment of bsdHISBRF1wt or bsdHISBRF1S203A, respectively, into the SacI/SmaI site of pQE-30.
Recombinant proteins were produced as previously described (41). BRF1 S203 and phospho-S203 peptides, as well as the keyhole limpet hemocyanin- and ovalbumin-coupled peptides, were purchased from PickCell Laboratories BV.
Cell culture, transfection, and reagents.
HT1080, slowC (40), NIH 3T3 B2A2-23 (48), HIRc-B (30), mouse embryo fibroblast (MEF)-PKBα+/+, and MEF-PKBα−/− (51) cells were grown in Iscove's modified Dulbecco's medium supplemented with 10% fetal calf serum, 50 μM 2-mercaptoethanol, 2 mM glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin at 37° in 5% CO2. To reduce background PKB activity, cells were starved overnight in serum-free medium before stimulation with insulin (20 μg/ml; Sigma). Transfection was performed with Lipofectamine 2000 (Life Technologies) by following the manufacturer's protocol.
Protein purification.
Cell pellets were lysed in radioimmunoprecipitation assay buffer containing Complete Protease Inhibitors (Roche) and Phosphatase Inhibitor Cocktails I and II (Sigma). Subcellular proteome fractionation was performed with the Qproteome Cell Compartment Reagents (QIAGEN) as recommended by the manufacturer. The final fractions were methanol precipitated and resuspended in 80 μl radioimmunoprecipitation assay buffer for protein concentration measurement. Ten micrograms of lysate was run on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) for Western blotting. Recombinant BRF1 bearing a His tag was purified with a nickel column as previously described (39).
In vitro phosphorylation.
The in vitro phosphorylation reaction was performed with a mixture of 30 mM Tris-HCl (pH 7.5), 5 mM MgCl2, 1 mM dithiothreitol, and 0.2 mM [γ-32P]ATP with 4 ng/μl recombinant BRF1 peptides as the substrate and 10 ng/μl of either activated PKB (50), ERK1 (Upstate), p38, or MK2 (kindly provided by H. Gram, Novartis, Basel, Switzerland) for 30 min at 30°C.
ARE mRNA decay analysis.
Measurement of RNA decay in NIH 3T3 B2A2-23 cells of the tetracycline-sensitive Tet-β-globin-IL-3 3′ UTR reporter gene was performed as described previously (39). Reporter transcription was blocked by addition of doxycycline (2 μg/ml; Sigma). Results were quantified by phosphorimaging with QuantityOne software (Bio-Rad).
ARE-dependent in vitro decay assay was performed as previously described (39). Briefly, 5 ng of recombinant BRF1 was added to 5 μg of slowC cell S100 extract and incubated with a labeled ARE RNA probe for the indicated times. The reactions were stopped by addition of sample buffer, and the products were resolved by denaturing PAGE.
Western blot analysis.
Standard Western blot protocols were used. The protein samples were run on precast 4 to 20% gradient SDS-polyacrylamide gels (Anamed) for optimum resolution of BRF1 bands. The antibodies used for Western blot analysis were sc-5536 rabbit polyclonal antitubulin (Santa Cruz), 597F11 mouse monoclonal anti-phospho-Ser473-PKB (Cell Signaling Technology), rabbit polyclonal anti-PKB (Cell Signaling Technology), rabbit polyclonal anti-BRF1 (37), sc-629 rabbit anti-14-3-3β (Santa Cruz), and anti-glyceraldehyde-3-phosphate dehydrogenase (anti-GAPDH; Abcam), anti-histone H1 (Calbiochem) and secondary antibodies goat anti-rabbit IgG-alkaline phosphatase (AP; Southern Biotech), and goat anti-mouse IgG-horseradish peroxidase (HRP; DAKO). Bands were visualized with CDP-Star (Roche) or ECL Advance (Amersham), respectively.
The anti-phospho-S203BRF1 antibody was generated by immunization of three rabbits at PickCell Laboratories BV with a keyhole limpet hemocyanin-linked phosphopeptide (R-L-Q-H-S-F-pS-F-A-G-F-P-S, corresponding to amino acids [aa] 197 to 209 of BRF1). The sera from two positive rabbits were affinity purified with the immunizing peptide linked to activated CH Sepharose 4B (Pharmacia) according to the manufacturer's protocol. The purified antibody was preadsorbed with an unphosphorylated peptide (same sequence as the immunizing peptide) for 3 h on ice before addition to the blot.
Coprecipitation of BRF1-14-3-3 complexes.
His-BRF1 pull down was performed as previously described (39). Briefly, 40 ng of His-tagged recombinant BRF1 (wild type, S90/92A, S203A, and S90/92/203A) was added to 40 μg of slowC cell S100 extract and incubated at 30°C for 30 min. BRF1 was bound to a nickel column (QIAGEN) at 4°C for 1 h in 1× binding buffer (1 mM MgCOOH, 1.5 mM KCOOH, 150 mM NaCl, 10% glycerol) with 10 mM imidazole. The beads were washed (4°C, 2 × 2 h) in 1× binding buffer containing 20 mM imidazole. Bound protein was eluted by boiling in SDS sample buffer and immediately loaded onto gels.
RESULTS
Identification of another PKB regulatory site in addition to BRF1 Ser92.
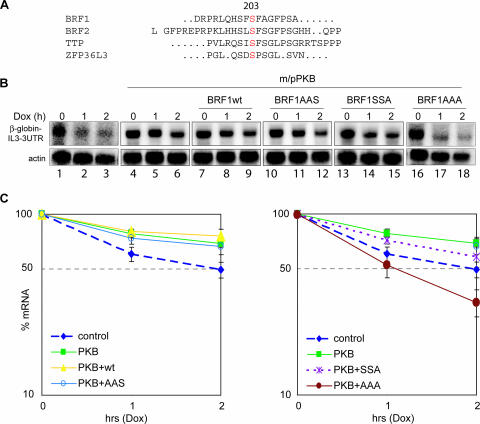
We previously showed by ARE-specific in vitro decay assay that BRF1 is inactivated by phosphorylation at Ser92 by PKB. A phospho-null BRF1 mutant protein with serine replaced at position 92 with alanine (S92A) was resistant to PKB mediated inactivation (39). To test whether this substitution is also sufficient to abolish PKB regulation in vivo, we cotransfected either wild-type BRF1 (BRF1wt) or the S92A mutant protein together with activated PKB into NIH 3T3 B2A2 mouse fibroblasts containing a tetracycline-inducible gene expression system (48). Expression of a β-globin reporter gene fused to the IL-3 3′ UTR is driven by a tetracycline-sensitive promoter in these cells. Addition of a tetracycline analogue, doxycycline, stops reporter gene expression, thereby circumventing the need for a general transcriptional repressor such as actinomycin D (18, 48). To measure the stability of the reporter mRNA in our cotransfection assays, transcription was stopped by addition of doxycycline and the rate of decay of the reporter transcript over time was determined by Northern hybridization. As expected, the reporter decayed rapidly because of the destabilizing presence of the ARE (Fig. 1B, lanes 1 to 3); however, cotransfection of activated PKB greatly increased the half-life of the transcript (lanes 4 to 6). Cotransfection of BRF1wt together with PKB did not antagonize stabilization (lanes 7 to 9), as BRF1 is inactivated by PKB. Surprisingly, and in disagreement with results obtained from the in vitro assay, the Ser90/92Ala mutant (BRF1AAS, lanes 10 to 12) was unable to evade stabilization by PKB. Ser90 and Ser92 were mutated together, as Ser90 can serve as an alternative phosphorylation site under in vitro conditions if Ser92 is mutated; nevertheless, a Ser90Ala mutation displays none of the biological properties observed with a Ser92Ala mutant (data not shown). Figure 1C (left side) shows quantified results from three independent experiments normalized to the actin control. These observations suggest that another regulatory site in addition to Ser92 is required for the in vivo regulation of BRF1 by PKB.
FIG. 1.
BRF1 activity is cooperatively regulated in vivo at serines 92 and 203. (A) Alignment of the known mammalian ZFP36/Tis11 family members BRF1, BRF2, TTP, and ZFP36L3 reveals a conserved serine at position 203 (BRF1 numbering). The human sequences are shown, except for ZFP36L3, which is mouse specific. The human and mouse BRF1 proteins share perfect homology over the sequence shown. (B) The Tet-β-globin-IL-3 3′ UTR reporter gene was transfected alone (lanes 1 to 3) or in combination with constitutively activated m/pPKB (lanes 4 to 18) and BRF1wt, (lanes 7 to 9), BRF1S90A/S92A (BRF1AAS, lanes 10 to 12), BRF1S203A (BRF1SSA, lanes 13 to 15), or BRF1S90A/S92A/S203A (BRF1AAA, lanes 16 to 18). After 48 h, transcription was stopped by addition of doxycycline (2 μg/ml). Cytoplasmic RNA was isolated at the indicated time points and processed for Northern blotting. Panels for lanes 1 to 3 and lanes 16 to 18 are longer exposures due to lower starting levels of the mRNA reporter. (C) Graphs showing quantification of the reporter transcript signal normalized to the actin control from three independent experiments. Reprobing of the blots for GAPDH and normalization of the reporter probe decay against GAPDH as an independent control gave results similar to those obtained with actin (data not shown).
Serine 203 in BRF1 is conserved and involved in regulation.
A search of the BRF1 protein sequence for target motifs of kinases identified Ser203 as a putative phosphorylation site. Serine 203 is homologous to serine 178 in TTP, another member of the ZFP36/Tis11 protein family, that is phosphorylated by MK2 (13). Notably, the serine 203 site is conserved in every member of the family (Fig. 1A). To determine whether this serine is indeed an additional regulatory site in BRF1, we constructed a Ser203Ala mutant (BRF1SSA) and a triple mutant Ser90/92/203Ala (BRF1AAA) and tested their behavior in vivo by using cotransfection assays. Mutation of serine 203 alone was unable to overcome the stabilizing effect of PKB (Fig. 1B, lanes 13 to 15). However, the combined mutations conferred resistance to PKB-mediated inactivation of BRF1 and the ARE reporter RNA was rapidly degraded (lanes 16 to 18). Quantified results from three independent experiments are shown in Fig. 1C (right side). Notably, transfection of the AAA mutant even in the presence of activated PKB destabilizes the reporter to even lower levels than in the control cells, suggesting that BRF1 activity in this case is completely deregulated from the PKB control. The lower baseline levels of the reporter transcript present in the untransfected cells (lanes 1 to 3) and BRF1AAA-transfected cells (lanes 16 to 18) presumably reflect the low stability of the reporter mRNA in these situations. These data suggest that Ser203, in addition to Ser92, also regulates BRF1 in response to PKB.
BRF1 serine 203 is phosphorylated by PKB in vitro.
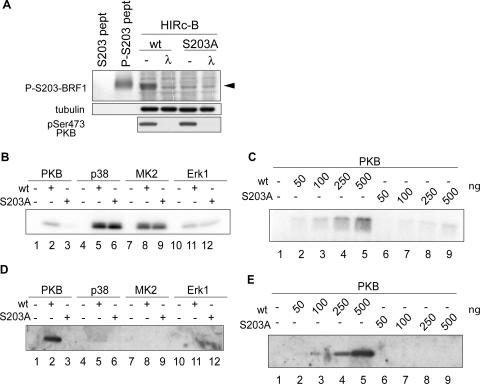
To test whether Ser203 is directly targeted by PKB, we prepared in vitro kinase reactions with activated forms of PKB, p38, MK2, and Erk1. In order to reduce the background signal from phosphorylation at other serines or threonines, we used a truncated BRF1 peptide spanning serine 203 (aa 143 to 233) with a corresponding fragment bearing the Ser203Ala substitution as a specificity control. While all of the other kinases tested were able to phosphorylate both the wild-type and mutated peptides (Fig. 2B), only PKB showed differential phosphorylation and was unable to phosphorylate the S203A-bearing fragment. Titration of increasing amounts of peptide into an in vitro PKB kinase reaction mixture showed the same difference between phosphorylation of the wild-type- and mutant-bearing fragments over a wide concentration range for the substrates (Fig. 2C). In order to confirm that the results observed thus far are specific to Ser203 phosphorylation and to exclude the possibility that the introduced mutation instead destroys the recognition site for another serine/threonine, we corroborated these experiments serologically with a phospho-Ser203-specific antibody. Rabbits were immunized with a phosphopeptide spanning Ser203 (R-L-Q-H-S-F-pS-F-A-G-F-P-S; aa 197 to 209 of BRF1), and the specificity of the sera was characterized as shown in Fig. 2A. We repeated the in vitro kinase assays and probed for a phospho-Ser203-specific signal with the phosphospecific antibody. A strong signal was obtained with the wild-type peptide incubated with PKB, but no corresponding phosphorylation could be observed in the mutant peptide (Fig. 2D, lanes 2 and 3, and E). No phospho-Ser203-specific signal was detected with p38, MK2, and Erk1, arguing for phosphorylation of the fragment at other sites by these kinases. This is notable, as the site homologous to serine 203 in TTP (serine 178) is phosphorylated by MK2. Although the phosphoregulatory site is conserved between the two proteins, different signaling pathways appear to be involved.
FIG. 2.
PKB phosphorylates BRF1 at serine 203. (A) Characterization of an anti-phospho-Ser203 antibody. HIRc-B cells were transfected with BRF1wt (wt) or the BRF1S203A mutant protein (S203A) and stimulated for 30 min with insulin to activate PKB. Cell lysates were run alongside phosphorylated and unphosphorylated control peptides (pept) to see if the antibody could detect phospho-Ser203 from total cell lysates. Where indicated, the extracts were treated with λ protein phosphatase (30°C, 30 min) before loading. The phosphospecific signal is indicated by the arrowhead. (B) A recombinant BRF1 peptide (aa 143 to 233) containing the wild-type sequence (wt) or the S203A mutation (S203A) was incubated with the activated kinases PKB, p38, MK2, and Erk1 together with radioactive ATP. The samples were resolved, and peptide labeling was visualized by autoradiography. (C) The same conditions for PKB phosphorylation were employed as in panel B with titration of increasing amounts of peptide. (D and E) The preceding experiments were repeated nonradioactively, and the reaction products were subjected to Western blotting with the anti-phospho-Ser203 antibody. Phosphospecific signals were only detected after PKB phosphorylation and were absent from reaction mixtures with the S203A peptide.
BRF1 is phosphorylated at Ser203 in vivo.
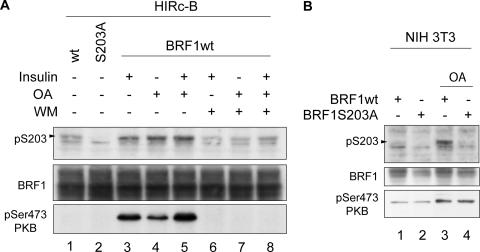
To confirm that BRF1 Ser203 is indeed phosphorylated in vivo, we used HIRc-B cells, which overexpress the insulin receptor and respond strongly to insulin (30). These cells were previously used to demonstrate insulin-stimulated activation of PKB and consequent phosphorylation at Ser92 of BRF1 (39). HIRc-B cells were transfected with BRF1wt and the BRF1S203A mutant and stimulated with either insulin or okadaic acid (OA). OA is a protein phosphatase 2A inhibitor that broadly activates multiple signaling pathways and stabilizes IL-3 mRNA (5). Lysates from treated cells were probed with the phospho-Ser203 antibody. Both stimuli were able to activate PKB and increase basal phosphorylation levels of BRF1 (Fig. 3A, compare lane 1 to lanes 3, 4, and 5). Crucially, pretreatment of the cells with wortmannin (WM), a PI3-K inhibitor that blocks insulin-induced activation of the PI3-K downstream target PKB, resulted in a marked decrease in BRF1 phosphorylation (lanes 6 to 8) that correlated with lack of PKB activation (lowest part). These data support the involvement of PKB in phosphorylating BRF1 at serine 203 in vivo.
FIG. 3.
BRF1 is phosphorylated in vivo at serine 203. (A) HIRc-B cells were transfected with BRF1wt or BRF1S203A and stimulated with insulin (20 μg/ml) or 400 nM OA for 30 min at 48 h after transfection. Where indicated, cells were pretreated with WM (200 nM) 15 min before stimulation. Cell lysates were probed with the anti-phospho-Ser203 BRF1 antibody and against anti-phospho-Ser473 PKB to show the correlation between BRF1 Ser203 phosphorylation and PKB activity. (B) NIH 3T3 cells were transfected with BRF1wt or BRF1S203A and stimulated with 400 nM OA for 30 min. The phospho-S203-specific signal is indicated on the left of each panel.
We next wanted to see if this also applies in NIH 3T3 B2A2 cells, where a functional role had already been demonstrated for PKB phosphorylation of BRF1, namely, in regulating its mRNA decay activity. BRF1wt and BRF1S203A constructs were transfected, and the cells were stimulated with OA rather than insulin because of a weak insulin response (data not shown). PKB activation levels were increased by OA, resulting in higher phosphorylation at BRF1 Ser203 (Fig. 3B, compare lanes 1 and 3). We conclude that BRF1 is phosphorylated at Ser203 in vivo in response to both physiological and nonphysiological stimuli that activate PKB.
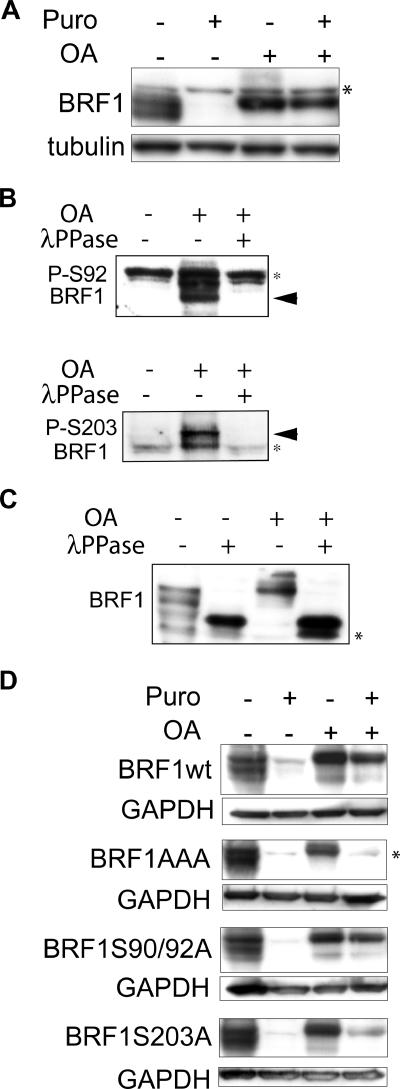
BRF1 protein is rapidly degraded and stabilized by phosphorylation.
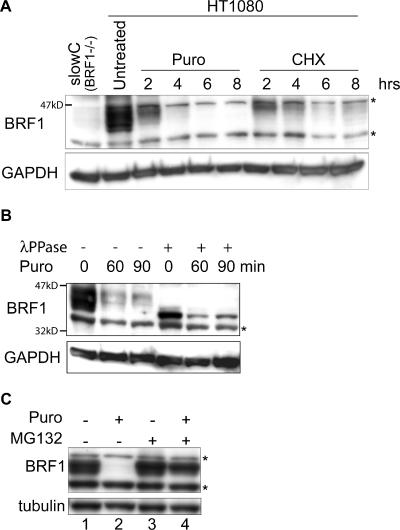
In parallel with these experiments, we were also interested in the stability of the BRF1 protein. As BRF1 is circadianly expressed (34, 43) it should, by prediction, be rapidly degraded in order to reflect this mode of expression. Translation was arrested in human HT1080 cells with puromycin and cycloheximide. Cells were harvested over a time course, and total cell lysates were subjected to Western blotting with an anti-BRF1 antibody. As seen in Fig. 4A, BRF1 is rapidly degraded with a half-life of <3 h. BRF1 migrates anomalously on SDS-PAGE (apparent size of 40 to 47 kDa versus a theoretical size of 36 kDa) and displays a characteristic pattern of broadly spread bands that are indicative of the state of phosphorylation and possibly other posttranslational modifications. Dephosphorylation in vitro with λ phosphatase results in the collapse of all of the bands into a single high-mobility band (Fig. 4B). Notably, the slower-migrating and presumably more highly phosphorylated form of BRF1 appears to decay less rapidly (Fig. 4B). Most proteins are degraded via the ubiquitin-proteasome pathway. When 26S proteasome activity was blocked with the specific inhibitor MG132, preexisting BRF1 protein was stabilized even in the absence of ongoing translation, arguing that it is indeed degraded via the proteasome (Fig. 4C, lane 4).
FIG. 4.
BRF1 protein is labile. (A) Translation was blocked in HT1080 cells with puromycin (Puro; 50 μg/ml) or 50 μM cycloheximide (CHX), and cells were harvested at the indicated times. Detection with an anti-BRF1 antibody reveals the rapid decay of the protein. The blot was stripped and reprobed against GAPDH as a loading control. Lysate from slowC cells (HT1080 BRF1−/−) was run alongside as a negative control for BRF1. (B) Translation was arrested with puromycin, and cells were harvested at earlier time points. The lysates were also treated with λ phosphatase and run alongside for comparison. (C) Cells were treated for 6 h with puromycin and MG132 (10 μM), a proteasome inhibitor. Stabilization of BRF1 in the presence of MG132 indicates that it is degraded via the proteasome. Asterisks denote nonspecific bands.
Phosphorylation at Ser92 and Ser203 is involved in BRF1 protein stability.
The turnover rate of many proteins can be influenced by phosphorylation at critical sites. Therefore, we used OA to hyperphosphorylate BRF1 while simultaneously blocking translation with puromycin. Persistence of preexisting BRF1, even after a 6-h block with puromycin (more than two half-lives), indicated that it is indeed stabilized by phosphorylation (Fig. 5A).
FIG. 5.
Phosphorylated BRF1 is stable. (A) HT1080 cells were treated with puromycin (Puro; 50 μg/ml) and OA (400 nM) either alone or in combination for 6 h, and lysates were probed with an anti-BRF1 antibody. The persistence of BRF1 in OA-induced cells, even when translation is blocked, indicates that it is stabilized by phosphorylation. Note the upward shift in BRF1 migration characteristic of the hyperphosphorylated form. (B) Probing of lysates from OA-induced cells (400 nM, 3 h) with phosphospecific antibodies directed against phospho-Ser92 and phospho-Ser203 confirms that these sites are phosphorylated in HT1080 cells. Phosphospecific signals (arrowheads) are detected after OA treatment and are sensitive to λ phosphatase (λPPase). (C) Lysates from panel B were probed with the anti-BRF1 antibody. The various BRF1 bands are shifted upward after OA treatment and revert to a single high-mobility band after dephosphorylation by λ phosphatase. (D) A series of BRF1 phospho-null mutants were stably expressed in slowC cells. Translation arrest and OA-induced phosphorylation of BRF1 were performed as described for panel A. Constitutive lability of BRF1AAA protein despite OA-induced hyperphosphorylation suggests that these sites are critical for phosphorylation-dependent stabilization. Unlike the endogenous protein, transfected BRF1 migrates slightly higher than the nonspecific band after OA treatment (compare with panel A) because of the presence of His and Xpress tags. Asterisks indicate nonspecific bands.
As a regulatory role for phosphorylation at Ser92 and Ser203 has already been demonstrated, we were curious as to whether these sites may also play a wider biological role. slowC cells, a derivative of HT1080 that is BRF1−/− (40), allowed expression of a panel of BRF1 constructs mutated at serines 90, 92, and 203 without confounding signals from endogenous BRF1. To confirm that BRF1 is phosphorylated at these sites in response to OA, we tested phosphospecific antibodies directed against phospho-Ser92 and phospho-Ser203 and found a specific signal of the expected size that disappears after treatment with λ phosphatase, indicating that these sites are indeed phosphorylated in HT1080 cells upon OA treatment (Fig. 5B). BRF1 mobility is reduced after OA treatment, and the various bands are shifted upward. After dephosphorylation, these revert to a single high-mobility band that migrates at approximately the theoretically predicted size (Fig. 5C). To test the effects of the various phospho-null BRF1 mutants on protein stability, the respective constructs were stably transfected into slowC cells and clones having roughly similar levels of expression were selected for analysis. As shown in Fig. 5D (top), BRF1wt behaves in a manner identical to that of the endogenous protein (Fig. 5A) and is stabilized by OA-induced phosphorylation. Ser-to-Ala substitutions at positions 90 and 92 did not affect protein stabilization, while a similar substitution at position 203 only slightly impaired stabilization. Strikingly, the triple mutant containing alanine substitutions at all three sites is incapable of phosphorylation-dependent stabilization, suggesting that the combined effort of both sites is required for it to exert its biological role. As BRF1 is evidently still phosphorylated on other sites because of its altered migration, it appears that it is sites 90, 92, and 203 that are critical for phosphorylation-dependent protein stabilization.
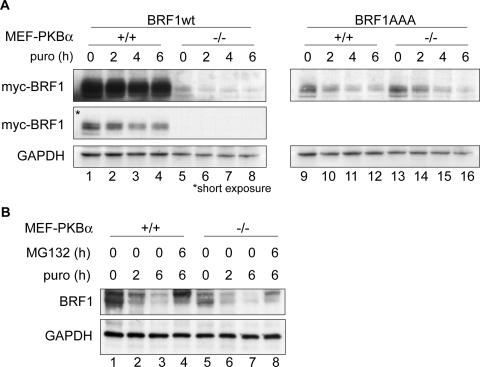
BRF1 protein is destabilized in PKBα−/− cells.
As PKB is responsible for phosphorylation at these sites, we investigated its effect in stabilizing the BRF1 protein. Myc-tagged constructs of BRF1wt and the triple mutant were transfected into MEFs derived from a PKBα+/+ or a PKBα−/− background (51). Protein synthesis was blocked by puromycin, and BRF1 protein levels were determined by immunoblotting (Fig. 6A). Interestingly, BRF1wt protein steady-state levels were much higher in the PKBα+/+ cells than in the knockout background and the half-life of the protein was also increased (compare lanes 1 to 4 to lanes 5 to 8). Notably, the mutant protein exhibited similar stability regardless of the presence (lanes 9 to 12) or absence (lanes 13 to 16) of PKBα. In addition, the lower levels of the mutated protein compared to the wild-type protein suggest that lack of phosphorylation at these sites has an adverse effect on BRF1 protein stability. A transcriptional effect of PKB can be excluded, as the same promoter was used to drive the expression of both BRF1 constructs.
FIG. 6.
PKB activity affects BRF1 levels. (A) Myc-tagged BRF1wt or the S90/92/203A mutant (BRF1AAA) was transfected into PKBα+/+ and PKBα−/− MEFs. Translation was stopped by the addition of puromycin (puro; 50 μg/ml) for the indicated time periods, and protein levels were determined by Western blotting against the myc tag and GAPDH (loading control). Two different exposures of the Western blot are shown for BRF1wt-transfected cells. (B) Levels of endogenous BRF1 in the MEF cells were determined after translation arrest (puromycin) or addition of the proteasome inhibitor MG132 (10 μM).
When we compared the levels of endogenous BRF1 in these cells, we observed higher levels of the protein (Fig. 6B, compare lanes 1 and 5) and a slower decay rate in PKBα+/+ cells than in PKB−/− cells (compare lanes 2 and 3 to lanes 6 and 7). In both cell types, BRF1 degradation could be inhibited by addition of MG132. These results further support the conclusion that phosphorylation of BRF1 at serines 92 and 203 by PKBα inhibits the proteasomal degradation of BRF1.
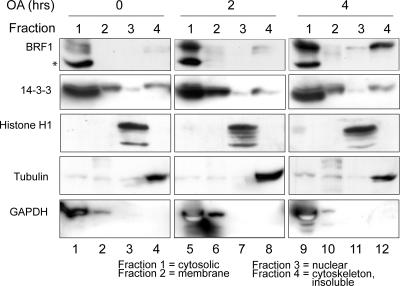
BRF1 localization is altered by phosphorylation.
Our previous results identified Ser92 phosphorylation as a trigger for recruitment of BRF1 to 14-3-3. Similar findings of phosphorylation-dependent binding to 14-3-3 have been reported for other members of the ZFP36/Tis11 family, and in the case of TTP, this was shown to be essential in determining its intracellular localization (21). Protein fractions from various cell compartments were extracted from control and OA-treated cells. Western blotting revealed that a significant fraction of BRF1, mainly in the highly modified low-mobility form, was increasingly found in the cytoskeletal/insoluble pellet after prolonged OA treatment (Fig. 7, lane 12). The blots were reprobed against 14-3-3, which was predominantly detected in the cytosolic fraction but was also well represented in the membrane and cytoskeletal/insoluble fractions. Perhaps because of an already substantial presence in the various intracellular compartments, where it participates in multiple activities, no gross changes in 14-3-3 localization were observed upon OA treatment. The blot was reprobed against known marker proteins for the various subcellular fractions that confirmed the integrity of the purification performed.
FIG. 7.
Fractionation of proteins from the major subcellular compartments in control and OA-induced HT1080 cells. Fractionation was performed with 106 cells, and 10 μg of protein was loaded per fraction. Localization of BRF1 and 14-3-3 was determined with the respective antibodies. Reprobing of this blot and a parallel blot made from the same extracts against known marker proteins histone H1 (nuclear), tubulin (cytoskeletal), and GAPDH (cytosolic/membrane) confirms the integrity of the fractionation. The asterisk denotes a nonspecific band.
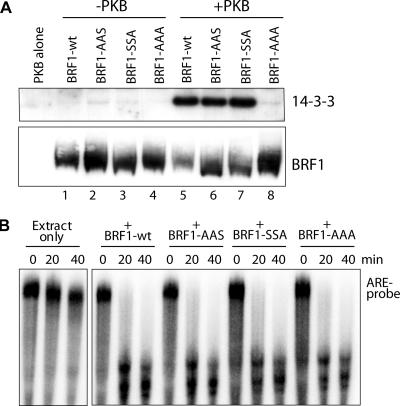
Phosphorylation-dependent binding of BRF1 to 14-3-3 is abolished by mutation at these sites.
We previously showed that Ser92 was important for phosphorylation-dependent binding of BRF1 to 14-3-3 (39). Mutation of Ser92 to alanine impaired but did not abolish this association. To investigate the role of the Ser203 site alone and in conjunction with Ser92, we purified recombinant His-tagged BRF1 bearing the various Ser-to-Ala substitutions. Recombinant BRF1 was added to slowC cell S100 extract either as unmodified protein or previously phosphorylated in vitro with PKBα. The BRF1 bait was bound to a nickel column, washed, and eluted in SDS sample buffer for SDS-PAGE. Western blotting of the eluted fractions against 14-3-3 revealed that interaction between BRF1wt and 14-3-3 was highly phosphorylation dependent (Fig. 8A, compare lanes 1 and 5). The interaction was unperturbed by mutations at Ser90 and Ser92 or at Ser203 (lanes 6 and 7). However, mutation at all of these sites completely abolished the BRF1-14-3-3 interaction (lane 8), indicating that a high degree of cooperation is somehow required among all of these sites to facilitate phosphorylation-dependent binding. The plot was reprobed against BRF1 (panel A, bottom), confirming that roughly equivalent amounts of BRF1 were recovered.
FIG. 8.
Coprecipitation of BRF1-14-3-3 complexes. (A) Recombinant His-BRF1 (40 ng), either unmodified or phosphorylated in vitro by PKBα, was added to 40 μg of slowC cell S100 extract. After binding to nickel beads and washing, the eluted fraction was run on SDS-PAGE and probed against 14-3-3 and BRF1 (as a precipitation control). (B) The various recombinant BRF1 proteins were tested for biological activity by an in vitro RNA decay assay with an ARE RNA probe (39). All proteins could promote rapid decay of the probe, indicating proper conformation and activity.
The introduced mutations may conceivably interfere with the proper folding of BRF1 and therefore might be relevant to the interpretation of the 14-3-3 pull-down data. Therefore, the purified protein was used in an in vitro ARE RNA decay assay (Fig. 8B) in order to assess its biological activity. The ability of the added protein to facilitate decay of the ARE RNA probe indicates that the purified recombinant protein was active and had most probably assumed the proper physiological conformation.
DISCUSSION
In our previous work, we showed by in vitro ARE-specific decay assays that BRF1 activity was regulated at Ser92 by PKB. However, the in vivo experiments reported here reveal that the mechanism is more complex in intact cells in that the PKB-dependent regulation of BRF1 activity could not be evaded by a Ser92 mutation (Fig. 1B), calling for the existence of at least one additional PKB regulatory site on BRF1. This is in keeping with the fact that the Ser92-based mechanism involves binding to 14-3-3, which usually binds to its target proteins at two separate locations (1, 49). We focused on Ser203 as a candidate for the additional phosphoregulatory site because of its homology to TTP Ser178, which serves as one of two 14-3-3 binding sites (21). Although TTP Ser178 is phosphorylated by MK2 (21, 42), we found no evidence that BRF1 Ser203 is phosphorylated in vitro by this kinase or by p38 or ERK1. Instead, the site is strongly phosphorylated by PKB (Fig. 2). Taken together with the in vivo data, where the phospho-Ser203-specific antibody revealed a WM-sensitive increase in Ser203 phosphorylation in extracts from cells with stimulated PKB activity (Fig. 3A), a strong argument can be made for PKB as the kinase responsible for Ser203 phosphorylation. Interestingly, this site differs markedly from the PKB consensus target sequence (BRF1, R-L-Q-H-201S-F-203-S-F-A-G; PKB consensus, R/K-X-R/K-X-X-S/TP). In contrast, the Ser92 site gives an ideal fit to both the PKB and 14-3-3 consensus binding sequences (BRF1, R-D-R-90S-F-92S-E-G; 14-3-3 consensus, R/K-X-X-SP-X-P). However, there are examples of noncanonical PKB and 14-3-3 binding sites in the literature (reviewed in references 1 and 15). An intriguing possibility is raised by the presence of a serine residue two positions before the phosphorylation site in both Ser92 and Ser203 and poses the question of whether this may generate a situation similar to that in the p53 protein. In p53, phosphorylation at Ser376 precludes phosphorylation at Ser378, and removal of Ser376 phosphorylation is required for Ser378 phosphorylation and subsequent 14-3-3 binding (45). However, we have not observed any biological effect with Ser90Ala and Ser201Ala mutant forms of BRF1 that are comparable to that of Ser92Ala, Ser203Ala, or both mutations (data not shown).
We tested the in vivo behavior of the Ser92 and Ser203 mutations on three different aspects of BRF1 biology, all of which fall under regulation by PKB: (i) negative regulation of ARE mRNA decay-promoting activity, (ii) phosphorylation-dependent regulation of protein turnover, and (iii) binding to 14-3-3. The Ser92Ala mutation was inert with respect to all three processes in vivo, while the Ser203Ala mutation was able to slightly impair PKB regulation and protein stabilization (Fig. 1B and 5D). Although the Ser92 site appears to be redundant in vivo, its effect is highly synergistic when it is mutated along with Ser203, leading to strong impairment of all three of the processes studied.
It appears surprising that protein stability should be conferred by the same sites that were originally identified as regulatory sites for BRF1 mRNA decay-promoting activity. BRF1 protein bearing mutations at these sites is still hyperphosphorylated by OA treatment, as shown by its altered migration on SDS-PAGE; nevertheless, it is incapable of being stabilized, pointing to the critical role played by Ser92 and Ser203 (Fig. 5D). When the protein is expressed in PKBα−/− and PKBα+/+ murine embryonic fibroblasts, a dramatic difference in steady-state BRF1wt levels is observed (Fig. 6), with greater protein stability in PKBα+/+ cells. This observation was further supported by the similar behavior of endogenous BRF1 in these backgrounds. However, expression of the BRF1 triple mutant, which cannot be phosphorylated by PKB at the mutated sites, yields equal BRF1 protein levels in both PKBα wild-type and knockout cells. These data suggest that basal levels of PKB phosphorylation on BRF1 may play a role in modulating the rate of degradation and consequently BRF1 protein levels.
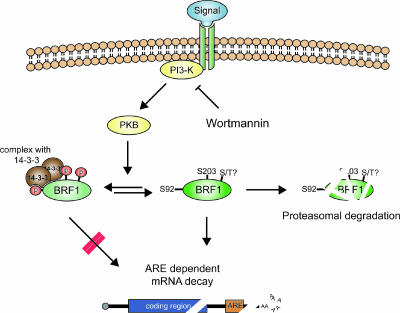
We propose a model (Fig. 9) whereby both of these seemingly unrelated aspects of BRF1, i.e., protein turnover and the regulation of its mRNA decay activity, may be accounted for. In the default state, BRF1 is unphosphorylated at the critical Ser92 and Ser203 sites and is active in promoting the decay of its target ARE transcripts. The protein also undergoes rapid turnover as a consequence of the circadian nature of BRF1 gene expression. Upon receipt of a stabilizing signal, the protein is phosphorylated at Ser92 and Ser203, binds to 14-3-3, and is prevented from participating in mRNA decay (which requires BRF1 binding to the ARE and interaction with components of the decay machinery), in addition to being protected from degradation (possibly by preventing ubiquitinylation and targeting to the proteasome). This could be achieved by either translocation or sequestration of the BRF1-14-3-3 complex and is consistent with our cell fractionation experiments in which BRF1 in OA-stimulated cells was shifted into another compartment (Fig. 7). As this fraction consists of both cytoskeletal and insoluble proteins, we cannot definitively state if phospho-BRF1 becomes associated with the cytoskeleton or with an as-yet-unidentified subcellular compartment such as the stress granule (22). The BRF1-14-3-3 coprecipitation data (Fig. 8A) indicate that each site is able to strongly bind 14-3-3 by itself; however, a Ser92 single mutant significantly reduces binding in vivo (39), suggesting that both sites are required together for 14-3-3 association in intact cells. Accordingly, mutation of both of these critical sites appears to fully abolish BRF1-14-3-3 association, with the attending biological consequences. The residual effects observed with the Ser203 mutation in vivo (Fig. 1B and 5D) raise the possibilities that formation of the phospho-BRF1-14-3-3 complex could also include input from subsidiary regulatory sites or accessory proteins and that the variable responses elicited by both single mutations may reflect a different sensitivity of the entire complex to the extent of 14-3-3 binding.
FIG. 9.
A model for coordinated regulation of BRF1 activity and protein stability. In the default state, BRF1 is unphosphorylated at Ser92 and Ser203 and is active in promoting ARE mRNA decay and at the same time is subject to rapid turnover due to proteasomal degradation. A specific mRNA-stabilizing signal results in PKB activation with consequent phosphorylation of these sites. This triggers phosphorylation-dependent BRF1 binding to 14-3-3 and translocation of the complex, which is now unable to promote mRNA decay and cannot be targeted to the proteasome for degradation. An open question is the existence of further phosphoregulatory sites on BRF1, which is extraordinarily serine/threonine rich, that may also regulate its activity by a 14-3-3-independent mechanism.
This model shares some features with that proposed by Hitti and others for TTP (8, 19). TTP is normally expressed at very low levels and is only highly expressed during the inflammatory response, where its main target transcript, TNF-α, is paradoxically also highly expressed. It appears likely that in this situation, TTP is synthesized but is kept quiescent by phosphorylation in order not to blunt the inflammatory response by degrading TNF-α mRNA. However, high levels of TTP are maintained by stabilization of the protein and upon the end of the inflammatory response, dephosphorylated TTP would be available to actively recruit TNF-α transcripts for degradation. This, however, is unlikely to apply to BRF1, which is highly expressed in many cell and tissue types, and poses the question of what biological rationale is served by the combined regulation of BRF1 activity and turnover. One possible reason could be constraints imposed by the circadian expression of BRF1 (34, 43). At the onset of mRNA stabilization, fluctuating levels of BRF1 protein do not present any difficulty as its activity is directly repressed via phosphorylation. Therefore, mRNA stabilization can be expected to proceed smoothly regardless of the actual amount of BRF1 present. However, reversal of stabilization upon dissipation of the stabilizing signal can pose a problem should this occur when BRF1 levels are at a low point in their daily oscillation. In this scenario, BRF1 target transcripts would have accumulated to high levels because of the preceding stabilization; however, there would then be insufficient BRF1 present to degrade this large pool of transcripts and reinstate the original default state of the cell. This dilemma is neatly resolved if the same signal that disables BRF1 activity also prevents its turnover, thereby ensuring that adequate levels of BRF1 will be present upon ending of stabilizing signals in order to resume rapid mRNA decay.
Circadian expression of BRF1 or other posttranscriptional regulators such as TTP (see the supplementary data in reference 43) raises the possibility that many genes exhibiting such expression do so in a secondary manner because they are under posttranscriptional regulation and not because their transcription is driven in a circadian fashion. As there are far more ARE-bearing transcripts than the few identified AUBPs, it is likely that each AUBP posttranscriptionally regulates and coordinates a set of target transcripts that can be functionally grouped in a posttranscriptional operon (reviewed in references 23 and 36); in this case, BRF1 could potentially impose a circadian pattern of expression on its target transcripts. A pertinent example is nocturnin, a deadenylase that displays high-amplitude circadian expression in the clock-containing photoreceptor cell layer in the Xenopus retina (3) and that is believed to deadenylate a subset of circadian-cycle-related transcripts. Circadian expression of an entire class of transcripts due to posttranscriptional regulation has also been reported in Arabidopsis (27).
Given the multiplicity of stimuli that a cell may have to simultaneously respond to, perhaps occasionally demanding contradictory responses, it appears evident that an AUBP is under regulation at multiple sites so that it can integrate the various signals in order to mount the appropriate response. When coupled with the complex spatiotemporal expression of the components participating in posttranscriptional regulation, together with the large number of potential target transcripts, this confronts us with a task of staggering magnitude, to comprehend the intricacies involved. Clearly, we have only just begun to understand AUBP regulation. For example, BRF1, which is 338 aa long, contains 49 serine and 21 threonine residues (the respective numbers for TTP are 57 serines and 17 threonines out of 326 aa), all of which may be regarded as potential phosphoregulatory sites, a sobering reminder of the daunting task ahead.
Acknowledgments
We thank Brian Hemmings for the PKBα wild-type and knockout MEFs.
This work was supported in part by a grant from the Schweizerische Nationalfonds zur Foerderung der wissenschaftlichen Forschung to C.M.
Footnotes
Published ahead of print on 9 October 2006.
REFERENCES
- 1.Aitken, A., H. Baxter, T. Dubois, S. Clokie, S. Mackie, K. Mitchell, A. Peden, and E. Zemlickova. 2002. Specificity of 14-3-3 isoform dimer interactions and phosphorylation. Biochem. Soc. Trans. 30:351-360. [DOI] [PubMed] [Google Scholar]
- 2.Andjelković, M., D. R. Alessi, R. Meier, A. Fernandez, N. J. Lamb, M. Frech, P. Cron, P. Cohen, J. M. Lucocq, and B. A. Hemmings. 1997. Role of translocation in the activation and function of protein kinase B. J. Biol. Chem. 272:31515-31524. [DOI] [PubMed] [Google Scholar]
- 3.Baggs, J. E., and C. B. Green. 2003. Nocturnin, a deadenylase in Xenopus laevis retina: a mechanism for posttranscriptional control of circadian-related mRNA. Curr. Biol. 13:189-198. [DOI] [PubMed] [Google Scholar]
- 4.Bakheet, T., M. Frevel, B. R. Williams, W. Greer, and K. S. Khabar. 2001. ARED: human AU-rich element-containing mRNA database reveals an unexpectedly diverse functional repertoire of encoded proteins. Nucleic Acids Res. 29:246-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benjamin, D., M. Colombi, and C. Moroni. 2004. A GFP-based assay for rapid screening of compounds affecting ARE-dependent mRNA turnover. Nucleic Acids Res. 32:e89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blackshear, P. J., R. S. Phillips, S. Ghosh, S. V. Ramos, E. K. Richfield, and W. S. Lai. 2005. Zfp36l3, a rodent X chromosome gene encoding a placenta-specific member of the tristetraprolin family of CCCH tandem zinc finger proteins. Biol. Reprod. 73:297-307. [DOI] [PubMed] [Google Scholar]
- 7.Briata, P., S. V. Forcales, M. Ponassi, G. Corte, C. Y. Chen, M. Karin, P. L. Puri, and R. Gherzi. 2005. p38-dependent phosphorylation of the mRNA decay-promoting factor KSRP controls the stability of select myogenic transcripts. Mol. Cell 20:891-903. [DOI] [PubMed] [Google Scholar]
- 8.Brook, M., C. R. Tchen, T. Santalucia, J. McIlrath, J. S. Arthur, J. Saklatvala, and A. R. Clark. 2006. Posttranslational regulation of tristetraprolin subcellular localization and protein stability by p38 mitogen-activated protein kinase and extracellular signal-regulated kinase pathways. Mol. Cell. Biol. 26:2408-2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cao, H., L. J. Deterding, J. D. Venable, E. A. Kennington, J. R. Yates III, K. B. Tomer, and P. J. Blackshear. 2006. Identification of the anti-inflammatory protein tristetraprolin as a hyperphosphorylated protein by mass spectrometry and site-directed mutagenesis. Biochem. J. 394:285-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carballo, E., W. S. Lai, and P. J. Blackshear. 1998. Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science 281:1001-1005. [DOI] [PubMed] [Google Scholar]
- 11.Carballo, E., H. Cao, W. S. Lai, E. A. Kennington, D. Campbell, and P. J. Blackshear. 2001. Decreased sensitivity of tristetraprolin-deficient cells to p38 inhibitors suggests the involvement of tristetraprolin in the p38 signaling pathway. J. Biol. Chem. 276:42580-42587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, C. Y., F. Del Gatto-Konczak, Z. Wu, and M. Karin. 1998. Stabilization of interleukin-2 mRNA by the c-Jun NH2-terminal kinase pathway. Science 280:1945-1949. [DOI] [PubMed] [Google Scholar]
- 13.Chrestensen, C. A., M. J. Schroeder, J. Shabanowitz, D. F. Hunt, J. W. Pelo, M. T. Worthington, and T. W. Sturgill. 2004. MK2 phosphorylates tristetraprolin on in vivo sites including S178, a site required for 14-3-3 binding. J. Biol. Chem. 279:10176-10184. [DOI] [PubMed] [Google Scholar]
- 14.Fan, X., and J. Steitz. 1998. Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J. 17:3448-3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fu, H., R. R. Subramanian, and S. C. Masters. 2000. 14-3-3 proteins: structure, function, and regulation. Annu. Rev. Pharmacol. Toxicol. 40:617-647. [DOI] [PubMed] [Google Scholar]
- 16.Gherzi, R., K. Y. Lee, P. Briata, D. Wegmuller, C. Moroni, M. Karin, and C. Y. Chen. 2004. A KH domain RNA binding protein, KSRP, promotes ARE-directed mRNA turnover by recruiting the degradation machinery. Mol. Cell 14:571-583. [DOI] [PubMed] [Google Scholar]
- 17.Gorospe, M., X. Wang, and N. J. Holbrook. 1998. p53-dependent elevation of p21Waf1 expression by UV light is mediated through mRNA stabilization and involves a vanadate-sensitive regulatory system. Mol. Cell. Biol. 18:1400-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gossen, M., and H. Bujard. 1992. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl. Acad. Sci. USA 89:5547-5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hitti, E., T. Iakovleva, M. Brook, S. Deppenmeier, A. D. Gruber, D. Radzioch, A. R. Clark, P. J. Blackshear, A. Kotlyarov, and M. Gaestel. 2006. Mitogen-activated protein kinase-activated protein kinase 2 regulates tumor necrosis factor mRNA stability and translation mainly by altering tristetraprolin expression, stability, and binding to adenine/uridine-rich element. Mol. Cell. Biol. 26:2399-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang, Z. F., J. B. Massey, and D. P. Via. 2000. Differential regulation of cyclooxygenase-2 (COX-2) mRNA stability by interleukin-1 beta (IL-1β) and tumor necrosis factor-alpha (TNF-α) in human in vitro differentiated macrophages. Biochem. Pharmacol. 59:187-194. [DOI] [PubMed] [Google Scholar]
- 21.Johnson, B. A., J. R. Stehn, M. B. Yaffe, and T. K. Blackwell. 2002. Cytoplasmic localization of tristetraprolin involves 14-3-3-dependent and independent mechanisms. J. Biol. Chem. 277:18029-18036. [DOI] [PubMed] [Google Scholar]
- 22.Kedersha, N., G. Stoecklin, M. Ayodele, P. Yacono, J. Lykke-Andersen, M. J. Fitzler, D. Scheuner, R. J. Kaufman, D. E. Golan, and P. Anderson. 2005. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 169:871-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keene, J. D., and P. J. Lager. 2005. Post-transcriptional operons and regulons co-ordinating gene expression. Chromosome Res. 13:327-337. [DOI] [PubMed] [Google Scholar]
- 24.Lai, W. S., E. Carballo, J. M. Thorn, E. A. Kennington, and P. J. Blackshear. 2000. Interactions of CCCH zinc finger proteins with mRNA. Binding of tristetraprolin-related zinc finger proteins to Au-rich elements and destabilization of mRNA. J. Biol. Chem. 275:17827-17837. [DOI] [PubMed] [Google Scholar]
- 25.Laroia, G., R. Cuesta, G. Brewer, and R. J. Schneider. 1999. Control of mRNA decay by heat shock-ubiquitin-proteasome pathway. Science 284:499-502. [DOI] [PubMed] [Google Scholar]
- 26.Laroia, G., B. Sarkar, and R. J. Schneider. 2002. Ubiquitin-dependent mechanism regulates rapid turnover of AU-rich cytokine mRNAs. Proc. Natl. Acad. Sci. USA 99:1842-1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lidder, P., R. A. Gutierrez, P. A. Salome, C. R. McClung, and P. J. Green. 2005. Circadian control of messenger RNA stability. Association with a sequence-specific messenger RNA decay pathway. Plant Physiol. 138:2374-2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lindstein, T., C. H. June, J. A. Ledbetter, G. Stella, and C. B. Thompson. 1989. Regulation of lymphokine messenger RNA stability by a surface-mediated T cell activation pathway. Science 244:339-343. [DOI] [PubMed] [Google Scholar]
- 29.Ma, W. J., S. Cheng, C. Campbell, A. Wright, and H. Furneaux. 1996. Cloning and characterization of HuR, a ubiquitously expressed Elav-like protein. J. Biol. Chem. 271:8144-8151. [DOI] [PubMed] [Google Scholar]
- 30.McClain, D. A., H. Maegawa, J. Lee, T. J. Dull, A. Ulrich, and J. M. Olefsky. 1987. A mutant insulin receptor with defective tyrosine kinase displays no biologic activity and does not undergo endocytosis. J. Biol. Chem. 262:14663-14671. [PubMed] [Google Scholar]
- 31.Ming, X. F., M. Kaiser, and C. Moroni. 1998. c-jun N-terminal kinase is involved in AUUUA-mediated interleukin-3 mRNA turnover in mast cells. EMBO J. 17:6039-6048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nair, A. P., S. Hahn, R. Banholzer, H. H. Hirsch, and C. Moroni. 1994. Cyclosporin A inhibits growth of autocrine tumour cell lines by destabilizing interleukin-3 mRNA. Nature 369:239-242. [DOI] [PubMed] [Google Scholar]
- 33.Neininger, A., D. Kontoyiannis, A. Kotlyarov, R. Winzen, R. Eckert, H. D. Volk, H. Holtmann, G. Kollias, and M. Gaestel. 2002. MK2 targets AU-rich elements and regulates biosynthesis of tumor necrosis factor and interleukin-6 independently at different post-transcriptional levels. J. Biol. Chem. 277:3065-3068. [DOI] [PubMed] [Google Scholar]
- 34.Panda, S., M. P. Antoch, B. H. Miller, A. I. Su, A. B. Schook, M. Straume, P. G. Schultz, S. A. Kay, J. S. Takahashi, and J. B. Hogenesch. 2002. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell 109:307-320. [DOI] [PubMed] [Google Scholar]
- 35.Paulding, W. R., and M. F. Czyzyk-Krzeska. 2000. Hypoxia-induced regulation of mRNA stability. Adv. Exp. Med. Biol. 475:111-121. [DOI] [PubMed] [Google Scholar]
- 36.Puig, S., E. Askeland, and D. J. Thiele. 2005. Coordinated remodeling of cellular metabolism during iron deficiency through targeted mRNA degradation. Cell 120:99-110. [DOI] [PubMed] [Google Scholar]
- 37.Raineri, I., D. Wegmueller, B. Gross, U. Certa, and C. Moroni. 2004. Roles of AUF1 isoforms, HuR and BRF1 in ARE-dependent mRNA turnover studied by RNA interference. Nucleic Acids Res. 32:1279-1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sarkar, B., Q. Xi, C. He, and R. J. Schneider. 2003. Selective degradation of AU-rich mRNAs promoted by the p37 AUF1 protein isoform. Mol. Cell. Biol. 23:6685-6693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmidlin, M., M. Lu, S. Leuenberger, G. Stoecklin, M. Mallaun, B. Gross, R. Gherzi, D. Hess, B. A. Hemmings, and C. Moroni. 2004. The ARE-dependent mRNA-destabilizing activity of BRF1 is regulated by protein kinase B. EMBO J. 23:4760-4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stoecklin, G., X. F. Ming, R. Looser, and C. Moroni. 2000. Somatic mRNA turnover mutants implicate tristetraprolin in the interleukin-3 mRNA degradation pathway. Mol. Cell. Biol. 20:3753-3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stoecklin, G., M. Colombi, I. Raineri, S. Leuenberger, M. Mallaun, M. Schmidlin, B. Gross, M. Lu, T. Kitamura, and C. Moroni. 2002. Functional cloning of BRF1, a regulator of ARE-dependent mRNA turnover. EMBO J. 21:4709-4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stoecklin, G., T. Stubbs, N. Kedersha, S. Wax, W. F. Rigby, T. K. Blackwell, and P. Anderson. 2004. MK2-induced tristetraprolin:14-3-3 complexes prevent stress granule association and ARE-mRNA decay. EMBO J. 23:1313-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Storch, K. F., O. Lipan, I. Leykin, N. Viswanathan, F. C. Davis, W. H. Wong, and C. J. Weitz. 2002. Extensive and divergent circadian gene expression in liver and heart. Nature 417:78-83. [DOI] [PubMed] [Google Scholar]
- 44.Stumpo, D. J., N. A. Byrd, R. S. Phillips, S. Ghosh, R. R. Maronpot, T. Castranio, E. N. Meyers, Y. Mishina, and P. J. Blackshear. 2004. Chorioallantoic fusion defects and embryonic lethality resulting from disruption of Zfp36L1, a gene encoding a CCCH tandem zinc finger protein of the tristetraprolin family. Mol. Cell. Biol. 24:6445-6455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Waterman, M. J., E. S. Stavridi, J. L. Waterman, and T. D. Halazonetis. 1998. ATM-dependent activation of p53 involves dephosphorylation and association with 14-3-3 proteins. Nat. Genet. 19:175-178. [DOI] [PubMed] [Google Scholar]
- 46.Wilson, G. M., J. Lu, K. Sutphen, Y. Sun, Y. Huynh, and G. Brewer. 2003. Regulation of A+U-rich element-directed mRNA turnover involving reversible phosphorylation of AUF1. J. Biol. Chem. 278:33029-33038. [DOI] [PubMed] [Google Scholar]
- 47.Wodnar-Filipowicz, A., C. H. Heusser, and C. Moroni. 1989. Production of the haemopoietic growth factors GM-CSF and interleukin-3 by mast cells in response to IgE receptor-mediated activation. Nature 339:150-152. [DOI] [PubMed] [Google Scholar]
- 48.Xu, N., P. Loflin, C. Y. Chen, and A. B. Shyu. 1998. A broader role for AU-rich element-mediated mRNA turnover revealed by a new transcriptional pulse strategy. Nucleic Acids Res. 26:558-565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yaffe, M. B., and E. A. Elia. 2001. Phosphoserine/threonine-binding domains. Curr. Opin. Cell Biol. 13:131-138. [DOI] [PubMed] [Google Scholar]
- 50.Yang, J., P. Cron, V. Thompson, V. M. Good, D. Hess, B. A. Hemmings, and D. Barford. 2002. Molecular mechanism for the regulation of protein kinase B/Akt by hydrophobic motif phosphorylation. Mol. Cell 9:1227-1240. [DOI] [PubMed] [Google Scholar]
- 51.Yang, Z. Z., O. Tschopp, M. Hemmings-Mieszczak, J. Feng, D. Brodbeck, E. Perentes, and B. A. Hemmings. 2003. Protein kinase B alpha/Akt1 regulates placental development and fetal growth. J. Biol. Chem. 278:32124-32131. [DOI] [PubMed] [Google Scholar]