Abstract
Epigenetic programming is critical for normal development of mammalian embryos. Errors cause misexpression of genes and aberrant development (E. Li, C. Beard, and R. Jaenisch, Nature 366:362-365, 1993). Imprinted genes are important targets of epigenetic regulation, but little is known about how the epigenetic patterns are established in the parental germ lines and maintained in the embryo. Paternal allele-specific expression at the imprinted Rasgrf1 locus in mice is controlled by paternal allele-specific methylation at a differentially methylated domain (DMD). DMD methylation is in turn controlled by a direct repeat sequence immediately downstream of the DMD which is required for establishing Rasgrf1 methylation in the male germ line (B. J. Yoon et al., Nat. Genet. 30:92-96, 2002). To determine if these repeats have a role in methylation maintenance, we developed a conditional deletion of the repeat sequence in mice and showed that the repeats are also required during a narrow interval to maintain paternal methylation of Rasgrf1 in developing embryos. Removing the repeats upon fertilization caused a total loss of methylation by the morula stage, but by the epiblast stage, the repeats were completely dispensable for methylation maintenance. This developmental interval coincides with genome-wide demethylation and remethylation in mice which most imprinted genes resist. Our data show that the Rasgrf1 repeats serve at least two functions: first, to establish Rasgrf1 DNA methylation in the male germ line, and second, to resist global demethylation in the preimplantation embryo.
Studies of epigenetic programming in mammals have identified changes in epigenetic modifications during development and loci that are targeted for these modifications. After fertilization in mice, there is a wave of genome-wide demethylation followed by remethylation ending around the blastocyst stage (14, 22, 23, 25, 28). Imprinted loci exhibit allele-specific expression and DNA methylation, yet they resist this wave of reprogramming (3, 33) and maintain their parent-of-origin specific imprint marks. This has made them attractive models for characterizing how epigenetic states are programmed. Imprinting of Rasgrf1 is controlled by a binary switch located 30 kbp 5′ of the promoter. One component of the switch is a repeated DNA element that regulates establishment of methylation at an adjacent sequence on the paternal allele. The second component of the switch is the sequence acquiring methylation (the differentially methylated domain [DMD]). The unmethylated maternal DMD can bind the enhancer blocking protein CTCF, block enhancer to promoter interactions, and silence the maternal allele. On the paternal allele, the repeats impart methylation to the DMD, ablating CTCF binding and thus allowing enhancer to promoter interactions and expression of the paternal allele (9, 39, 40). The establishment of methylation at Rasgrf1 is completed in the germ line, with mature sperm being fully methylated (21, 30).
Rasgrf1 is one of the few loci at which the cis-acting DNA sequences controlling establishment of germ line DNA methylation have been described previously. Transgenic studies showed that sequences from Igf2r (2) and Snrpn (1, 15) may control establishment of maternal allele methylation. However, the role of the Igfr2 sequences in the germ line or at the endogenous locus is unknown. Similarly, the importance of the Snrpn sequences at the endogenous locus has not been tested. In gene targeting experiments in which the mouse Snrpn imprinting center was replaced by orthologous sequences from human SNRPN, results showed that the human sequences on the maternal mouse chromosome enabled establishment of maternal methylation in oocytes; however, methylation was not maintained in somatic tissue (12). This suggested that sequences important for methylation establishment on the maternal allele of Snrpn are conserved between mice and humans; however, the human sequences lack features present in the mouse that are needed for methylation maintenance. The dichotomy between mechanisms controlling methylation establishment and maintenance was also demonstrated at H19. Promoter-proximal sequences that acquired DNA methylation in the paternal germ line did not require all sequences within the DMD for their establishment; however, an intact DMD was needed for efficient maintenance of these marks (35, 36). Also, methylation that spread from the DMD to the paternal promoter in zygotes required the DMD for its maintenance in more-differentiated tissue (35). In transgenic mice with a 100-kbp human H19 transgene, germ line DNA methylation was properly established, but it was not maintained in somatic tissue (13). A negatively acting signal that includes CTCF binding sites at the endogenous H19 locus maintains the maternal allele in an unmethylated state (26, 29). A comparable negative signal from Snrpn may maintain the paternal allele in the unmethylated state (16), but its activity at the endogenous locus is not known.
The relationship between mechanisms that establish and maintain DMD methylation remains unknown. Although we showed that the Rasgrf1 repeats regulate establishment of DNA methylation in the male germ line (40), nothing is known about their role, if any, in maintaining methylation during somatic development. To address this question, we designed a conditional deletion of the repeats by flanking the repeat sequence at Rasgrf1 with LoxP sites. By crossing these mice with various cre transgenics to delete the repeats, we identified distinct roles for the repeats at different times during development.
MATERIALS AND METHODS
Generation of mutant mice.
The Rasgrf1tm4Pds targeting vector included a 2.0-kbp 5′ arm extending from a BamHI site to the repeats, an 1,841-nucleotide (nt)-repeat-containing BceAI-to-EcoRV fragment modified to include a LoxP site-containing oligonucleotide at the BceAI and EcoRV sites, an Frt-flanked Pgk-neo cassette, and a 2.7-kbp EcoRV-to-SmaI 3′ arm. Homologous recombination was verified at the 5′ end using the 1,046-nt PstI-to-BamHI probe lying between kbp −3.0 and −2.0 5′ of the repeats as described before (9, 39, 40). At the 3′ end homologous recombination was verified using a neo cassette-specific primer and a primer 3′ of the 3′ homologous arm, followed by sequencing of the PCR product. We prepared Rasgrf1tm4Pds mice using standard embryonic stem cell culture and blastocyst injection methods and excised the Frt-flanked neo marker by crossing them with FLPe transgenic mice (5) to produce the Rasgrf1tm4.1Pds allele, referred to as flox-R. Zp3-cre transgenics used to produce the Rasgrf1tm4.2Pds allele, referred to as Δ-R, were produced by Gail Martin (19) and provided, with her permission, by Barbara Knowles. Other cre transgenics were from Jackson Laboratories (stock numbers 003771, 003574, and 003755). All mice were maintained on a C57BL/6 background, except for allelic expression analysis, where F1 mice from a C57BL/6 and PWK cross were used (see Fig. 2c only). Primers for DNA analysis were P1 (ATGATTGAACAGATGGATTGCAC), P2 (TTCGTCCAGATCATCCTGATCGAC), P3 (CTGCACCGCTGCCGCTAAGC), P4 (CCTGCAGGTCGACATAACTTC), P5 (GCACTTCGCTACCGTTTCGC), P6 (TTTCTGCCATCATCCCAGCC), P7 (TGTCCTCCACCCCTCCACC), P10 (ATACTTTCTCGGCAGGAGCA), P11 (GGGTGGGCTTTGAGTGTTTA), Timp1F (GTCATAAGGGCTAAATTCATGGG), and Timp1R (ACTCTTCACTGCGGTTCTGGGAC).
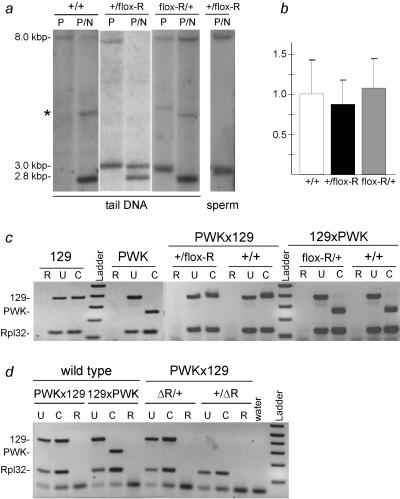
FIG. 2.
flox-R allows normal imprinting like the wild-type Rasgrf1 allele. (a) Southern blot analysis of DMD methylation at the NotI (N) site using tail or sperm DNA digested with PstI (P) or PstI and NotI (P/N). Mice were wild type (+/+) or heterozygous for a paternal (+/flox-R) or maternal (flox-R/+) flox-R allele. Restriction site locations and probes are as in Fig. 1. The asterisk denotes nonspecific hybridization occasionally seen when hybridization and wash stringency are reduced. (b) Quantitative RT-PCR for Rasgrf1 in neonatal brains of +/+, +/flox-R, and flox-R/+ mice. The y axis shows the fluorescence signal normalized to +/+. There were no significant differences. (c) Allele-specific RT-PCR of Rasgrf1 in mice with a flox-R allele and wild-type littermates. (d) Allele-specific RT-PCR of Rasgrf1 in mice with a Δ-R allele and wild-type littermates. For panels c and d, RNA was from neonatal brains of strain 129SvS4Jae (129) and PWK mice or from progeny of PWK mothers and 129 fathers (PWKx129) or the reciprocal cross (129xPWK). In crosses, the 129 partner was heterozygous for the mutated allele. Reactions included RNA (R) or cDNA (U and C) and primers P8 and P9 (Fig. 1) or Rpl32 as control. Products were undigested (U) or digested with HhaI (C). An HhaI site in the PWK RT-PCR product is absent from 129.
RNA analysis.
We prepared RNA and carried out allele-specific reverse transcription-PCR (RT-PCR) assays for Rasgrf1 on oligo(dT)-primed cDNA as previously described (40) and did real-time RT-PCR using cDNA made with random primers and the Applied Biosystems predesigned Rasgrf1 TaqMan gene expression assay (assay identification Mm00441097_m1) multiplexed with TaqMan Ribosomal Control primers (product number 4308329) amplified using the ABI 7500 Realtime PCR system. Other primers were P8 (CTTGGTGTTCATCGAGGAGG), P9 (ATATTCTCGGGGAAGCACAC), Rpl32F (CATGCACACAAGCCATCTACTCA), and Rpl32R (TGCTCACAATGTGTCCTCTAAGAAC).
Methylation analysis.
We used proteinase K to extract genomic DNA from tail, neonatal brain, liver, sperm, and preimplantation embryos and included glycogen (15 μg) to precipitate the embryonic DNAs. Methylation analysis of the DMD using restriction enzymes was as previously described (40). Primers used, in addition to those described above, were PGKF (CTTTGCTCCTTCGCTTTCTG) and PGKR (ACGTCCAGCTTGTCCAAAGT). Bisulfite sequencing was carried out as described previously (10) with some modifications, using fresh solutions. Briefly, we denatured 1.0 μg of DNA in 50 μl Tris-EDTA by adding 5 μl 3 M NaOH and incubating the mixture at 37°C for 10 min. After adding 30 μl 10 mM hydroquinone and 510 μl 3.9 M sodium bisulfite, we allowed deamination to proceed at 55°C for 16 h. We purified treated DNA using a QIAquick gel extraction kit according to the manufacturer's instructions (QIAGEN) and eluted DNA from the kit column in 50 μl elution buffer. The bisulfite reaction was completed by adding 5 μl 3 M NaOH and incubating the mixture at room temperature for 5 min. We purified DNA again using a QIAquick gel extraction kit and eluted it in 30 μl. For PCR, we amplified 1 μl of DNA using primers MF (GAGTATGTAAAGTTAGAGTT) and MR (ATAAACTACTACAACAACTT) for 40 cycles, gel purified all PCR products, and then cloned them using the Topo pCR2.1 cloning kit (Invitrogen). Clones with the correct insert size were sequenced using M13F and M13R primers. Embryonic bisulfite DNAs were amplified with a nested PCR. First-round primers used were MF (as above) and nMR (ACTATCCTCCACCCCTCCAC); a second round of PCR used MF and MR.
RESULTS
Development of a conditional allele at Rasgrf1.
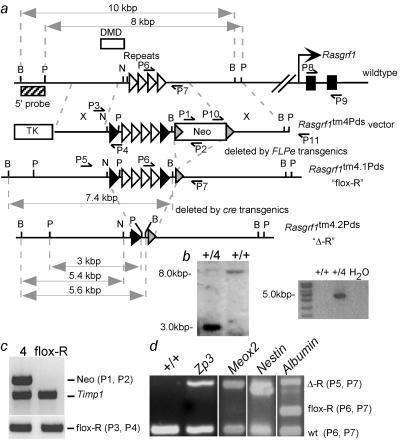
To study the relationship between establishment and maintenance of methylation and to identify the critical developmental interval when maintenance of epigenetic programming occurs, we first identified the sequences that maintain Rasgrf1 methylation. We considered the possibility that the repeats which control establishment may also control maintenance. To test this, we generated mice in which the repeats were flanked by LoxP sites (floxed). The LoxP sites facilitated deletion of the repeats by cre recombinase transgenic mice after methylation was established and allowed us to determine if methylation could be maintained without the repeats. Depending on the cre transgenic used, we could delete the repeats at different developmental times and determine if there was a developmental interval during which the repeats must be present to maintain the DNA methylation. A targeting vector with floxed repeats and a neomycin phosphotransferase cassette flanked by FRT sites was used to prepare mice by gene targeting in embryonic stem cells (Fig. 1a). Homologous recombination was confirmed at the 5′ end by Southern blot assays (Fig. 1b) and at the 3′ end by PCR. The primer pair used in the PCR assay included one specific to the targeting vector and one beyond the 3′ boundary of the 3′ homologous arm. Amplification of DNA from targeted cells, but not wild-type cells, resulted in a band of the predicted size (4,650 nt). It was cloned, and sequencing revealed that the predicted recombination had occurred at the 3′ end (Fig. 1b and data not shown). The drug resistance marker was removed by crossing heterozygotes with FLPe transgenic mates (Fig. 1c) (5). We refer to the alleles as Rasgrf1tm4Pds prior to drug resistance marker removal and as the Rasgrf1tm4.1Pds or flox-R allele afterward (Fig. 1).
FIG. 1.
Controlled deletion of the Rasgrf1 repeats. (a) Wild-type Rasgrf1 locus, Rasgrf1tm4Pds targeting vector, and Rasgrf1tm4.1Pds (flox-R) and Rasgrf1tm4.2Pds (Δ-R) alleles. The 350-bp DMD is upstream of the 1.6-kb repeat sequence (white triangles). Removing the neo cassette flanked by FRT sites (gray triangles) in FLPe transgenic mice produced the flox-R allele with LoxP site (black triangles)-flanked repeats. Removing the repeats in cre transgenic mice produced the Δ-R allele. Restriction sites are PstI (P), BanII (B), and NotI (N). P1 through P9 are PCR primers. (b) Southern blot analysis of PstI-digested DNA isolated from parental J1 embryonic stem cells (+/+) and a Rasgrf1tm4Pds clone (+/4) using the probe in panel a (left panel). The probe lies 5′ of the 5′ homologous arm. PCR analysis across the 3′ junction used primers P10 and P11 (right). P11 lies 3′ of the 3′ homologous arm of the vector. (c) PCR analysis of DNA from progeny of a flox-R-containing dam and a FLPe transgenic sire used primers in panel a to detect Neo (P1 and P2), the flox-R allele (P3 and P4), or Timp1 as a control. (d) PCR of DNAs from wild-type control animals (+/+) or pups born to flox-R sires and cre transgenic dams expressing cre from the Zp3, Meox2, Nestin, or Albumin promoters. Primers in panel a detected the Δ-R allele (P5 and P7) or the wild-type (wt) and flox-R alleles (P6 and P7), which generated different-sized products. The progeny of Albumin-cre dams are mosaics. The figure is not to scale.
Normal DNA imprinting can occur in mice with the conditional allele.
We tested animals with a flox-R allele transmitted maternally or paternally, to determine if floxing the repeats impaired normal imprinted methylation and expression. We monitored DMD methylation status in somatic tissue by Southern blot analysis of tail DNA digested with PstI and the methylation-sensitive enzyme NotI. PstI digestion discriminated between the two alleles, with the wild-type allele producing an 8.0-kbp band and the mutated flox-R allele producing a 3.0-kbp band. Methylation of either allele prevents further digestion by NotI; absence of methylation permits further digestion, producing a 2.8-kbp band. Maternal transmission of the floxed repeats (flox-R/+) did not affect normal Rasgrf1 methylation—the paternal allele remained methylated and the maternal allele remained unmethylated (Fig. 2a). When paternally transmitted (+/flox-R), the floxed repeats imparted normal DMD methylation in somatic tissue of 38 out of 42 mice tested; four mice were hypomethylated at NotI and HhaI sites in the paternal DMD as assayed by PCR and Southern blotting (Fig. 2a and data not shown). Methylation was detected in the sperm DNA of +/flox-R mice, indicating that the floxed paternal allele underwent proper establishment of germ line methylation and remained methylated at the point of fertilization (Fig. 2a). We verified that the mutated paternal allele in +/flox-R animals was methylated across the entire DMD using bisulfite sequencing (Fig. 3c). The analysis revealed that all 26 CpGs assayed were extensively methylated to levels comparable to that of the wild-type paternal allele (40). The bisulfite data confirmed our previous studies showing that the methylation state of the NotI site accurately predicted the methylation of the remaining CpGs (9, 40). We next characterized Rasgrf1 expression using an allele-specific RT-PCR assay to determine if Rasgrf1 expression in the neonatal brain was still exclusively from the paternal allele in mice with floxed repeats. Also, we used real-time RT-PCR to see if floxing the repeats affected the levels of accumulated Rasgrf1 RNA. As was true with DNA methylation, Rasgrf1 expression was unaffected by floxing of the repeats (Fig. 2b and c).
FIG. 3.
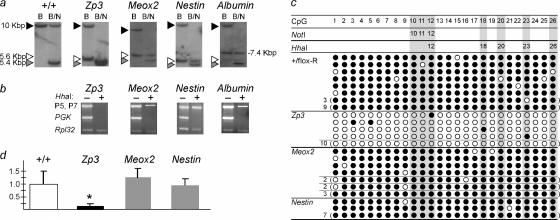
The Rasgrf1 repeats required for methylation maintenance after fertilization are dispensable by e5.5. (a) Southern blot analysis of methylation at the DMD NotI site after repeat loss. DNAs were from Rasgrf1 heterozygotes born to sires heterozygous for flox-R and transgenic dams expressing cre from Zp3, Meox2, Nestin, or Albumin promoters. DNAs were from tail (+/+, Zp3, and Meox2), brain (Nestin), or liver (Albumin). Probe and restriction enzymes BanII (B) and NotI (N) distinguished methylated forms of +/+ (10-kbp), flox-R (7.4-kbp), and Δ-R (5.6-kbp) alleles with unmethylated alleles producing a 5.4-kbp band (Fig. 1). (b) PCR analysis of methylation at the DMD HhaI sites. DNAs were those used in panel a. Primers flanked HhaI sites in the DMD (P5 and P7), an unmethylated promoter (Pgk), or a sequence lacking HhaI sites (Rpl32). Before amplification, templates were HhaI digested (+) or left undigested (−). (c) Bisulfite analysis of DMD methylation. DNAs were from four +/flox-R animals (+/flox-R) or four progeny of the above cre crosses. The 26 CpGs assayed were specific to the mutated allele containing the repeats (+/flox-R) or the recombined Δ-R allele in progeny from cre crosses and included three CpGs at the single NotI site and five CpGs at the five HhaI sites (shaded areas). Filled and open circles represent methylated and unmethylated CpGs, respectively. The numbers to the left of each line indicate the number of clones having identical sequences. Where no number is shown, a unique sequence was obtained. (d) Real-time RT-PCR measurements of Rasgrf1 in brains of mice from panels a and b. The y axis shows expression normalized to wild-type levels. Expression was significantly lower (*) in progeny of Zp3-cre mice where DMD methylation was lost.
Methylation establishment and maintenance are controlled by Rasgrf1 repeats.
Having determined that the floxed repeats behave just as the wild-type allele does, we crossed male mice with a flox-R allele to females expressing cre. Recombination by the maternal cre deletes the paternal repeats forming the Rasgrf1tm4.2Pds allele. For simplicity, we refer to the repeat-deficient allele as Δ-R. The transgenes that we used express cre in different tissues at different developmental times and included Zp3-cre (oocytes [19]), Meox2-cre (embryonic ectoderm of the day 5.5 [e5.5] epiblast [34]), Nestin-cre (e11 central nervous system [7]), and Albumin-cre (by postpartum day 1 [P1] liver [27]). To confirm that the maternal cre deleted the paternal repeats in the progeny of these crosses, we used a PCR assay that detected the recombination product (Fig. 1d) and Southern blot assays that distinguished the wild-type and mutated alleles, both recombined (Δ-R) and unrecombined (flox-R) (Fig. 3). The sources of DNAs for these tests were the tails of neonatal progeny of Zp3-cre and Meox2-cre females and brain and liver, respectively, for progeny of Nestin-cre and Albumin-cre females. Each cre transgene was able to delete the repeats in the tissues tested with efficiencies ranging from virtually 100% for Zp3- and Nestin-cre to approximately 90% and 20% for Meox2-cre and Albumin-cre, respectively (Fig. 1d and 3).
Our Southern blot assays used NotI- and BanII-digested DNAs. Unlike PstI digestion, this combination allowed us to differentiate, in mosaic animals, alleles with excised repeats (Δ-R) from those that cre failed to touch (flox-R). At the same time, this digest evaluated the methylation status of the NotI site. BanII digestion alone produces a 10-kbp wild-type band and a 5.6-kbp mutant band in mice carrying the Δ-R allele. Further digestion with NotI will present a 5.4-kbp band in the absence of methylation (Fig. 1 and 3). In DNAs from progeny of Zp3-cre dams and flox-R sires, both the 10-kbp maternal and the 5.6-kbp paternal bands were digested to 5.4 kbp by NotI. This indicated that the paternal allele methylation that had been established on the flox-R allele (Fig. 2a) could not be maintained in neonates after Zp3-cre-mediated recombination converted it to the repeat-deficient Δ-R allele in the zygote (Fig. 3a). This implicates the repeats as playing a role in methylation maintenance as well as in methylation establishment as we previously showed (40). In sharp contrast, when maternal cre deleted the paternal repeats at later times in the epiblast, central nervous system, or postpartum liver as directed by the Meox2-cre, Nestin-cre, or Albumin-cre transgenes, respectively, the Δ-R allele consistently maintained a high level of methylation (Fig. 3a). This indicated that the role of the Rasgrf1 repeats in DNA methylation maintenance depends on the developmental stage of the embryo.
We extended our methylation analysis beyond the NotI site detected by Southern blots to the HhaI sites in the DMD by using a methylation-sensitive PCR assay. DNAs used in Southern blot assays shown in Fig. 3a were digested with the methylation-sensitive enzyme HhaI or left undigested and then amplified with primers that were specific for the Δ-R allele. HhaI digestion prevented any detectable amplification when assays were done using DNA from progeny of Zp3-cre females, but amplification products were readily detected in DNAs from other cre transgenic females (Fig. 3b). This was consistent with the Southern blot results and showed that additional CpGs in the DMD failed to maintain methylation after the repeats were deleted in the zygote.
Finally, we confirmed the Southern and PCR results with bisulfite sequencing (Fig. 3c). As predicted from the restriction analysis (Fig. 3a and b), repeat-deficient DNAs from progeny of the Zp3-cre cross had lost virtually all their DMD methylation from the paternal allele while DNAs from progeny of the Meox2-cre and Nestin-cre crosses had maintained virtually all of their methylation (Fig. 3c). This further confirmed our previous observations that the methylation states of the NotI and HhaI sites accurately predict the methylation status of at least 26 CpGs in the DMD (9, 40).
Our interpretation of these data is that not only are the Rasgrf1 repeats required for establishing DNA methylation at the paternal DMD in the male germ line as we previously showed (40) and they are also required to maintain the established methylation after fertilization. Furthermore, the interval between fertilization and the epiblast stage is the critical period when the repeats must be present in cis for maintaining paternal allele methylation at Rasgrf1 in mice. The shared requirement for the repeats during methylation establishment and maintenance suggests that a common mechanism is used for the two processes at Rasgrf1.
Loss of methylation maintenance silences Rasgrf1.
To evaluate the effects of the cre-mediated deletions on Rasgrf1 expression, we performed real-time RT-PCR using neonatal brain cDNAs from progeny of the Zp3-, Meox2-, and Nestin-cre crosses. Rasgrf1 is not expressed in liver or in early embryos, and so progeny of the Albumin-cre crosses and preimplantation embryos were not analyzed. We detected wild-type levels of Rasgrf1 RNA in progeny of the Meox2- and Nestin-cre crosses that maintained DMD methylation and markedly reduced levels in Zp3-cre progeny that lost DMD methylation (Fig. 3d). This was consistent with our previous data showing that DMD methylation is needed for expression of Rasgrf1 in neonatal brain (9, 40). Loss of imprinted expression in neonates did not produce overt developmental changes.
The critical interval for methylation maintenance is between fertilization and the epiblast stage.
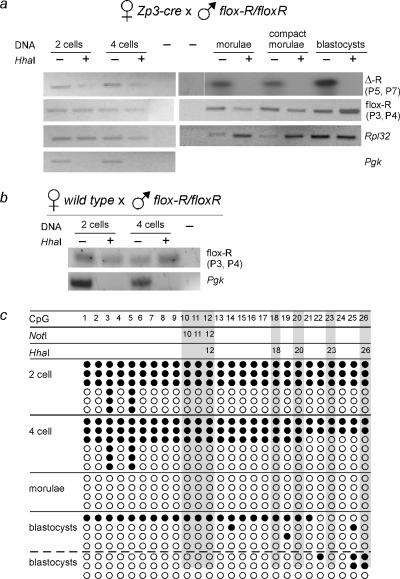
Our methylation analysis of the Δ-R allele in progeny of Zp3-cre dams and flox-R sires was done using tissue taken from neonates. The lack of Δ-R allele methylation in these mice may have been due to a gradual loss of methylation between fertilization, the earliest time when the deletion could occur, and the perinatal period, when tissues for methylation assays were taken. Alternatively, there may have been a rapid loss of methylation immediately after repeat deletion and prior to implantation, a period when the genome undergoes extensive global changes in the methylation state (14, 22, 23, 25, 28). These are not mutually exclusive possibilities. To determine how quickly methylation could be lost from the Δ-R allele after being formed by zygotic cre, we examined the methylation states of DNAs from two-cell, four-cell, morula-, and blastocyst-stage embryos isolated from Zp3-cre transgenic dams that were mated with flox-R sires. The sires had normal methylation at their flox-R allele (not shown). We used a restriction enzyme-based assay (Fig. 4a) and bisulfite PCR (Fig. 4c). Both assays showed that loss of methylation could occur on the Δ-R allele as soon as the two-cell stage. However, two- and four-cell embryos also provided evidence of full methylation on the Δ-R allele, raising the possibility that failure of methylation maintenance was by a passive mechanism. By the morula and blastocyst stages, loss of methylation from the Δ-R allele was nearly complete. All embryonic stages assayed were mosaic for the recombined Δ-R and unrecombined flox-R allele, allowing us to monitor flox-R methylation. Importantly, the unrecombined allele remained methylated, presumably because of the maintenance function provided by the still-present repeats. Methylation of the paternal flox-R allele was also present in two- and four-cell embryos taken from wild-type female dams (Fig. 4c). Only the Δ-R allele, without repeats, failed to maintain the methylation that had been established in the paternal germ line.
FIG. 4.
Zygotic deletion of repeats leads to loss of methylation maintenance. (a) Preimplantation embryos came from crosses between flox-R homozygous sires and Zp3-cre dams. DMD methylation was detected using the digestion PCR assay described in the Fig. 3b legend. Primers detected Δ-R (P5 and P7) and flox-R (P3 and P4) present in mosaics. Rpl32 primers amplified sequences lacking HhaI sites and served as a positive control. The unmethylated Pgk promoter served as a digestion control. The products of P5 and P7 amplification were detected by Southern blot hybridization. (b) As a control for panel a, methylation of the flox-R allele was detected in two- and four-cell embryos from crosses between flox-R homozygous sires and wild-type dams lacking the cre transgene. (c) Bisulfite PCR of the Δ-R allele in DNAs from panel a. One preparation of pooled morulae and two preparations of pooled blastocysts were used. Filled circles represent methylated residues. Each line represents an independent DNA sequence. Filled and open circles represent methylated and unmethylated CpGs, respectively. CpGs 12, 18, 20, 23, and 26 are within the five HhaI sites.
DISCUSSION
Rasgrf1 imprinting is controlled by a binary switch that includes the DMD and the repeats. The DMD is a methylation-sensitive enhancer blocker that binds CTCF on the maternal allele, causing silencing. Methylation established on the paternal DMD by the repeats prevents CTCF binding and allows expression of the paternal allele (39). We have previously shown that the repeats function to establish Rasgrf1 methylation in the male germ line (40), which others have shown is complete in mature sperm (21). Here we show that the Rasgrf1 repeats also regulate methylation maintenance in the embryo and identify the zygote-to-epiblast stage as the critical interval when maintenance mechanisms operate. Failure of maintenance in the zygote led to a rapid and nearly complete loss of DNA methylation by the uncompacted morula state. This critical period during which the Rasgrf1 repeats are needed for methylation maintenance coincides with drastic changes in methylation patterns in the two parental genomes in mice. Immediately after fertilization, the paternal genome undergoes rapid, possibly active demethylation, followed shortly by the maternal genome, which may be passively demethylated through replication without remethylation (25, 28). The apparently gradual loss of methylation as zygotes became two-cell and then four-cell embryos is consistent with a passive mechanism of methylation loss. However, these assays may not precisely report kinetics of methylation loss and cannot rule out an active mechanism.
The paternal flox-R allele was detected in blastocysts taken from Zp3-cre females, indicating that zygotic cre had not completely converted the flox-R allele to the Δ-R form at that stage. But by the end of gestation, we saw no mosaicism by PCR (Fig. 1d) or Southern blotting (Fig. 3a). We also saw no evidence of remaining DNA methylation at the end of gestation (Fig. 3). This indicates that failure of maintenance occurred if repeats were removed after the blastocyst stage. However, removing the repeats at the epiblast stage was not sufficient to disrupt methylation.
Our results are comparable to those showing that the paternal DMD at H19 is needed in the zygote for expansion of its germ line methylation mark into the promoter region but is dispensable in differentiated somatic tissue for maintenance of the expanded methylation (31, 32). However, unlike the Rasgrf1 repeats, the H19 DMD is not required for establishing methylation at the H19 promoter (35).
How the Rasgrf1 repeats regulate epigenetic programming at the locus either during establishment in the male germ line or during maintenance in the early embryo is unknown. The mechanism differs from those apparently operating at other imprinted loci because the same repeated sequences regulate both processes at Rasgrf1. Regulation by the repeats may involve small RNAs of the kind that can direct DNA methylation in plant and mammalian systems (17, 24, 38) and histone modifications in Schizosaccharomyces pombe (8, 37). Additionally, the repeats may attract chromatin-modifying (18) or remodeling (6) factors or other unknown components that, in turn, influence DMD methylation. None of these possibilities is mutually exclusive. It is not known how well mechanisms regulating epigenetic programming at imprinted loci reflect mechanisms operating at nonimprinted loci. It is clear, however, that imprinted loci undergo reprogramming failure in animals prepared by somatic cell nuclear transfer to eggs which may contribute to abnormal growth phenotypes of cloned animals (11). Understanding the mechanisms that regulate the epigenetic state may help to minimize epigenetic perturbations in manipulated embryos.
Acknowledgments
We thank Colleen Kane, Diane Poslinski, and Aimee Stablewski of the Roswell Park Cancer Institute Gene Targeting and Transgenic Core for technical support and Laurie Jackson-Grusby for the FRT-flanked neo cassette.
This work was supported by grants to P.D.S. from the National Cancer Institute, the National Eye Institute, and the U.S. Army and by a Cancer Center Support Grant to Roswell Park Cancer Institute.
Footnotes
Published ahead of print on 9 October 2006.
REFERENCES
- 1.Bielinska, B., S. M. Blaydes, K. Buiting, T. Yang, M. Krajewska-Walasek, B. Horsthemke, and C. I. Brannan. 2000. De novo deletions of SNRPN exon 1 in early human and mouse embryos result in a paternal to maternal imprint switch. Nat. Genet. 25:74-78. [DOI] [PubMed] [Google Scholar]
- 2.Birger, Y., R. Shemer, J. Perk, and A. Razin. 1999. The imprinting box of the mouse Igf2r gene. Nature 397:84-88. [DOI] [PubMed] [Google Scholar]
- 3.Brandeis, M., T. Kafri, M. Ariel, J. R. Chaillet, J. McCarrey, A. Razin, and H. Cedar. 1993. The ontogeny of allele-specific methylation associated with imprinted genes in the mouse. EMBO J. 12:3669-3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reference deleted.
- 5.Farley, F. W., P. Soriano, L. S. Steffen, and S. M. Dymecki. 2000. Widespread recombinase expression using FLPeR (flipper) mice. Genesis 28:106-110. [PubMed] [Google Scholar]
- 6.Gibbons, R. J., T. L. McDowell, S. Raman, D. M. O'Rourke, D. Garrick, H. Ayyub, and D. R. Higgs. 2000. Mutations in ATRX, encoding a SWI/SNF-like protein, cause diverse changes in the pattern of DNA methylation. Nat. Genet. 24:368-371. [DOI] [PubMed] [Google Scholar]
- 7.Haigh, J. J., P. I. Morelli, H. Gerhardt, K. Haigh, J. Tsien, A. Damert, L. Miquerol, U. Muhlner, R. Klein, N. Ferrara, E. F. Wagner, C. Betsholtz, and A. Nagy. 2003. Cortical and retinal defects caused by dosage-dependent reductions in VEGF-A paracrine signaling. Dev. Biol. 262:225-241. [DOI] [PubMed] [Google Scholar]
- 8.Hall, I. M., G. D. Shankaranarayana, K. Noma, N. Ayoub, A. Cohen, and S. I. Grewal. 2002. Establishment and maintenance of a heterochromatin domain. Science 297:2232-2237. [DOI] [PubMed] [Google Scholar]
- 9.Herman, H., M. Lu, M. Anggraini, A. Sikora, Y. Chang, B. J. Yoon, and P. D. Soloway. 2003. Trans allele methylation and paramutation-like effects in mice. Nat. Genet. 34:199-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Herman, J. G., J. R. Graff, S. Myohanen, B. D. Nelkin, and S. B. Baylin. 1996. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc. Natl. Acad. Sci. USA 93:9821-9826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Humpherys, D., K. Eggan, H. Akutsu, A. Friedman, K. Hochedlinger, R. Yanagimachi, E. S. Lander, T. R. Golub, and R. Jaenisch. 2002. Abnormal gene expression in cloned mice derived from embryonic stem cell and cumulus cell nuclei. Proc. Natl. Acad. Sci. USA 99:12889-12894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnstone, K. A., A. J. Dubose, C. R. Futtner, M. D. Elmore, C. I. Brannan, and J. L. Resnick. 2006. A human imprinting centre demonstrates conserved acquisition but diverged maintenance of imprinting in a mouse model for Angelman syndrome imprinting defects. Hum. Mol. Genet. 15:393-404. [DOI] [PubMed] [Google Scholar]
- 13.Jones, B. K., J. Levorse, and S. M. Tilghman. 2002. A human H19 transgene exhibits impaired paternal-specific imprint acquisition and maintenance in mice. Hum. Mol. Genet. 11:411-418. [DOI] [PubMed] [Google Scholar]
- 14.Kafri, T., M. Ariel, M. Brandeis, R. Shemer, L. Urven, J. McCarrey, H. Cedar, and A. Razin. 1992. Developmental pattern of gene-specific DNA methylation in the mouse embryo and germ line. Genes Dev. 6:705-714. [DOI] [PubMed] [Google Scholar]
- 15.Kantor, B., Y. Kaufman, K. Makedonski, A. Razin, and R. Shemer. 2004. Establishing the epigenetic status of the Prader-Willi/Angelman imprinting center in the gametes and embryo. Hum. Mol. Genet. 13:2767-2779. [DOI] [PubMed] [Google Scholar]
- 16.Kantor, B., K. Makedonski, Y. Green-Finberg, R. Shemer, and A. Razin. 2004. Control elements within the PWS/AS imprinting box and their function in the imprinting process. Hum. Mol. Genet. 13:751-762. [DOI] [PubMed] [Google Scholar]
- 17.Kawasaki, H., and K. Taira. 2004. Induction of DNA methylation and gene silencing by short interfering RNAs in human cells. Nature 431:211-217. [DOI] [PubMed] [Google Scholar]
- 18.Lehnertz, B., Y. Ueda, A. A. Derijck, U. Braunschweig, L. Perez-Burgos, S. Kubicek, T. Chen, E. Li, T. Jenuwein, and A. H. Peters. 2003. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr. Biol. 13:1192-1200. [DOI] [PubMed] [Google Scholar]
- 19.Lewandoski, M., K. M. Wassarman, and G. R. Martin. 1997. Zp3-cre, a transgenic mouse line for the activation or inactivation of loxP-flanked target genes specifically in the female germ line. Curr. Biol. 7:148-151. [DOI] [PubMed] [Google Scholar]
- 20.Reference deleted.
- 21.Li, J. Y., D. J. Lees-Murdock, G. L. Xu, and C. P. Walsh. 2004. Timing of establishment of paternal methylation imprints in the mouse. Genomics 84:952-960. [DOI] [PubMed] [Google Scholar]
- 22.Mayer, W., A. Niveleau, J. Walter, R. Fundele, and T. Haaf. 2000. Demethylation of the zygotic paternal genome. Nature 403:501-502. [DOI] [PubMed] [Google Scholar]
- 23.Monk, M., M. Boubelik, and S. Lehnert. 1987. Temporal and regional changes in DNA methylation in the embryonic, extraembryonic and germ cell lineages during mouse embryo development. Development 99:371-382. [DOI] [PubMed] [Google Scholar]
- 24.Morris, K. V., S. W. Chan, S. E. Jacobsen, and D. J. Looney. 2004. Small interfering RNA-induced transcriptional gene silencing in human cells. Science 305:1289-1292. [DOI] [PubMed] [Google Scholar]
- 25.Oswald, J., S. Engemann, N. Lane, W. Mayer, A. Olek, R. Fundele, W. Dean, W. Reik, and J. Walter. 2000. Active demethylation of the paternal genome in the mouse zygote. Curr. Biol. 10:475-478. [DOI] [PubMed] [Google Scholar]
- 26.Pant, V., S. Kurukuti, E. Pugacheva, S. Shamsuddin, P. Mariano, R. Renkawitz, E. Klenova, V. Lobanenkov, and R. Ohlsson. 2004. Mutation of a single CTCF target site within the H19 imprinting control region leads to loss of Igf2 imprinting and complex patterns of de novo methylation upon maternal inheritance. Mol. Cell. Biol. 24:3497-3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Postic, C., and M. A. Magnuson. 2000. DNA excision in liver by an albumin-Cre transgene occurs progressively with age. Genesis 26:149-150. [DOI] [PubMed] [Google Scholar]
- 28.Santos, F., B. Hendrich, W. Reik, and W. Dean. 2002. Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev. Biol. 241:172-182. [DOI] [PubMed] [Google Scholar]
- 29.Schoenherr, C. J., J. M. Levorse, and S. M. Tilghman. 2003. CTCF maintains differential methylation at the Igf2/H19 locus. Nat. Genet. 33:66-69. [DOI] [PubMed] [Google Scholar]
- 30.Shibata, H., Y. Yoda, R. Kato, T. Ueda, M. Kamiya, N. Hiraiwa, A. Yoshiki, C. Plass, R. S. Pearsall, W. A. Held, M. Muramatsu, H. Sasaki, M. Kusakabe, and Y. Hayashizaki. 1998. A methylation imprint mark in the mouse imprinted gene Grf1/Cdc25Mm locus shares a common feature with the U2afbp-rs gene: an association with a short tandem repeat and a hypermethylated region. Genomics 49:30-37. [DOI] [PubMed] [Google Scholar]
- 31.Srivastava, M., E. Frolova, B. Rottinghaus, S. P. Boe, A. Grinberg, E. Lee, P. E. Love, and K. Pfeifer. 2003. Imprint control element-mediated secondary methylation imprints at the Igf2/H19 locus. J. Biol. Chem. 278:5977-5983. [DOI] [PubMed] [Google Scholar]
- 32.Srivastava, M., S. Hsieh, A. Grinberg, L. Williams-Simons, S. P. Huang, and K. Pfeifer. 2000. H19 and Igf2 monoallelic expression is regulated in two distinct ways by a shared cis acting regulatory region upstream of H19. Genes Dev. 14:1186-1195. [PMC free article] [PubMed] [Google Scholar]
- 33.Stoger, R., P. Kubicka, C. G. Liu, T. Kafri, A. Razin, H. Cedar, and D. P. Barlow. 1993. Maternal-specific methylation of the imprinted mouse Igf2r locus identifies the expressed locus as carrying the imprinting signal. Cell 73:61-71. [DOI] [PubMed] [Google Scholar]
- 34.Tallquist, M. D., and P. Soriano. 2000. Epiblast-restricted Cre expression in MORE mice: a tool to distinguish embryonic vs. extra-embryonic gene function. Genesis 26:113-115. [DOI] [PubMed] [Google Scholar]
- 35.Thorvaldsen, J. L., K. L. Duran, and M. S. Bartolomei. 1998. Deletion of the H19 differentially methylated domain results in loss of imprinted expression of H19 and Igf2. Genes Dev. 12:3693-3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thorvaldsen, J. L., A. M. Fedoriw, S. Nguyen, and M. S. Bartolomei. 2006. Developmental profile of H19 differentially methylated domain (DMD) deletion alleles reveals multiple roles of the DMD in regulating allelic expression and DNA methylation at the imprinted H19/Igf2 locus. Mol. Cell. Biol. 26:1245-1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Volpe, T. A., C. Kidner, I. M. Hall, G. Teng, S. I. Grewal, and R. A. Martienssen. 2002. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297:1833-1837. [DOI] [PubMed] [Google Scholar]
- 38.Wassenegger, M., S. Heimes, L. Riedel, and H. L. Sanger. 1994. RNA-directed de novo methylation of genomic sequences in plants. Cell 76:567-576. [DOI] [PubMed] [Google Scholar]
- 39.Yoon, B.-J., H. Herman, B. Hu, Y. J. Park, A. M. Lindroth, A. Bell, A. G. West, Y. Chang, A. Stablewski, J. C. Piel, D. I. Loukinov, V. Lobanenkov, and P. D. Soloway. 2005. Rasgrf1 imprinting is regulated by a CTCF-dependent methylation-sensitive enhancer blocker. Mol. Cell. Biol. 25:11184-11190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoon, B. J., H. Herman, A. Sikora, L. T. Smith, C. Plass, and P. D. Soloway. 2002. Regulation of DNA methylation of Rasgrf1. Nat. Genet. 30:92-96. [DOI] [PMC free article] [PubMed] [Google Scholar]