Abstract
Mucopolysaccharidosis IIIC (MPS IIIC, or Sanfilippo C syndrome) is a lysosomal storage disorder caused by the inherited deficiency of the lysosomal membrane enzyme acetyl–coenzyme A:α-glucosaminide N-acetyltransferase (N-acetyltransferase), which leads to impaired degradation of heparan sulfate. We report the narrowing of the candidate region to a 2.6-cM interval between D8S1051 and D8S1831 and the identification of the transmembrane protein 76 gene (TMEM76), which encodes a 73-kDa protein with predicted multiple transmembrane domains and glycosylation sites, as the gene that causes MPS IIIC when it is mutated. Four nonsense mutations, 3 frameshift mutations due to deletions or a duplication, 6 splice-site mutations, and 14 missense mutations were identified among 30 probands with MPS IIIC. Functional expression of human TMEM76 and the mouse ortholog demonstrates that it is the gene that encodes the lysosomal N-acetyltransferase and suggests that this enzyme belongs to a new structural class of proteins that transport the activated acetyl residues across the cell membrane.
Heparan sulfate is a polysaccharide found in proteoglycans associated with the cell membrane in nearly all cells. The lysosomal membrane enzyme, acetyl–coenzyme A (CoA):α-glucosaminide N-acetyltransferase (N-acetyltransferase) is required to N-acetylate the terminal glucosamine residues of heparan sulfate before hydrolysis by the α-N-acetyl glucosaminidase. Since the acetyl-CoA substrate would be rapidly degraded in the lysosome,1 N-acetyltransferase employs a unique mechanism, acting both as an enzyme and a membrane channel, and catalyzes the transmembrane acetylation of heparan sulfate.2 The mechanism by which this is achieved has been the topic of considerable investigation, but, for many years, the isolation and cloning of N-acetyltransferase has been hampered by its low tissue content, instability, and hydrophobic nature.3–5
Genetic deficiency of N-acetyltransferase causes mucopolysaccharidosis IIIC (MPS IIIC [MIM 252930], or Sanfilippo syndrome C), a rare autosomal recessive lysosomal disorder of mucopolysaccharide catabolism.6–8 MPS IIIC is clinically similar to other subtypes of Sanfilippo syndrome.9 Patients manifest symptoms during childhood with progressive and severe neurological deterioration causing hyperactivity, sleep disorders, and loss of speech accompanied by behavioral abnormalities, neuropsychiatric problems, mental retardation, hearing loss, and relatively minor visceral manifestations, such as mild hepatomegaly, mild dwarfism with joint stiffness and biconvex dorsolumbar vertebral bodies, mild coarse faces, and hypertrichosis.7 Most patients die before adulthood, but some survive to the 4th decade and show progressive dementia and retinitis pigmentosa. Soon after the first 3 patients with MPS IIIC were described by Kresse et al.,6 Klein et al.8,10 reported a similar deficiency in 11 patients who had received the diagnosis of Sanfilippo syndrome, therefore suggesting that the disease is a relatively frequent subtype. The birth prevalence of MPS IIIC in Australia,11 Portugal,12 and the Netherlands13 has been estimated to be 0.07, 0.12, and 0.21 per 100,000, respectively.
The putative chromosomal locus of the MPS IIIC gene was first reported in 1992. By studying two siblings who received the diagnosis of MPS IIIC and had an apparently balanced Robertsonian translocation, Zaremba et al.14 suggested that the mutant gene may be located in the pericentric region of either chromosome 14 or chromosome 21, but no further confirmation of this finding was provided. Previously, we performed a genomewide scan on 27 patients with MPS IIIC and 17 unaffected family members, using 392 highly informative microsatellite markers with an average interspacing of 10 cM. For chromosome 8, the scan showed an apparent excess of homozygosity in patients compared with their unaffected relatives.15 Additional genotyping of 38 patients with MPS IIIC for 22 markers on chromosome 8 identified 15 consecutive markers (from D8S1051 to D8S2332) in an 8.3-cM interval for which the genotypes of affected siblings were identical in state. A maximum multipoint LOD score of 10.6 was found at marker D8S519, suggesting that this region includes the locus for MPS IIIC.15 Recently, localization of the MPS IIIC causative gene on chromosome 8 was confirmed by microcell-mediated chromosome transfer in cultured skin fibroblasts of patients with MPS IIIC.16
Here, we report the results of linkage analyses that narrowed the candidate region for MPC IIIC to a 2.6-cM interval between D8S1051 and D8S1831 and the identification of the TMEM76 gene, located within the candidate region, as the gene that codes for the lysosomal N-acetyltransferase and, when mutated, is responsible for MPS IIIC.
Material and Methods
Families
In Montreal, 33 affected individuals and 35 unaffected relatives comprising 15 families informative for linkage were genotyped. The families came from Europe, North Africa, and North America. An additional 27 affected individuals and 9 unaffected relatives in uninformative pedigrees, as well as 40 controls, were also genotyped. Eleven of these families and the controls have been reported elsewhere.15 In addition, 54 individuals from four MPS IIIC–affected families from the Czech Republic were studied in Prague (fig. 1). One family had two affected brothers, whereas the remaining three families each had one affected individual. The families came from various regions of the Czech Republic and were not related within the four most-recent generations. The diagnosis for affected individuals was confirmed by the measurement of N-acetyltransferase activity in cultured skin fibroblasts or white blood cells.
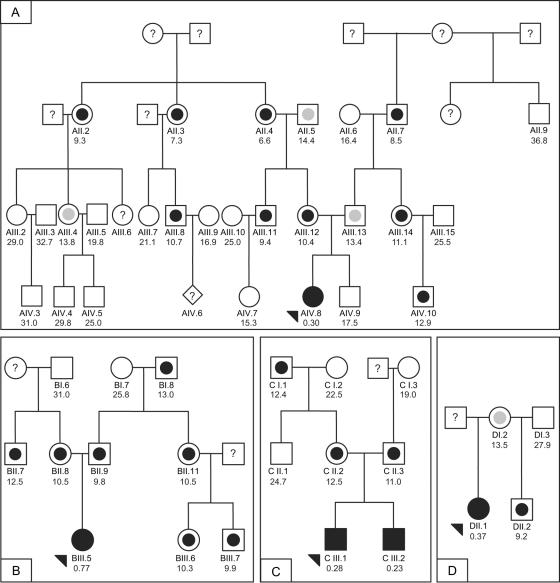
Figure 1. .
Four families from the Czech Republic used in the linkage and mutation analyses. Fully blackened symbols indicate individuals with MPS IIIC; arrowheads indicate probands. Measurements in seven obligate heterozygotes from these pedigrees (mean ± SD 11.6 ± 1.5 nmol/h/mg) and 89 controls not known to be related to members of the pedigree (mean ± SD 24.4 ± 5.7 nmol/h/mg) were used to establish N-acetyltransferase activity ranges for heterozygotes (symbols with blackened inner circle) and normal homozygotes (open symbols). An individual was assigned to a class if his or her enzyme activity was within 2 SDs of the class, unless the value was within the overlap of the upper end of the obligate heterozygotes and the lower end of the controls. Individuals with values within the open interval 13.0–14.6 nmol/h/mg were classified as unknown (symbols with gray inner circle). A symbol with a question mark (?) indicates that no material was available for the enzyme assay. DNA was available for individuals with ID numbers, and N-acetyltransferase activity measurements in white blood cells are shown below the ID numbers.
Genotyping
The samples in Montreal were genotyped for 22 microsatellite markers in the pericentromeric region of chromosome 8 spanning 8.9 cM on the Rutgers map, version 2.0.17 The genotyping was performed as described by Mira et al.18 at the McGill University and Genome Quebec Innovation Centre on an ABI 3730xl DNA Analyzer platform (Applied Biosystems). Alleles were assigned using Genotyper, version 3.6 (Applied Biosystems). The random-error model of SimWalk2, version 2.91,19,20 was used to detect potential genotyping errors, with an overall error rate of 0.025. Nine genotypes for which the posterior probability of being incorrect was >0.5 were removed before subsequent analyses. In addition, nine genotypes for one marker in one family were removed because of a suspected microsatellite mutation. The samples from the Czech Republic were genotyped in Prague for 18 microsatellite markers in an 18.7-cM region that includes the 8.9-cM region mentioned above. The genotyping was performed on an LI-COR IR2 sequencer by use of Saga genotyping software (Li-Cor) as described elsewhere.21 Genotypes were screened for errors by use of the PedCheck program.22
Linkage Analysis
For the families genotyped in Montreal, multipoint linkage analysis was performed using the Markov chain–Monte Carlo (MCMC) method implemented in SimWalk2, version 2.91,19 since one pedigree was too large to be analyzed by exact computation. A fully penetrant autosomal recessive parametric model was used with a disease-allele frequency of 0.0045. Marker-allele frequencies were estimated by counting alleles in the available parents of patients with MPS IIIC and in control individuals. To check the consistency of the results, the MCMC analysis was repeated four times.
N-acetyltransferase activity was measured in all participants of the four families from the Czech Republic.23 Individuals were classified as affected, carriers, or unaffected on the basis of the results of this assay. Mean affected and carrier activities were determined from the five affected individuals and their seven obligate heterozygote parents, respectively, whereas the mean control activity was determined from a sample of 89 unrelated individuals. Four individuals were unable to be classified because their values were within 2 SDs of the means of both the control and carrier groups. Multipoint linkage analysis was performed using a codominant model with a penetrance of 0.99 and a phenocopy rate of 0.01, to account for the possibility of misclassification or genotyping errors. The same disease-allele frequency of 0.0045 was used. Marker-allele frequencies were estimated by counting all genotyped individuals. Exact multipoint linkage analysis was run on 18 microsatellite markers by use of Allegro 1.2c,24 which was also used to infer haplotypes.
Gene-Expression Analysis
For each of 32 genes located in the candidate interval, a single 5′-amino–modified 40-mer oligonucleotide probe (Illumina) was spotted in quadruplicate on aminosilane-modified microscopic slides and was immobilized using a combination of baking and UV cross-linking. Total RNA (250–1,000 ng) from white blood cells of two patients with MPS IIIC (patients AIV.8 and BIII.5) and four healthy individuals were amplified using the SenseAmp plus RNA Amplification Kit (Genisphere) and were reverse transcribed using 300 ng of poly(A)-tailed mRNA. Reverse transcription and microarray detection were done using the Array 900 Expression Detection Kit (Genisphere) according to the manufacturer’s protocol. The two patient samples and four control samples were analyzed in dye-swap mode, in two replicates of each mode. The hybridized slides were scanned with a GenePix 4200A scanner (Molecular Devices), with photomultiplier gains adjusted to obtain the highest-intensity unsaturated images. Data analysis was performed in the R statistical environment (The R Project for Statistical Computing, version 2.2.1) by use of the Linear Models for Microarray Data package (Limma, version 2.2.0).25 Raw data were processed using loess normalization and a moving minimum background correction on individual arrays and quantile normalization between arrays. The correlation between four duplicate spots per gene on each array was used to increase the robustness. A linear model was fitted for each gene given a series of arrays by use of the lmFit function. The empirical Bayes method26 was used to rank the differential expression of genes by use of the eBayes function. Correction for multiple testing was performed using the Benjamini and Hochberg false-discovery–rate method.27 We considered genes to be differentially expressed if the adjusted P value was <.01.
DNA and RNA Isolation and Sequencing
Cultured skin fibroblasts from patients with MPS IIIC and normal controls were obtained from cell depositories (Hôpital Debrousse, France; NIGMS Human Genetic Mutant Cell Repository; Montreal Children’s Hospital, Canada; and Department of Clinical Genetics, Erasmus Medical Center, The Netherlands). Blood samples from patients with MPS IIIC, their relatives, and controls were collected with ethics approval from the appropriate institutional review boards. DNA from blood or cultured skin fibroblasts was extracted using the PureGene kit (Gentra Systems). Total RNA from cultured skin fibroblasts and pooled tissues (spleen, liver, kidney, heart, lung, and brain) of a C57BL/6J mouse was isolated using Trizol (Invitrogen), and first-strand cDNA synthesis was prepared with SuperScript II (Invitrogen). DNA fragments containing TMEM76 exons and adjacent regions (∼40 bp from each side; primer sequences are shown in appendix A) were amplified by PCR from genomic DNA and were purified with Montage PCR96 filter plates (Millipore). Each sequencing reaction contained 2 μl of purified PCR product, 5.25 μl of H2O, 1.75 μl of 5× sequencing buffer, 0.5 μl of 20 μM primer, and 0.5 μl of Big Dye Terminator v3.1 (all from Applied Biosystems). In Montreal, PCR products were analyzed using an ABI 3730xl DNA Analyzer (Applied Biosystems). In Prague, PCR products were analyzed on an ALFexpress DNA sequencer (Pharmacia), as described elsewhere.28 Included in the sequencing analysis were 30 probands with MPS IIIC who were considered unrelated and 105 controls. The controls were unrelated CEPH individuals, and amplified DNAs were combined in pools of two before sequencing.
Northern Blotting
A 12-lane multiple-tissue northern blot containing 1 μg of poly A+ RNA per lane from various human tissues (BD Biosciences Clontech) was hybridized with the 220-bp cDNA fragment corresponding to exons 8–10 of the human TMEM76 gene or the entire cDNA of human β-actin labeled with [32P]-dCTP by random priming with the MegaPrime labeling kit (Amersham). Prehybridization of the blot was performed at 68°C for 30 min in ExpressHyb (Clontech). The denatured probes were added directly to the prehybridization solution and were incubated at 68°C for 1 h. The blots were washed twice for 30 min at room temperature with 2× sodium chloride–sodium citrate (SSC) solution and 0.05% SDS and once for 40 min at 50°C with 0.1× SSC and 0.1% SDS and were exposed to a BioMax film for 48 h.
Mouse and Human TMEM76 cDNA Cloning
Mouse coding sequence was amplified by PCR (forward primer 5′-GAATTCATGACGGGCGGGTCGAGC-3′; reverse primer 5′-ATATGTCGACGATTTTCCAAAACAGCTTC-3′) and was cloned into pCMV-Script, pCMV-Tag4A (Stratagene), and pEGFP-N3 (BD Biosciences Clontech) vectors by use of the EcoRI and SalI restriction sites of the primers. The cloned sequence was identical to GenBank accession number AK152926.1, except that an “AT” was needed to complete an alternate ATG initiation codon. GenBank accession number AK149883.1 provides what we consider to be the complete clone and encodes a 656-aa protein. The GenBank sequences differ by 1 aa and three silent substitutions.
A 1,907-bp fragment of the human TMEM76 cDNA (nt +75 to +1992) was amplified using Platinum High Fidelity Taq DNA polymerase (Invitrogen), a sense primer with an HindIII site (5′-AAGCTTGGCGGCGGGCATGAG-3′), and an antisense primer with an SalI site (5′-GTCGACCTCAGTGGGAGCCATCAGATTTT-3′) and was cloned into pCMV-Script expression vector (Stratagene). Since high GC content (85%) of the 5′ region of human TMEM76 cDNA prevented its amplification by PCR, a synthetic 186-bp codon-optimized double-stranded oligonucleotide fragment (5′-AAGCTTATGACCGGAGCGAGGGCAAGCGCCGCCGAACAAAGAAGAGCCGGACGGTCCGGCCAGGCTAGGGCCGCAGAGCGAGCTGCTGGCATGTCAGGTGCAGGGCGCGCACTTGCCGCCTTGCTGCTCGCCGCGAGTGTGCTGAGCGCTGCCCTCCTGGCTCCCGGAGGCTCTTCCGGGCGGGAC-3′) corresponding to nt +1 to +186 of human TMEM76 cDNA was purchased from BioS&T. A 177-bp 5′ fragment was combined with rest of the cDNA by use of HindIII and SapI sites. The cloned sequence is identical to GenBank accession number XM_372038.4 from nt 131 to nt 1946, except for the presence of SNP rs1126058.
Cell Culture and Transfection
Skin fibroblasts and COS-7 cells were cultured in Eagle’s minimal essential medium (Invitrogen) supplemented with 10% (v/v) fetal bovine serum (Invitrogen) and were transfected with the full-size mouse Tmem76 (Hgsnat) coding sequence subcloned into pCMV-Script, pCMV-Tag4A, and pEGFP-N3 vectors or with the full-size human TMEM76 coding sequence subcloned into pCMV-Script vector by use of Lipofectamine Plus (Invitrogen) according to the manufacturer’s protocol. The cells were harvested 48 h after transfection, and N-acetyltransferase activity was measured in the homogenates of TMEM76-transfected and mock-transfected cells (i.e., transfected with only the cloning vector).
Enzyme Assay
N-acetyltransferase enzymatic activity was measured using the fluorogenic substrate 4-methylumbelliferyl β-d-glucosaminide (Moscerdam) as described elsewhere.23 Protein concentration was measured according to the method of Bradford.29 This assay was used for the activity measurements in cultured skin fibroblasts or white blood cells from patients and all participating members of the Czech families and for the functional expression experiments.
Confocal Microscopy
To establish colocalization of the tagged protein with the lysosomal compartment, the skin fibroblasts expressing mouse TMEM76-EGFP were treated with 50 nM LysoTracker Red DND-99 dye (Molecular Probes), were washed twice with ice-cold PBS, and were fixed with 4% paraformaldehyde in PBS for 30 min. Slides were studied on an LMS 510 Meta inverted confocal microscope (Zeiss).
Results
Linkage Analysis
Previously, we performed a genomewide linkage study that indicated that the locus for MPS IIIC is mapped to an 8.3-cM interval in the pericentromeric region of chromosome 8.15 To reduce this interval, we genotyped the families from that study as well as newly obtained MPS IIIC–affected families for 22 microsatellite markers (Montreal data). Linkage analysis under an autosomal recessive model resulted in LOD scores >14 in the 4.2-cM region spanning D8S1051 to D8S601, which included the centromere (fig. 2). The results of multiple MCMC runs showed consistent trends. Linkage was also performed in four families from the Czech Republic by use of an autosomal codominant model (Prague data). For these data, linkage analysis produced a maximum LOD score of 7.8 at 66.4 cM at D8S531 and reduced the linked region for the Montreal data to a 2.6-cM interval between D8S1051 and D8S1831. This region was defined by inferred recombinants at D8S1051 in one family in each of the Montreal and Prague data sets, and a recombinant at D8S1831 in an additional family in the Prague data set. This interval contains 32 known or predicted genes and ORFs.
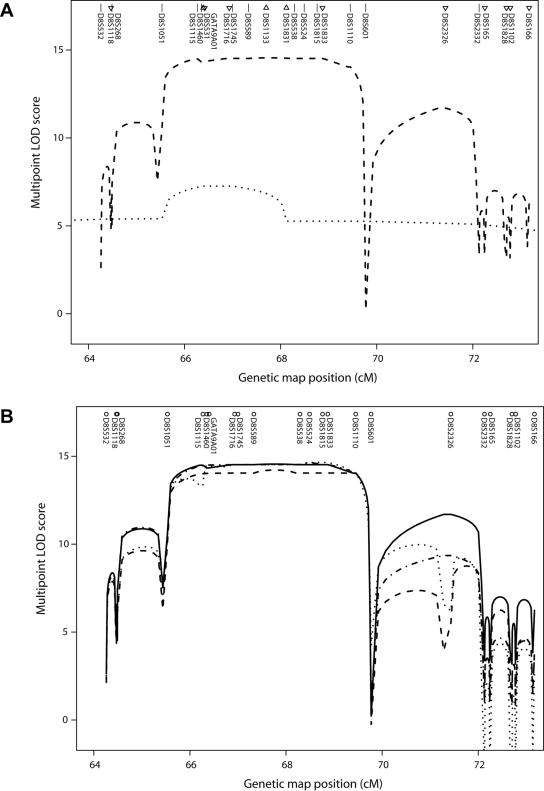
Figure 2. .
Multipoint linkage analysis of MPS IIIC on chromosome 8. A, Multipoint LOD scores in an 8.9-cM interval from two sets of families. Symbols above the marker names indicate the map position. Marker names are listed in the correct order but may be displaced from the symbols for visibility. The dashed line is based on families genotyped in Montreal, and the dotted line on families genotyped in Prague. Straight lines next to marker names indicate that the markers were typed in both data sets. Triangles pointing down indicate markers typed only in the Montreal data set, and triangles pointing up indicate markers typed only in the Prague data set. For the Montreal data, the SimWalk2 run with the highest likelihood is shown. TMEM76 lies between D8S1115 and D8S1460, and, according to the March 2006 freeze of the human genome sequence from the University of California–Santa Cruz Genome Browser,30 the order is D8S1115–(500 kb)–TMEM76–(800 kb)–centromere–(200 kb)–D8S1460. B, Multipoint LOD scores from the Montreal data from four runs of SimWalk2, version 2.91,19 showing the variation between runs.
Identification of a Candidate Gene
On the basis of our previous studies that defined the molecular properties of the lysosomal N-acetyltransferase,31 we searched the candidate region for a gene encoding a protein with multiple transmembrane domains and a molecular weight of ∼100 kDa, which allowed us to exclude the majority of the genes in the region. In contrast, the predicted protein product of the TMEM76 gene has multiple putative transmembrane domains. The predicted coding region in GenBank accession number XM_372038.4 was extended by 28 residues at the 5′ end on the basis of the transcript in GenBank accession number DR000652.1 (which includes 14 of the 28 residues), examination of the genomic sequence in NT_007995.14, and comparison with mouse sequence AK149883.1. We predict that the modified TMEM76 contains 18 exons, corresponding to an ORF of 1,992 bp, and codes for a 73-kDa protein. A comparison of human TMEM76 with five vertebrate orthologs is shown in figure 3. Furthermore, of all the genes present in the candidate interval, only TMEM76 showed a statistically significant reduction of the transcript level in the cells of two patients with MPS IIIC (AIV.8 and BIII.5; adjusted P values < .001) in the custom oligonucleotide-based microarray assay (fig. 4). Further, we showed that both patients carried nonsense mutations presumably causing mRNA decay (R534X and L349X; see table 2).
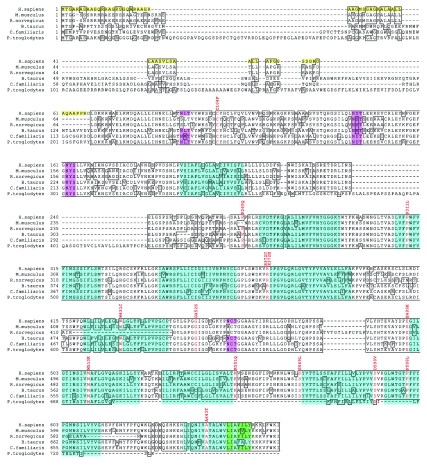
Figure 3. .
Predicted amino acid sequence of the TMEM76 protein. Amino acid sequence alignment of Homo sapiens TMEM76 with orthologs from Mus musculus (cloned sequence), Canis familiaris (GenBank accession number XP_539948.2), Bos taurus (XP_588978.2), Rattus norvegicus (XP_341451.2), and Pan troglodytes (XP_519741.1) by use of BLAST. All cDNA sequences are predicted except the sequence for M. musculus. The identical residues are boxed, the residues with missense mutations in patients with MPS IIIC are shown in red, and the amino acid changes are indicated above the sequence. The first 67 aa of the human sequence shown as black on yellow comprise the predicted signal peptide. The predicted transmembrane domains in the human sequence are shown as black on turquoise. The topology model5–7 strongly predicts that the N-terminus is inside the lysosome and the C-terminus is outside. Four predicted N-glycosylation sites are shown as black on pink, and the predicted motifs for the lysosomal targeting, as black on green.
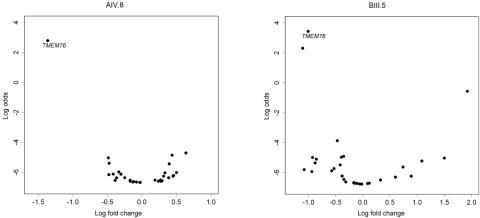
Figure 4. .
Volcano plot of genes located within the MPS IIIC candidate region, showing significantly reduced expression of the TMEM76 gene in white blood cells of two patients with MPS IIIC: AIV.8 and BIII.5. The natural logarithm of the probability that the gene is differentially expressed (Log odds) is plotted as a function of the logarithm of the gene-expression log2 fold change (Log fold change) between the patient and control samples.
Table 2. .
TMEM76 Predicted Mutations in Probands from 30 Families with MPS IIIC
| Patient Group and Mutation 1 | Mutation 2 | No. of Patients | Geographic Origin of Patient(s) |
| Patients from Czech families: | |||
| p.I373SfsX3 | p.R534X | 1 | Czech Republic |
| p.L349X | p.M510K | 1 | Czech Republic |
| p.F313X | p.R412X | 1 | Czech Republic |
| p.R372H | p.P599L | 1 | Czech Republic |
| Patients homozygous for TMEM76 mutations: | |||
| p.[D68VfsX19; P265Q] | p.[D68VfsX19; P265Q] | 3 | Morocco, Morocco, and Spain |
| p.[P193HfsX20; K551Q] | p.[P193HfsX20; K551Q] | 1 | France |
| p.P311L | p.P311L | 1 | United Kingdom |
| p.W344X | p.W344X | 1 | France |
| p.R372C | p.R372C | 1 | United Kingdom |
| p.R412X | p.R412X | 2 | Turkey and Poland |
| p.[W431C; A643T] | p.[W431C; A643T] | 1 | France |
| p.G452S | p.G452S | 1 | Canada |
| p.E499K | p.E499K | 1 | Canada |
| p.S567NfsX14 | p.S567NfsX14 | 1 | Turkey |
| Patients compound heterozygous for TMEM76 mutations: | |||
| p.C104F | … | 1 | Belarus |
| p.E499K | p.D590V | 1 | France |
| p.P193HfsX20 | p.R412X | 1 | Canada |
| p.P311L | p.R372C | 1 | France |
| p.R412X | … | 1 | Poland |
| p.R412X | p.G446X | 1 | Poland |
| p.S569L | … | 2 | France and Portugal |
| p.S569L | p.L69EfsX32 | 1 | United States |
| p.V488GfsX22 | p.S569L | 1 | Poland |
| p.V612SfsX16 | … | 1 | Finland |
| Families with no mutations identified to date | … | 2 | North Africa and Portugal |
Analysis of the TMEM76 Transcript by Northern Blot and RT-PCR
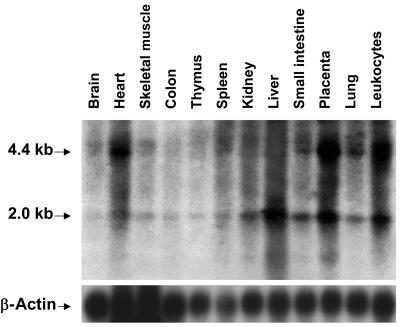
Northern-blot analysis identified two major TMEM76 transcripts of 4.5 and 2.1 kb ubiquitously expressed in various human tissues (fig. 5). The highest expression was detected in leukocytes, heart, lung, placenta, and liver, whereas the gene was expressed at a much lower level in the thymus, colon, and brain, which is consistent with the expression patterns of lysosomal proteins. Consistent with the northern-blot results, a full-length 4.5-kb cDNA containing 1,992 bp of coding sequence and two polyadenylation signals as well as two shorter transcripts were amplified by RT-PCR from the total RNA of normal human skin fibroblasts, white blood cells, and skeletal muscle. In one transcript, exons 9 and 10 were spliced out, leading to an in-frame deletion of 64 aa, which contains the predicted transmembrane domains III and IV. Most likely, this transcript does not encode an active enzyme, since it was also detected in the RNA of two patients with MPS IIIC (patients CIII.1 and CIII.2) who had almost complete loss of N-acetyltransferase activity. Another transcript lacked exons 3, 9, and 10.
Figure 5. .
Northern-blot analysis of TMEM76 mRNA in human tissues. A 12-lane blot containing 1 μg of poly A+ RNA per lane from various adult human tissues was hybridized with a [32P]-labeled 220-bp cDNA fragment corresponding to exons 8–10 of the TMEM76 gene or β-actin, as described in the Material and Methods section.
The deduced amino acid sequence predicts 11 transmembrane domains and four potential N-glycosylation sites (fig. 3), consistent with the molecular properties of lysosomal N-acetyltransferase.31 The first 67 aa may comprise the signal peptide, with length and composition resembling those of lysosomal proteins. According to the predictions made by empirical computer algorithms,32–34 the C-terminus of the TMEM76 protein is exposed to the cytoplasm and contains conserved Tyr-X-X-Θ and Leu-Leu sequence motifs involved in the interaction with the adaptor proteins responsible for the lysosomal targeting of membrane proteins.35
Mutation Analysis
We identified 27 TMEM76 mutations in the DNA of 30 MPS IIIC–affected families (table 1) that were not found in DNA from 105 controls. Among the identified mutations, there were 4 nonsense mutations, 14 missense mutations, 3 predicted frameshift mutations due to deletions or duplications, and 6 splice-site mutations. All the missense mutations occur at residues conserved among five species with the most homologous TMEM76 sequences (fig. 3), except for P265Q, which is not conserved in the mouse, and W431C, which is not conserved in the rat. There were three instances of two mutations on the same allele that were found in patients who were homozygous, and these are designated as complex mutations in table 1. cDNA sequencing of one of the patients homozygous for the splice-site mutation in intron 2 and a missense mutation (P265Q) demonstrated that the splice-site mutation disrupts the consensus splice-site sequence between exon 2 and intron 2 and causes exon 2 skipping and a frameshift (not shown).
Table 1. .
Mutations in TMEM76 Identified in Patients from 30 Families with MPS IIIC
| Mutation Group and Mutationa |
Predicted Effect on Protein |
No. of Alleles | Location in TMEM76 |
| Nonsense mutations: | |||
| c.1031G→A | p.W344X | 2 | Exon 10 |
| c.1046T→G | p.L349X | 1 | Exon 10 |
| c.1234C→T | p.R412X | 8 | Exon 12 |
| c.1600C→T | p.R534X | 1 | Exon 15 |
| Missense mutations: | |||
| c.311G→T | p.C104F | 1 | Exon 2 |
| c.932C→T | p.P311L | 3 | Exon 9 |
| c.1114C→T | p.R372C | 3 | Exon 11 |
| c.1115G→A | p.R372H | 1 | Exon 11 |
| c.1354G→A | p.G452S | 2 | Exon 13 |
| c.1495G→A | p.E499K | 3 | Exon 14 |
| c.1529T→A | p.M510K | 1 | Exon 14 |
| c.1706C→T | p.S569L | 4 | Exon 17 |
| c.1769A→T | p.D590V | 1 | Exon 17 |
| c.1796C→T | p.P599L | 1 | Exon 17 |
| Frameshift mutations: | |||
| c.1118_1133del | p.I373SfsX3 | 1 | Exon 11 |
| c.1420_1456dup | p.V488GfsX22 | 1 | Exon 13 |
| c.1834delG | p.V612SfsX16 | 1 | Exon 18 |
| Splice-site mutations: | |||
| c.202+1G→A | p.L69EfsX32b | 1 | Intron 1 |
| c.577+1G→A | p.P193HfsX20b | 1 | Intron 4 |
| c.935+5G→A | p.F313X | 1 | Intron 9 |
| c.1334+1G→A | p.G446Xb | 1 | Intron 12 |
| c.1810+1G→A | p.S567NfsX14 | 2 | Intron 17 |
| Complex mutations: | |||
| c.[318+1G→A; 794C→A] | p.[D68VfsX19; P265Q] | 6 | Intron 2; exon 7 |
| c.[577+1G→A; 1650A→C] | p.[P193HfsX20; K551Q] | 2 | Intron 4; exon 16 |
| c.[1293G→T; 1927G→A] | p.[W431C; A643T] | 2 | Exon 12; exon 18 |
Mutation names were assigned according to the guidelines of the Human Genome Variation Society and on the basis of the cDNA sequence from GenBank accession number NT_007995.14, except that the first exon includes 84 nt 5′ of the stated ATG initiation codon. Thus, +1 corresponds to the A of the ATG at nt 13315945 (instead of nt 13316029).
The mutations were named under the assumption that no exon skipping takes place; cDNA sequencing was not done.
Consanguinity was reported in 4 of the 13 families in which the patients were homozygous for TMEM76 mutations: the two Moroccan families, the French family with two missense mutations (W431C and A643T), and the Turkish family with the splice-site mutation in intron 17. The two Moroccan families were not known to be related to each other or to the Spanish patient homozygous for the same mutations (table 2). The parents of the French patient are second cousins in two ways (see family F1 in the work of Ausseil et al.15).
The splice-site mutation in the above-mentioned Turkish family disrupts the consensus splice-site sequence between exon 17 and intron 17 and causes exon 17 skipping and a frameshift in all transcripts, as detected by sequencing of multiple RT-PCR clones (not shown). The two affected siblings in this family (family F8 in the work of Ausseil et al.15) had a severe form of MPS IIIC and showed almost complete loss of N-acetyltransferase activity in cultured skin fibroblasts. Among other severely affected patients with MPS IIIC, a patient of French origin was homozygous for a nonsense mutation (W344X) in exon 10, which may result in the synthesis of a truncated protein or RNA decay. A patient of Polish origin was a compound heterozygote for a 37-bp duplication in exon 13 and a missense mutation (S569L) in exon 17 (table 2). The duplication results in a frameshift, whereas the substitution of a strictly conserved small polar Ser for a bulky hydrophobic Leu may have a significant structural impact (fig. 3).
The five patients from four Czech families are all compound heterozygotes for eight different mutations (table 2). Five of the eight mutations are predicted to result in truncated products (three nonsense mutations, one 16-bp deletion, and one splice-site mutation leading to the inclusion of 89 bases from the 5′ end of intron 9 and the splicing out of exon 10 in the transcript, and the remaining three are missense mutations affecting residues conserved among multiple species and located either in the predicted transmembrane regions (fig. 3) or in their close vicinity, suggesting that they may have a serious structural impact. In the Czech families, the mutations completely segregated with reduced enzyme activity. That is, all individuals assigned to be heterozygotes on the basis of the enzyme assay as well as the four individuals who were within 2 SD of the lower end of the controls (symbols with gray inner circle in fig. 1) were found to carry TMEM76 mutations.
Functional Expression Studies
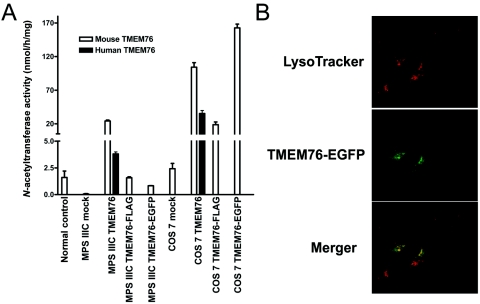
The fibroblast cell line from a patient homozygous for a splice-site mutation in intron 17 with negligible N-acetyltransferase activity was transfected with plasmids containing human TMEM76 cDNA or cDNA of the mouse ortholog of TMEM76 carrying a FLAG tag on the C-terminus or of a fusion protein of mouse TMEM76 with enhanced green fluorescent protein (EGFP). All constructs increased the N-acetyltransferase activity in the mutant fibroblast cells to approximately normal level (fig. 6A). Significant increase in activity was also observed in transfected COS-7 cells, confirming that the TMEM76 protein by itself has N-acetyltransferase activity. Confocal fluorescent microscopy shows that TMEM76-EGFP (fig. 6B) or TMEM-FLAG (not shown) peptides are targeted in human fibroblasts to cytoplasmic organelles, colocalizing with the lysosomal-endosomal marker LysoTracker Red.
Figure 6. .
Functional expression of human and mouse TMEM76 protein. A, The full-size human and mouse TMEM76 coding sequences subcloned into pCMV-Script, pCMV-Tag4A, and pEGFP-N3 vectors were expressed in COS-7 cells and in cultured skin fibroblasts from a patient with MPS IIIC. The cells were harvested 48 h after transfection, and N-acetyltransferase activity was measured in the homogenates of TMEM76-transfected and mock-transfected fibroblast or COS-7 cells by use of the artificial fluorometric substrate 4-methylumbelliferyl-β-d-glucosaminide.23 Values represent means ± SD of four independent experiments. B, The intracellular localization of TMEM76 was studied by expressing the fusion protein of the mouse TMEM76 with EGFP. Before fixation, the cells were treated for 45 min with 50 nM lysosomal marker, LysoTracker Red DND-99 dye. Slides were analyzed on an LMS 510 Meta confocal microscope (Zeiss). Magnification ×1000. The image was randomly selected from 30 studied panels, all of which showed a similar localization of TMEM76-EGFP. The fluorescence of EGFP was not quenched as it would have been if the fluorophore had been exposed to the acidic lysosomal microenvironment, confirming that the C-terminus of TMEM76 faces the cytoplasmic side of the lysosomal membrane.
Discussion
Degradation of heparan sulfate occurs within the lysosomes by the concerted action of a group of at least eight enzymes: four sulfatases, three exo-glycosydases, and one N-acetyltransferase, which work sequentially at the terminus of heparan sulfate chains, producing free sulfate and monosaccharides. The inherited deficiencies of four enzymes involved in the degradation of heparan sulfate cause four subtypes of MPS III: MPS IIIA (heparan N-sulfatase deficiency [MIM 252900]), MPS IIIB (α-N-acetylglucosaminidase deficiency [MIM 252920]), MPS IIIC (acetyl-CoA:α-glucosaminide acetyltransferase deficiency), and MPS IIID (N-acetylglucosamine 6-sulfatase deficiency [MIM 252940]). Since the clinical phenotypes of all these disorders are similar, precise diagnosis relies on the determination of enzymatic activities in patients’ cultured skin fibroblasts or leukocytes. The biochemical defect in MPS IIIC was identified 30 years ago as a deficiency of an enzyme that transfers an acetyl group from cytoplasmically derived acetyl-CoA to terminal α-glucosamine residues of heparan sulfate within the lysosomes, resulting in the accumulation of heparan sulfate. Therefore, for identification of the molecular basis of this disorder, we used two complementary approaches. First, we performed a partial purification of human and mouse lysosomal N-acetyltransferase, which suggested that the enzyme has properties of an oligomeric transmembrane glycoprotein, with an ∼100-kDa polypeptide containing the enzyme active site.31 Second, by linkage analysis, we narrowed the locus for MPC IIIC to a 2.6 cM-interval (D8S1051–D8S1831) and, third, compared the level of transcripts of the genes present in the candidate region between normal control cells and those from patients with MPS IIIC. Thus, an integrated bioinformatic search and gene-expression analysis both pinpointed a single gene, TMEM76, which encodes a 73-kDa protein with predicted multiple transmembrane domains and glycosylation sites. DNA mutation analysis showed that patients with MPS IIIC harbor TMEM76 mutations incompatible with the normal function of the predicted protein, whereas expression of human TMEM76 and the mouse ortholog proved that the protein has N-acetyltransferase activity and lysosomal localization, providing evidence that TMEM76 is the gene that codes for the lysosomal N-acetyltransferase.
The TMEM76 protein does not show a structural similarity to any known prokaryotic or eukaryotic N-acetyltransferases or to other lysosomal proteins, on the basis of sequence homology searches. Thus, we think that it belongs to a new structural class of proteins capable of transporting the activated acetyl residues across the cell membrane. Moreover, TMEM76 shares homology with a conserved family of bacterial proteins COG4299 (uncharacterized protein conserved in bacteria) (Entrez Gene GeneID 138050). All 146 members of this family are predicted proteins from diverse bacterial species, including Proteobacteria, Cyanobacteria, and Deinococci. Since many of these bacteria are capable of synthesizing heparan sulfate and other structurally related glycosaminoglycans and perform reactions of transmembrane acetylation, it is tempting to speculate that this activity may also be performed by the proteins of the COG4299 family. Previous studies suggested two contradictory mechanisms of transmembrane acetylation. Bame and Rome2,36,37 proposed that it is performed via a ping-pong mechanism. First, the acetyl group of acetyl-CoA is transferred to an His residue in the active site of the enzyme. This induces a conformational change that results in the translocation of the protein domain containing the acetylated residue to the lysosome, where the acetyl residue is transferred to the glucosamine residue of heparan sulfate. In contrast, Meikle et al.38 were unable to demonstrate any specific acetylation of the lysosomal membranes and proposed an alternative mechanism that involved the formation of a tertiary complex of the enzyme, acetyl-CoA, and heparan sufate. Identification of N-acetyltransferase as a 73-kDa protein with multiple transmembrane domains, together with our previous data that showed that N-acetyltransferase is acetylated by [14C]acetyl-CoA in the absence of glucosamine,31 strongly supports the ping-pong mechanism of transmembrane acetylation.
For 23 of the 30 probands included in this study for mutation analysis, TMEM76 mutations were identified in both alleles. Five probands were heterozygous for a missense mutation, with a second mutation yet to be identified. In two probands from North Africa and Portugal, we did not identify any mutations in the coding regions or immediate flanking regions. These patients are homozygous for the microsatellite markers throughout the entire MPS IIIC locus and may be homozygous for a yet-to-be-identified TMEM76 mutation; however, we cannot formally exclude defects in other genes. Additional studies have been initiated to search for mutations in the introns and promoter regions. The patients with MPS IIIC with the identified frameshift and nonsense mutations all have a clinically severe early-onset form. The almost complete deficiency of N-acetyltransferase activity in cultured skin fibroblasts from these patients is consistent with the predicted protein truncations and/or nonsense-mediated mRNA decay. Further expression studies are necessary to confirm the impact of the identified substitutions of the conserved amino acids on enzyme activity. Nevertheless, the identification of the lysosomal N-acetyltransferase gene which, when mutated, accounts for the molecular defect in patients with MPS IIIC sets the stage for DNA-based diagnosis and genotype-phenotype correlation studies and marks the end of the gene-discovery phase for lysosomal genetic enzymopathies.
Acknowledgments
We thank the patients, their families, and the Czech Society for Mucopolysaccharidosis, for participating in our study, and members of the sequencing and genotyping facilities at the McGill University and Genome Quebec Innovation Centre, for their technical support. We also acknowledge Nina Gusina, Joe Clarke, and Tony Rupar, for providing cell lines from patients with MPS IIIC; Mila Ashmarina, Milan Elleder, J. Loredo-Osti, and Johanna Rommens, for helpful discussions; Karine Landry, for technical support; and Maryssa Canuel, for help with confocal microscopy. The Montreal study was supported by operating grants from the Sanfilippo Children’s Research Foundation (to A.V.P.) and by the Canadian Networks of Centres of Excellence Program—the Mathematics of Information Technology and Complex System network (to K.M.). The Prague study was supported by grants NR8069-1 and 1A/8239-3 from the Grant Agency of the Ministry of Health of the Czech Republic. Institutional support was provided by Ministry of Education of the Czech Republic grant MSM0021620806. A.V.P. is a National Investigator of the Fonds de la Recherche en Santé du Québec.
Appendix A
Table A1. .
Exon-Flanking Primers Used for PCR Amplification and Sequencing of the Exons in the Human TMEM76 Gene[Note]
| Primer | Sequence (5′→3′) |
| TMEM76_Exon1_F | CTCCCGAAGACAAACACTCC |
| TMEM76_Exon1_R | GCGAAGTCGCAGCAACAGC |
| TMEM76_Exon2_F | AAGCTTTTGAGAAGCACTACTGG |
| TMEM76_Exon2_R | GAAGGGCTTTAGACATGAGAGC |
| TMEM76_Exon3_F | GGAAAAGTCATGTCAGGATCTCC |
| TMEM76_Exon3_R | GAATAATACATGTTCCTGGGTACG |
| TMEM76_Exon4_F | TTATTCTGCCTCCATGATATTAGC |
| TMEM76_Exon4_R | CTACAGAAAGCGTCATGGACTGC |
| TMEM76_Exon5_F | GGAAATTCAGCATGAGAATATAGG |
| TMEM76_Exon5_R | GCCACTTGAGGGTGACAGC |
| TMEM76_Exon6_F | GAATATGAGCTTTAATTTTATTTCC |
| TMEM76_Exon6_R | TTAGGAATACGGGAGCTACAACC |
| TMEM76_Exon7_F | CAAAATGAAATTTACCCCTTAGC |
| TMEM76_Exon7_R | ACATCCAAGAAATCCTTCCTAGC |
| TMEM76_Exon8_F | CCTTCCTTTTCACATAGCAAACC |
| TMEM76_Exon8_R | GCTCTGTGAAGGACGTATATAAGC |
| TMEM76_Exon9_F | CCCCTGGGTTTACTTTCTATACC |
| TMEM76_Exon9_R | CCAGCATCATCTGAAAAACAGG |
| TMEM76_Exon10_F | GGGGCTATATTCTGAACTCTTCC |
| TMEM76_Exon10_R | ACCTGAGATGGAGGAATTGC |
| TMEM76_Exon11_F | CTGGGATGAGAGGAGAAGTCC |
| TMEM76_Exon11_R | ACTTGAAGCCAGGAGTGAGG |
| TMEM76_Exon12_F | CCTTCTATTTGCATTTAGTTCACC |
| TMEM76_Exon12_R | GAGAATTCCTCTGACTCGAGACC |
| TMEM76_Exon13_F | TTTTATTCTTGTCCCTCTGTTCG |
| TMEM76_Exon13_R | CACTTCTGAAAGCCTGAGTTCC |
| TMEM76_Exon14_F | TTGGTCTAGGAGCTGTTTGTACG |
| TMEM76_Exon14_R | CCATAGCACAAGAGAGAATATGC |
| TMEM76_Exon15_F | TCTTTGTCAGGTAGTTAAGACAGTGG |
| TMEM76_Exon15_R | GTGAAGGAAAGGAATTTTAGC |
| TMEM76_Exon16_F | ACAAGTTTCAGCCCTCTCTACG |
| TMEM76_Exon16_R | GTGGAGGAGACGTTTCAGTGC |
| TMEM76_Exon17_F | ATGCTGAAATTGGATTTGTTCC |
| TMEM76_Exon17_R | ACCAAGGATGCTCCAGAGG |
| TMEM76_Exon18_F | AGTAGCCAACAATGGAAGTGC |
| TMEM76_Exon18_R | GAGCCGTGTCACAGTTAACC |
Note.— For bidirectional sequencing on the ALFexpress DNA sequencer, all primers have the universal overhang synthesized on the 5′ end (AATACGACTCACTATAG for forward [F] primers and CAGGAAACAGCTATGAC for reverse [R] primers).
Footnotes
Footnote added in proof: the gene name has been changed to HGSNAT.
Web Resources
Accession numbers and URLs for data presented herein are as follows:
- BLAST, http://www.ncbi.nlm.nih.gov/blast/ (used to identify ortholog protein sequences)
- Entrez Gene, http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=gene (for GeneID 138050)
- GenBank, http://www.ncbi.nih.gov/Genbank/ (for accession numbers AK152926.1, AK149883.1, DR000652.1, XM_372038.4, NT_007995.14, XP_539948.2, XP_588978.2, XP_341451.2, and XP_519741.1)
- Human Genome Variation Society, http://www.hgvs.org/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for MPS IIIA, IIIB, IIIC, and IID)
- The R Project for Statistical Computing, http://www.r-project.org/
References
- 1.Rome LH, Hill DF, Bame KJ, Crain LR (1983) Utilization of exogenously added acetyl coenzyme A by intact isolated lysosomes. J Biol Chem 258:3006–3011 [PubMed] [Google Scholar]
- 2.Bame KJ, Rome LH (1985) Acetyl-coenzyme A:α-glucosaminide N-acetyltransferase: evidence for a transmembrane acetylation mechanism. J Biol Chem 260:11293–11299 [PubMed] [Google Scholar]
- 3.Pohlmann R, Klein U, Fromme HG, von Figura K (1981) Localisation of acetyl-CoA: α-glucosaminide N-acetyltransferase in microsomes and lysosomes of rat liver. Hoppe Seylers Z Physiol Chem 362:1199–1207 [DOI] [PubMed] [Google Scholar]
- 4.Hopwood JJ, Freeman C, Clements PR, Stein R, Miller AL (1983) Cellular location of N-acetyltransferase activities toward glucosamine and glucosamine-6-phosphate in cultured human skin fibroblasts. Biochem Int 6:823–830 [PubMed] [Google Scholar]
- 5.Meikle PJ, Whittle AM, Hopwood JJ (1995) Human acetyl-coenzyme A:α-glucosaminide N-acetyltransferase: kinetic characterization and mechanistic interpretation. Biochem J 308:327–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kresse H, von Figura K, Bartsocas C (1976) Clinical and biochemical findings in a family with Sanfilippo disease, type C. Clin Genet 10:364 [Google Scholar]
- 7.Bartsocas C, Grobe H, van de Kamp JJ, von Figura K, Kresse H, Klein U, Giesberts MA (1979) Sanfilippo type C disease: clinical findings in four patients with a new variant of mucopolysaccharidosis III. Eur J Pediatr 130:251–258 10.1007/BF00441361 [DOI] [PubMed] [Google Scholar]
- 8.Klein U, Kresse H, von Figura K (1978) Sanfilippo syndrome type C: deficiency of acetyl-CoA:α-glucosaminide N-acetyltransferase in skin fibroblasts. Proc Natl Acad Sci USA 75:5185–5189 10.1073/pnas.75.10.5185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sanfilippo SJ, Podosin R, Langer LO Jr, Good RA (1963) Mental retardation associated with acid mucopolysacchariduria (heparitin sulfate type). J Pediatr 63:837–838 10.1016/S0022-3476(63)80279-6 [DOI] [Google Scholar]
- 10.Klein U, van de Kamp JJP, von Figura K, Pohlmann R (1981) Sanfilippo syndrome type C: assay for acetyl-CoA:α-glucosaminide N-acetyltransferase in leukocytes for detection of homozygous and heterozygous individuals. Clin Genet 20:55–59 [DOI] [PubMed] [Google Scholar]
- 11.Meikle PJ, Hopwood JJ, Clague AE, Carey WF (1999) Prevalence of lysosomal storage disorders. JAMA 281:249–254 10.1001/jama.281.3.249 [DOI] [PubMed] [Google Scholar]
- 12.Pinto R, Caseiro C, Lemos M, Lopes L, Fontes A, Ribeiro H, Pinto E, Silva E, Rocha S, Marcao A, Ribeiro I, Lacerda L, Ribeiro G, Amaral O, Sa Miranda MC (2004) Prevalence of lysosomal storage diseases in Portugal. Eur J Hum Genet 12:87–92 10.1038/sj.ejhg.5201044 [DOI] [PubMed] [Google Scholar]
- 13.Poorthuis BJ, Wevers RA, Kleijer WJ, Groener JE, de Jong JG, van Weely S, Niezen-Koning KE, van Diggelen OP (1999) The frequency of lysosomal storage diseases in The Netherlands. Hum Genet 105:151–156 [DOI] [PubMed] [Google Scholar]
- 14.Zaremba J, Kleijer WJ, Juijmans JG, Poorthuis B, Fidzianska E, Glogowska I (1992) Chromosomes 14 and 21 as possible candidates for mapping the gene for Sanfilippo disease type IIIC. J Med Genet 29:514 [PMC free article] [PubMed] [Google Scholar]
- 15.Ausseil J, Loredo-Osti JC, Verner A, Darmond-Zwaig C, Maire I, Poorthuis B, van Diggelen OP, Hudson TJ, Fujiwara TM, Morgan K, Pshezhetsky AV (2004) Localization of a gene for mucopolysaccharidosis IIIC to chromosome region 8p11-8q11. J Med Genet 41:941–945 10.1136/jmg.2004.021501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seyrantepe V, Tihy F, Pshezhetsky AV (2006) The microcell-mediated transfer of human chromosome 8 restores the deficient N-acetylytransferase activity in skin fibroblasts of mucopolysaccharidosis type IIIC patients. Hum Genet 120:293–296 10.1007/s00439-006-0211-4 [DOI] [PubMed] [Google Scholar]
- 17.Kong X, Murphy K, Raj T, He C, White PS, Matise TC (2004) A combined linkage-physical map of the human genome. Am J Hum Genet 75:1143–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mira MT, Alcais A, Nguyen VT, Moraes MO, Di Flumeri C, Vu HT, Mai CP, Nguyen TH, Nguyen NB, Pham XK, Sarno EN, Alter A, Montpetit A, Moraes ME, Moraes JR, Dore C, Gallant CJ, Lepage P, Verner A, Van De Vosse E, Hudson TJ, Abel L, Schurr E (2004) Susceptibility to leprosy is associated with PARK2 and PACRG. Nature 427:636–640 10.1038/nature02326 [DOI] [PubMed] [Google Scholar]
- 19.Sobel E, Lange K (1996) Descent graphs in pedigree analysis: applications to haplotyping, location scores, and marker sharing statistics. Am J Hum Genet 58:1323–1337 [PMC free article] [PubMed] [Google Scholar]
- 20.Sobel E, Papp JC, Lange K (2002) Detection and integration of genotyping errors in statistical genetics. Am J Hum Genet 70:496–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hodanova K, Majewski J, Kublova M, Vyletal P, Kalbacova M, Stiburkova B, Hulkova H, Chagnon YC, Lanouette CM, Marinaki A, Fryns JP, Venkat-Raman G, Kmoch S (2005) Mapping of a new candidate locus for uromodulin-associated kidney disease (UAKD) to chromosome 1q41. Kidney Int 68:1472–1482 10.1111/j.1523-1755.2005.00560.x [DOI] [PubMed] [Google Scholar]
- 22.O’Connell JR, Weeks DE (1998) PedCheck: a program for identifying genotype incompatibilities in linkage analysis. Am J Hum Genet 63:259–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Voznyi YV, Karpova EA, Dudukina TV, Tsvetkova IV, Boer AM, Janse HC, van Diggelen OP (1993). A fluorimetric enzyme assay for the diagnosis of Sanfilippo disease C (MPS III C). J Inher Metab Dis 16:465–472 10.1007/BF00710299 [DOI] [PubMed] [Google Scholar]
- 24.Gudbjartsson DF, Jonasson K, Frigge M, Kong A (2000) Allegro, a new computer program for multipoint linkage analysis. Nat Genet 25:12–13 10.1038/75514 [DOI] [PubMed] [Google Scholar]
- 25.Smyth GK (2005) Limma: linear models for microarray data. In: Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W (eds) Bioinformatics and computational biology solutions using R and Bioconductor. Springer, New York, pp 397–420 [Google Scholar]
- 26.Smyth GK (2004) Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3:article 3 [DOI] [PubMed] [Google Scholar]
- 27.Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 57:289–300 [Google Scholar]
- 28.Kmoch S, Hartmannova H, Stiburkova B, Krijt J, Zikanova M, Sebesta I (2000) Human adenylosuccinate lyase (ADSL), cloning and characterization of full-length cDNA and its isoform, gene structure and molecular basis for ADSL deficiency in six patients. Hum Mol Genet 9:1501–1513 10.1093/hmg/9.10.1501 [DOI] [PubMed] [Google Scholar]
- 29.Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254 10.1016/0003-2697(76)90527-3 [DOI] [PubMed] [Google Scholar]
- 30.Hinrichs AS, Karolchik D, Baertsch R, Barber GP, Bejerano G, Clawson H, Diekhans M, et al (2006) The UCSC Genome Browser Database: update 2006. Nucleic Acids Res 34:D590–D598 10.1093/nar/gkj144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ausseil J, Landry K, Seyrantepe V, Trudel S, Mazur A, Lapointe F, Pshezhetsky AV (2006) An acetylated 120-kDa lysosomal transmembrane protein is absent from mucopolysaccharidosis IIIC fibroblasts: a candidate molecule for MPS IIIC. Mol Genet Metab 87:22–31 10.1016/j.ymgme.2005.09.021 [DOI] [PubMed] [Google Scholar]
- 32.Kahsay RY, Gao G, Liao L (2005) An improved hidden Markov model for transmembrane protein detection and topology prediction and its applications to complete genomes. Bioinformatics 21:1853–1858 10.1093/bioinformatics/bti303 [DOI] [PubMed] [Google Scholar]
- 33.Jensen LJ, Gupta R, Blom N, Devos D, Tamames J, Kesmir C, Nielsen H, Staerfeldt HH, Rapacki K, Workman C, Andersen CA, Knudsen S, Krogh A, Valencia A, Brunak S (2002) Prediction of human protein function from post-translational modifications and localization features. J Mol Biol 319:1257–1265 10.1016/S0022-2836(02)00379-0 [DOI] [PubMed] [Google Scholar]
- 34.Blom N, Sicheritz-Ponten T, Gupta R, Gammeltoft S, Brunak S (2004) Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics 4:1633–1649 10.1002/pmic.200300771 [DOI] [PubMed] [Google Scholar]
- 35.Bonifacino JS, Traub LM (2003) Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu Rev Biochem 72:395–447 10.1146/annurev.biochem.72.121801.161800 [DOI] [PubMed] [Google Scholar]
- 36.Bame KJ, Rome LH (1986a) Acetyl-coenzyme A:α-glucosaminide N-acetyltransferase: evidence for an active site histidine residue. J Biol Chem 261:10127–10132 [PubMed] [Google Scholar]
- 37.Bame KJ, Rome LH (1986b) Genetic evidence for transmembrane acetylation by lysosomes. Science 233:1087–1089 [DOI] [PubMed] [Google Scholar]
- 38.Meikle PJ, Whittle AM, Hopwood JJ (1995) Human acetyl-coenzyme A:α-glucosaminide N-acetyltransferase: kinetic characterization and mechanistic interpretation. Biochem J 308:327–333 [DOI] [PMC free article] [PubMed] [Google Scholar]