Abstract
Mutations in FOXP2 cause developmental verbal dyspraxia (DVD), but only a few cases have been described. We characterize 13 patients with DVD—5 with hemizygous paternal deletions spanning the FOXP2 gene, 1 with a translocation interrupting FOXP2, and the remaining 7 with maternal uniparental disomy of chromosome 7 (UPD7), who were also given a diagnosis of Silver-Russell Syndrome (SRS). Of these individuals with DVD, all 12 for whom parental DNA was available showed absence of a paternal copy of FOXP2. Five other individuals with deletions of paternally inherited FOXP2 but with incomplete clinical information or phenotypes too complex to properly assess are also described. Four of the patients with DVD also meet criteria for autism spectrum disorder. Individuals with paternal UPD7 or with partial maternal UPD7 or deletion starting downstream of FOXP2 do not have DVD. Using quantitative real-time polymerase chain reaction, we show the maternally inherited FOXP2 to be comparatively underexpressed. Our results indicate that absence of paternal FOXP2 is the cause of DVD in patients with SRS with maternal UPD7. The data also point to a role for differential parent-of-origin expression of FOXP2 in human speech development.
Forkhead-box P2 (FOXP2) was the first gene discovered to be involved in speech and language disorder, through genetic mapping and mutational analysis in a three-generation pedigree (referred to as the “KE” family) with an autosomal dominant form of the condition.1–3 Characterization of a translocation breakpoint in a patient (called “CS”) with a similar clinical presentation added further support.2 The KE phenotype is described as severe developmental verbal apraxia with impairment in both expressive and receptive language skills,3 categorized as developmental verbal dyspraxia (DVD [MIM 602081]). Disruption of both copies of Foxp2 in mice causes severe motor impairment, premature death, and absence of ultrasonic vocalizations normally elicited when pups are separated from their mothers.4 In the heterozygous-knockout mice, there is modest developmental delay but a significant alteration in ultrasonic vocalizations.
The FOXP2 protein is an evolutionarily conserved transcriptional repressor containing a zinc-finger motif, a forkhead DNA-binding domain, and a polyglutamine tract.5 There are several isoforms of the gene, with the longest characterized transcript encompassing 25 exons spanning >600 kb of DNA (fig. 1)5; the gene is expressed in a wide range of tissues throughout development. On the basis of structural and functional studies in the KE family, FOXP2 has been suggested to be involved in the development of brain regions affecting motor control and neural structures that mediate speech and language.6 The KE family carries a heterozygous point mutation in exon 14 (amino acid 553, resulting in an arginine-to-histidine change) of the FOXP2 consensus. Transmission of mutated FOXP2 is maternal, except in one case of paternal transmission.3 The chromosome 7 translocation breakpoint (parental origin not described) in CS occurs between exons 3b and 4, disrupting all known isoforms of FOXP2. Recently, a screen for FOXP2 mutations in 49 probands with verbal dyspraxia as their primary phenotype detected a maternally inherited nonsense mutation yielding a truncated protein in one family, as well as two putative missense mutations in two other patients.7 These findings suggest that FOXP2 mutations are a relatively rare cause for speech and language impairment.
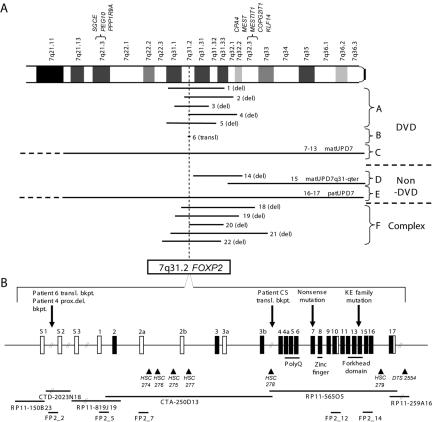
Figure 1. .
A, Summary of the chromosomal aberrations in patients with and without DVD. Patient numbers correspond to descriptions in table 1. The location of FOXP2 is denoted by the dotted line. Two known clusters of imprinted genes on 7q are shown above the chromosome. SGCE, PEG10, MEST, COPG2IT1, and MESTIT1 are paternally expressed, and PPPIR9A and CPA4 are maternally expressed. A new paternally imprinted gene, KLF14, is also now known to reside at 7q32 (L. Parker-Katiraee, personal communication). B, FOXP2 locus at 7q31.2. A consensus transcript encompassing all known exons of the gene is presented. Coding exons are shown in black. The translocation breakpoint (transl. bkpt.) for patient 6 and the proximal deletion breakpoint (prox. del. bkpt.) for patient 4 map between exons s1 and s2. The KE mutation and the CS translocation breakpoint,2 as well as a nonsense mutation in exon 7 that segregates with the DVD phenotype,7 are also shown. Known functional motifs are indicated below the exons coding for each. Mapping reagents, including BAC clones, long-PCR FISH probes, and microsatellite markers, are shown. Markers HSC274–HSC279 are in GenBank, under accession numbers BV123532, BV123533, BV123528, BV123529, BV123530, and BV123531, respectively. Additional probes and mapping reagents can be found at the Chromosome 7 Annotation Project Web site and will be distributed on request.
The distinctive clinical presentation of the KE family8 has served as a reference for us to identify patients with a similar phenotype. In addition, we have been collecting patients with chromosomal anomalies affecting chromosome 7,9 with a specific focus on patients with an affected FOXP2 locus. Thus, patients have been ascertained both via genotype and via phenotype. A specific subgroup of patients displaying DVD (in addition to other phenotypic characteristics) includes patients with maternal uniparental disomy (UPD) of chromosome 7 (matUPD7). We hypothesized that this can be attributed to FOXP2 and that there may be a parent-of-origin effect involved in FOXP2 regulation. The aim of this article is to describe the genetic and phenotypic similarities among patients exhibiting DVD, with FOXP2 as a main focus.
We characterized 22 samples in this study. In this article, we describe 13 individuals with DVD who can be divided into groups on the basis of their genetic characteristics (table 1)—namely, group A, consisting of 5 individuals with 7q31 deletions of FOXP2 on the paternally inherited chromosome; group B, consisting of 1 individual with a 7q31 translocation interrupting FOXP2; and group C, consisting of 7 patients with matUPD7. Table 1 also describes nine individuals without DVD. Group D includes two patients, one with with partial matUPD7 and one with a deletion, with rearrangements starting downstream of FOXP2; group E includes two patients with paternal UPD7 (patUPD7); and group F has five patients with FOXP2 deletions (on the paternally derived chromosome) and a more complex global developmental delay, including speech and language disorder.
Table 1. .
Summary of 22 Individuals with or without DVD
| Presence of Traitc |
||||||||||
| Group and Patient (Alias and Reference)a |
Karyotype (Deletion Size or Parental Source)b |
Late Talking |
Oromotor/ Verbal Dyspraxia |
Articulation Disorder |
Receptive Language Impairment |
Expressive Language Impairment |
Speech Therapy |
Below-Average Nonverbal/ Performance IQ |
Gross- and/or Fine- Motor Delays |
Phenotyped |
| Reference: | ||||||||||
| KE2 | Point mutation | + | + | + | + | + | + | +/− | − | DVD |
| CS2 | t(5;7)(q22q31.2) | + | + | + | + | + | NR | − | + | DVD |
| A: | ||||||||||
| 1 (13772) | 46,XX,del(7)(q31.1q31.3)pat (15 Mb) | + | + | + | + | + | + | + | + | DVD and DD |
| 2 (27162) | 46,XX,del(7)(q31.2q32)pat (13 Mb) | + | + | + | + | + | + | + | + | DVD |
| 3 (24784) | 46,XY,del(7)(q31.1q31.3)pat (11 Mb) | + | + | + | + | + | + | − | + | DVD and ASD |
| 4 (13583)11 | 46,XX,del(7)(q31.2q32.3)pat (15 Mb) | + | + | + | + | + | + | − | + | DVD and ASD-like |
| 5 (33466) | 46,XX,del(7)(q22q31.3)pat (15 Mb) | + | + | + | + | + | + | + | NR | DVD and DD |
| B: | ||||||||||
| 6 (28577) | 46,XX,t(3;7)(q23q31.2) | + | + | + | + | + | + | NR | + | DVD |
| C: | ||||||||||
| 712 | 46,XY,matUPD7 (iso) | + | + | + | − | + | + | − | + | DVD and SRS |
| 812 | 46,XY,matUPD7 (iso/hetero) | + | NR | + | − | − | + | NR | NR | DVD and SRS |
| 912 | 46,XX,matUPD7 (iso/hetero) | + | + | + | − | + | + | NR | − | DVD and SRS |
| 1012 | 46,XX,matUPD7 (iso/hetero) | + | − | + | NR | − | + | NR | + | DVD and SRS |
| 1113 | 46,XX,matUPD7 (iso) | + | NR | + | NR | NR | + | + | − | DVD and SRS |
| 12 (LGL12989) | 46,XX,matUPD7 (iso) | + | NR | + | + | + | + | NR | + | DVD and SRS |
| 13 (27297) | 46,XY,matUPD7 (iso) | + | + | + | + | + | + | + | + | DVD, SRS, and ASD |
| De: | ||||||||||
| 14 (C70001-3) | 46,XY,del(7)(q31.2q32.3)pat (15 Mb) | + | − | − | + | + | + | + | + | DD |
| 1514 | 46,XX,matUPD7q31-qter (iso) | − | − | − | − | − | − | − | − | SRS |
| E: | ||||||||||
| 1615 | 46,XX,patUPD7 (iso) | − | − | − | − | − | − | − | − | CLD |
| 17 (18406) | 46,XY,patUPD7 (unknown) | + | − | − | − | − | + | + | + | DD and CF |
| Ff: | ||||||||||
| 18 (23145) | 46,XY,del(7)(q31.2q32)pat (26 Mb) | NR | NR | NR | NR | NR | NR | + | + | DD, no speech, and ASD |
| 19 (6635) | 46,XY,del(7)(q22q31.33)pat (22 Mb) | NR | NR | NR | NR | NR | NR | NR | NR | NR |
| 20 (10203) | 46,XX,del(7)(q31.2q32.2)pat (14 Mb) | + | + | NR | NR | NR | NR | − | + | DD |
| 21 (11912) | 46,XX,del(7)(q31.1q33)pat (30 Mb) | NR | NR | NR | NR | NR | NR | + | + | DD and no speech |
| 22 (31065) | 46,XY,del(7)(q31.2q32)pat (21 Mb) | NR | NR | NR | NR | NR | NR | + | + | DD and no speech |
Literature and database searches revealed another 16 patients (not shown) whose karyotypes suggest deletion of FOXP2, but no molecular or parent-of-origin data are available. Moreover, most of these patients were described as infants, and a specific language phenotype was therefore not included. Additional details for these samples and for those described in the table can be found at the Chromosome 7 Annotation Project Web site. We note that we do not see any neutral copy-number variants in the FOXP2 region (see the Database of Genomic Variants,10).
For patients with UPD, the source of parental chromosomes—isodisomic (iso), heterodisomic (hetero), or both—is shown, if known.
Plus (+) indicates positive scoring on this trait, and minus (−) indicates negative scoring. NR = not recorded. IQ = intelligence quotient.
The final phenotype, as decided by multidisciplinary assessment, is shown. DD = developmental delay. CF = cystic fibrosis. CLD = congenital chloride diarrhea. Specific phenotypic details for any patients are available on request.
Representative patients with chromosome 7 aberrations, FOXP2 intact, and no DVD.
Patients with paternally inherited FOXP2 deletions but with incomplete clinical information or too complex a phenotype for the proper assessment of DVD.
The patients with matUPD7 have also received diagnoses of Silver-Russell Syndrome (SRS [MIM 180860]), a condition characterized by intrauterine growth retardation, postnatal short stature, and a characteristic, small triangular face. Dysmorphic features such as fifth-finger clinodactyly and skeletal asymmetry are also common in patients with SRS but are not required for diagnosis. Approximately 10% of patients with SRS have matUPD7.13,16,17 Whereas DVD is not a frequent symptom in patients with SRS in general, it is commonly found in the subgroup of patients with matUPD7.12 PatUPD7 has no apparent effect on growth and development, indicating that an imprinted gene on chromosome 7 may be involved in causing SRS.12,18
All individuals described in groups A, B, and C in table 1 had normal hearing. One of us (J.O.C.) had previously studied the prototypic KE family, which allowed direct comparisons of clinical descriptions to be made. Blood samples were collected and lymphoblast cell lines were established from a subset of the patients and their parents. The study was approved by the ethical review board of each hospital involved in this study. All patients and their families gave written consent. Karyotyping was performed on blood from all patients and, for the parents of patients with deletions, by analysis of G-banded metaphase chromosomes from cultured peripheral blood lymphocytes (table 1 and fig. 1). All parental karyotypes were normal, indicating that deletions were de novo. The site and/or extent of the chromosomal rearrangement(s) or UPDs and inheritance patterns were determined using a combination of genotyping, array comparative genomic hybridization, and FISH.
One patient (patient 6) carried a translocation interrupting the FOXP2 gene. To our knowledge, only two patients with a translocation or inversion breakpoint within FOXP2 have been described elsewhere.2,19 The translocation breakpoint in patient 6 is localized to the intron between the first two 5′ UTR exons (s1 and s2) (fig. 1) of the longest known isoform of FOXP2 (not characterized in the original mutation study). Parental samples were not available for analysis.
As summarized above, we observed that the remaining 12 individuals with confirmed DVD (5 with deletions and 7 with matUPD7) for whom inheritance could be established failed to have a paternally inherited FOXP2 gene. In contrast, patient 14, with an interstitial 7q31.2-q32.3 deletion starting just telomeric of FOXP2 (but leaving it intact), and patient 15, with partial matUPD7 (matUPD7q31-qter) (fig. 1)14 and both parental copies of FOXP2, did not exhibit any characteristics of DVD. In group E (patients 16 and 17), absence of maternally inherited alleles in patUPD7 also does not lead to DVD. Patient 16, affected with recessive congenital chloride diarrhea,15 had normal psychomotor and speech development without any articulation difficulties. Patient 17 (a patient with cystic fibrosis) had mild language delay, but this was likely due to general developmental delay, and he did not exhibit verbal dyspraxia. No data from either our current study or the literature yet document a maternal deletion of FOXP2; however, one maternally inherited translocation interrupting FOXP2 has been described.19 These observations suggest a role for differential parental regulation of FOXP2 expression and lead us to reconsider possible mechanisms of FOXP2 activity in the pathophysiology of DVD and in normal development.
Patients with an interstitial deletion encompassing FOXP2 (patients 1–5) (table 1) as well as with the t(3;7)(q23;q31.2) translocation interrupting FOXP2 (patient 6) all have a more severe form of DVD compared with the KE family members. Data on the patients' speech disorders were collected from evaluations by a phoniatric specialist, a speech-language pathologist, or a neuropsychologist. The majority of these patients have some extent of psychomotor delay and global cognitive impairment, but the speech and language deficits are much more severe. All areas of speech and language development are seriously affected; receptive language is less impaired than expressive language, and articulation is the most compromised. For example, at age 8 years, patient 3 had nonverbal cognitive functioning at a 6-year age level, receptive language at a 4.5-year level, expressive language at a 34-mo level, and articulation at a 25-mo level. First words were typically produced at age 3 (mean ± SD 36 ± 14 mo; n=6), whereas most children typically produce their first word (other than “mama” or “dada”) by the end of their 1st year. Two patients (3 and 4) started combining words (which typically begins at ∼19 mo) at ages 4 and 6 years, respectively, with the remainder having failed to attain this expressive language milestone. All patients have greatly limited oral vocabulary size. Because of the severe verbal dyspraxia, speech is restricted to the simplest of sounds and is often unintelligible. In their early years, these patients suffered from difficulties with chewing, gagging, and swallowing, often to the extent that aspiration occurred (table 1). Problems with coughing, throat clearing, and sneezing are observed in most patients, and some are unable to blow their noses. Oromotor problems include difficulties with lip protrusion, tongue elevation and lateralization, and rapid alternating movements.
In addition to the six patients with DVD with deletion or translocation rearrangements described above, we document another five patients (group F) who all carry de novo deletions of the paternally inherited chromosome that includes the FOXP2 region (table 1 and fig. 1). These individuals could not be given formal diagnoses of DVD since (i) we had limited clinical information about them (e.g., patients 19 and 20) or (ii) they suffered from global developmental delay (e.g., patients 18, 21, and 22) complicating phenotypic assessment. The more complex phenotype in patients 18, 21, and 22 may be due to the larger deletions (all >20 Mb in size) they harbor, compared with the smaller deletions (all <15 Mb in size) observed in the five individuals having confirmed DVD (group A) (table 1). Notwithstanding, we do know that all five group F patients have severe dyspraxia and language delay, with three patients (18, 21, and 22) remaining nonverbal at ages 10, 11, and 20, respectively.
In the patients with FOXP2 deletion, there are additional phenotypic complexities, and, although these complexities could be due to the size of the chromosomal lesion (see fig. 1 and table 1), no obvious features are consistently observed. For example, patient 3, who has the smallest deletion (11.3 Mb), meets the full criteria for autism spectrum disorder (ASD [MIM 209850]), on the basis of the Autism Diagnostic Observation Schedule (ADOS) and the Autism Diagnostic Interview-Revised (ADI-R) exams. Patient 18 was also given a diagnosis of ASD on the basis of similar criteria. Patient 4 (with a 15.4-Mb deletion) was initially given a diagnosis of ASD because she exhibited repetitive behaviors, such as hand flapping, and unusual sensory interests indicated by sniffing and squeezing. On reexamination, however, it was determined that she does not meet full ADOS or ADI-R criteria for ASD because of her strong social communication, which included spontaneous use of varied gestures to compensate for her language difficulties.11 There is a history of Williams-Beuren syndrome (WBS [MIM 194050]) in her family, and she carries the WBS-susceptibility inversion20; because of these complexities, we categorized her as “ASD-like.” Patients 1 and 5, who have deletions encompassing the region absent from patient 3, do not exhibit ASD. Importantly, other than DVD, none of the patients with the FOXP2 deletion exhibit any other features commonly observed in patients with SRS.
The patients with matUPD7 and SRS (patients 7–13) all have marked speech delay and difficulties in speech output, especially articulation. It is noteworthy that SRS is clinically and genetically heterogeneous, but mainly only patients with complete matUPD7 (∼10% of patients with SRS) exhibit DVD. The dyspraxia phenotype is very similar to that in individuals with FOXP2 deletions but is somewhat milder. First words were usually spoken at 1.5–2.5 years (mean ± SD 24 ± 5.9 mo; n=4). Whereas receptive language skills range from mildly delayed to average, all suffer from impairment in the expressive domain. Articulation difficulties with oromotor dyspraxia make the speech unclear and difficult to understand. This is combined with a limited vocabulary and problems with word finding. Although significant improvement is made over time with targeted speech therapy, the oldest patient (patient 7, age 24 years) is still slightly inarticulate and continues to have problems finding words. Similar to the patients with deletion of FOXP2, the patients with matUPD7 suffered from difficulties with chewing, swallowing, and feeding in the early years. Problems with laughing are observed in most patients, and some have difficulty coughing, sneezing, clearing their throats, and blowing their noses. Although all aspects of oromotor functioning are similar between patients with deletions and patients with matUPD7 in the first year of life, the phenotype in the patients with matUPD7 is milder, and its progression fares better. We note that patient 13 was first referred to the study on the basis of a DVD and ASD diagnosis; the SRS phenotype was assessed only after genotyping revealed matUPD7.
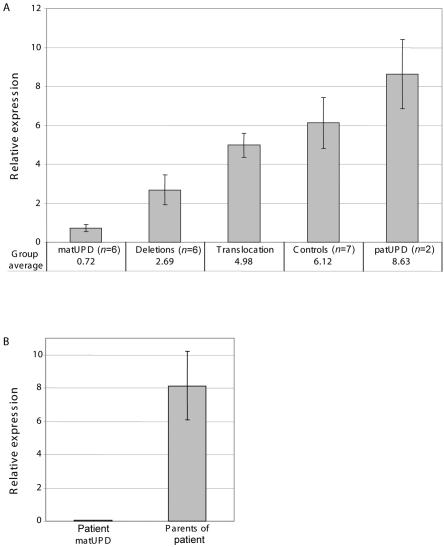
To assess how expression of FOXP2 was affected in the different patient groups, we performed quantitative real-time PCR (qPCR), on RNA obtained from lymphoblast cell lines, for a subset of patients (fig. 2). For the deletion group, we included patients 1–4, 18, and 21. The PCR was designed to amplify the forkhead-domain region of FOXP2 (exons 13–14), and GAPDH was used as internal control. Each sample was run in triplicate. The lowest expression was found in the patients with matUPD7, followed by the patients with deletions, the patient with the translocation, the controls, and the patients with patUPD7. The difference in expression among the groups was significant, as assessed by one-way ANOVA (P=.0003), and the post hoc test showed significant difference between patients with matUPD7 and controls and between patients with deletions and controls. There was no significant difference between patients with matUPD7 and those with the deletion. The patients with the translocation and patUPD7 were not included in the statistical analysis because of the small number of samples available in each group (one and two patients, respectively). Parental samples of patient 8 with matUPD7 were also tested, to compare expression levels between a child and the parents (fig. 2B). The expression levels of the parental samples (8.15 [SEM 2.06]) corresponded with the other controls and were, thus, significantly higher than those of their child (0.05 [SEM 0.01]; P=.029). These results suggest that the multitranscript FOXP2 locus is differentially regulated in a parent-of-origin–specific manner. The lack of coding variants in FOXP2 makes it difficult to study parent-of-origin–specific expression in individuals who are diploid for the locus.
Figure 2. .
Results of qPCR used to estimate FOXP2 expression in cell lines from patients with deletions, a translocation, matUPD7, and patUPD7 and from controls. GAPDH was used as reference. Analysis was performed with a subset of samples for which RNA was available. Taqman qPCR of FOXP2 exons 13–14 was run on (A) six patients with deletions (patients 1–4 and the two nonverbal patients, 18 and 21), one patient with a translocation (patient 6), six patients with matUPD7 (patients 7–12), two patients with patUPD7 (patients 16 and 17), seven control samples, and (B) patient 8 with matUPD7 and his parents. There was a significant difference between the groups (ANOVA P=.0003). There was a statistically significant difference between the patients with matUPD7 and controls and between patients with deletions and controls (ANOVA and Dunnett's post hoc test). The expression for patient 8 was also significantly lower than in his parents (two-tailed t test P=.03). Statistical analysis was not performed for the patients with the translocation and patUPD7 because of small sample size. All samples were run in triplicate, and the experiment was repeated twice, with consistent results. Bars indicate SEM. For qPCR, 2 μg of total RNA was reversely transcribed in a 100-μl reaction with random hexamers, with the use of Taqman Reverse Transcription Reagents kit, and real-time PCR was performed using an ABI7700 sequence-detection system. PCR was performed in 25-μl reactions with 45 cycles of amplification. Each plate contained a water control, a calibrator sample (control 1 cDNA), and serially diluted concentrations of control 3 cDNA (range 62.5–0.5 ng), from which a standard curve was generated for transcript quantification.
On the basis of our data, it is possible, for the first time, to attribute the speech and language phenotype of the patients with matUPD7—observed primarily in the patients with SRS (with matUPD7)—to absence of paternal FOXP2. This finding, coupled with the observation that all other non-DVD features of SRS are present in patient 15, who has partial matUPD7q31-qter, indicates that two or more genes likely contribute to the chromosome 7 form of this disorder. Moreover, none of the patients with the FOXP2 deletion exhibited typical SRS features. In our interpretation, FOXP2 accounts for the DVD, and one or more other genes mapping to the 7q32-qter cause the intrauterine and postnatal growth retardation symptoms. Whereas there is a cluster of imprinted genes localized to 7q32 (fig. 1A), the causative SRS gene(s) on chromosome 7 remains to be identified.
Interestingly, four patients with FOXP2 deletion (3, 4, 13, and 18) were also given diagnoses of ASD, providing a clinically homogeneous group distinguishable from patients with other forms of ASD. Since speech delay is a component of ASD, it has been speculated that FOXP2 itself may be implicated in autism.21,22 The same general 7q31 region has been shown, by linkage analysis of sib-pair families23,24 and chromosome rearrangement studies,25,26 to harbor a putative autism susceptibility locus. There are also reports describing an increased transmission of paternal alleles for markers associated with autism, suggesting the presence of a maternally imprinted gene at 7q31.23,27 Our results, however, do not provide any direct support that FOXP2 is etiologic in idiopathic ASD, which is consistent with other findings.21,22,28 Instead, it is possible that those rare patients with ASD and demonstrating DVD will have genetic lesions affecting FOXP2 in combination with variants in other susceptibility genes, some of which might also reside on chromosome 7.
In any scenario, our data emphasize the need to examine the entire >600 kb FOXP2 locus, its many transcripts including untranslated exons, and putative surrounding regulatory regions, when the gene or gene expression is being tested for involvement in disease. Since some isoforms of the gene (e.g., FOXP2-S5) may not include the forkhead domain (and may not be transcription factors) and/or potentially other motifs, accurate genotype and phenotype interpretations will require substantially greater sample size. This is further highlighted by the patient with a translocation breakpoint between exons s1 and s2. One or both of these untranslated exons have not been included in most mutation screens of FOXP2 in cohorts with DVD and autism.7,21,22
There are different models that can reconcile our current observations with data published elsewhere. We hypothesize that, just as has been shown with many of the FOX genes and proteins, proper dosage of FOXP2 at precise times and locations (at the cellular and tissue levels) is required for normal development.29 On the basis of our data, the majority of FOXP2 transcripts are of paternal origin, so individuals with chromosome deletions inherited from the father or with matUPD7 or other functionally equivalent mutations would display a more severe phenotype. There is no phenotypic information available that allows comparison of patients carrying mutations of maternal and paternal origin, respectively, in the KE family. Clearly, mutations in the maternal copy of FOXP2 is sufficient to cause a DVD phenotype, as reported in both the KE family3 and a multiplex family in which both the mother and her two children carry a nonsense mutation in exon 7, which segregates with the DVD phenotype.7 It is possible that these mutations could express themselves at the level of the protein in a dominant-negative manner consistent with FOXP2 requiring dimerization for binding30 and for other functions attributed to different isoforms of the gene. However, our interpretation is that the difference between maternal and paternal mutations/deletions will be manifested at the level of phenotypic severity.
Variability in expression levels between maternal and paternal chromosomes could reflect either imprinting of a chromosome 7 gene involved in regulation of FOXP2 transcription or imprinting affecting one or more isoforms of FOXP2. The difference in severity of DVD seen among the different types of aberrations—with an increase in severity for UPD, mutation, and deletion—is similar to the findings for the imprinted Angelman Syndrome/Prader-Willi Syndrome region on chromosome 15.31 However, no direct evidence of imprinting of FOXP2 itself has yet been found. The lack of coding variants in FOXP2 makes it difficult to study parent-of-origin–specific expression in individuals who are diploid for the locus. We have, therefore, searched for tissue-specific imprinting in mouse but have not yet observed it (unpublished data). An alternative explanation of our data is that (i) the preponderance of paternal deletions merely reflects a higher mutation rate occurring during spermatogenesis compared with oogenesis32 and/or that (ii) deletion of the maternally inherited allele is selected against during development. The description of the two patients with patUPD7 in our study and of one other in the literature33 argues against the latter scenario. It could also be possible that there is a maternally imprinted gene near FOXP2 within the minimal interval defined by patients 4 and 6 (defining a centromeric boundary) and patient 14 (defining a telomeric boundary) (fig. 1), but, so far, no such transcript has been found.
In summary, our findings suggest that DVD presenting either in isolation, as part of a syndrome such as SRS, or in a more complex setting like ASD can be attributable to lesions or imbalances affecting the FOXP2 locus. Whereas substantial attention has been focused on the nature of conservation of the protein through evolution and on human-specific variants,34 differential allelic expression of FOXP2 and, possibly, maternal imprinting may be equally important in human development and disease.
Acknowledgments
We are grateful to the families who participated in this study. This study was supported by the Academy of Finland, the Sigrid Juselius Foundation (to J.K.), the Swedish Medical Research Council (to J.K. and L.F.), and the Genome Canada/Ontario Genomics Institute and the Hospital for Sick Children Foundation (to S.W.S.). S.W.S. is an investigator of the Canadian Institutes of Health Research and an International Scholar of the Howard Hughes Medical Institute.
Web Resources
Accession numbers and URLs for data presented herein are as follows:
- Chromosome 7 Annotation Project, http://www.chr7.org/ (for clinical tables, see http://www.chr7.org/clinical.php)
- Database of Genomic Variants, http://projects.tcag.ca/variation/
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for markers HSC274–HSC279 [accession numbers BV123532, BV123533, BV123528, BV123529, BV123530, and BV123531, respectively])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for DVD, SRS, ADS, and WBS)
References
- 1.Lai CS, Fisher SE, Hurst JA, Levy ER, Hodgson S, Fox M, Jeremiah S, Povey S, Jamison DC, Green ED, Vargha-Khadem F, Monaco AP (2000) The SPCH1 region on human 7q31: genomic characterization of the critical interval and localization of translocations associated with speech and language disorder. Am J Hum Genet 67:357–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lai CS, Fisher SE, Hurst JA, Vargha-Khadem F, Monaco AP (2001) A forkhead-domain gene is mutated in a severe speech and language disorder. Nature 413:519–523 10.1038/35097076 [DOI] [PubMed] [Google Scholar]
- 3.Hurst JA, Baraitser M, Auger E, Graham F, Norell S (1990) An extended family with a dominantly inherited speech disorder. Dev Med Child Neurol 32:352–355 [DOI] [PubMed] [Google Scholar]
- 4.Shu W, Cho JY, Jiang Y, Zhang M, Weisz D, Elder GA, Schmeidler J, De Gasperi R, Sosa MA, Rabidou D, Santucci AC, Perl D, Morrisey E, Buxbaum JD (2005) Altered ultrasonic vocalization in mice with a disruption in the Foxp2 gene. Proc Natl Acad Sci USA 102:9643–9648 10.1073/pnas.0503739102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bruce HA, Margolis RL (2002) FOXP2: novel exons, splice variants, and CAG repeat length stability. Hum Genet 111:136–144 10.1007/s00439-002-0768-5 [DOI] [PubMed] [Google Scholar]
- 6.Lai CS, Gerrelli D, Monaco AP, Fisher SE, Copp AJ (2003) FOXP2 expression during brain development coincides with adult sites of pathology in a severe speech and language disorder. Brain 126:2455–2462 10.1093/brain/awg247 [DOI] [PubMed] [Google Scholar]
- 7.MacDermot KD, Bonora E, Sykes N, Coupe AM, Lai CS, Vernes SC, Vargha-Khadem F, McKenzie F, Smith RL, Monaco AP, Fisher SE (2005) Identification of FOXP2 truncation as a novel cause of developmental speech and language deficits. Am J Hum Genet 76:1074–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vargha-Khadem F, Gadian DG, Copp A, Mishkin M (2005) FOXP2 and the neuroanatomy of speech and language. Nat Rev Neurosci 6:131–138 10.1038/nrn1605 [DOI] [PubMed] [Google Scholar]
- 9.Scherer SW, Cheung J, MacDonald JR, Osborne LR, Nakabayashi K, Herbrick JA, Carson AR, et al (2003) Human chromosome 7: DNA sequence and biology. Science 300:767–772 10.1126/science.1083423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, Scherer SW, Lee C (2004) Detection of large-scale variation in the human genome. Nat Genet 36:949–951 10.1038/ng1416 [DOI] [PubMed] [Google Scholar]
- 11.Zeesman S, Nowaczyk MJ, Teshima I, Roberts W, Cardy JO, Brian J, Senman L, Feuk L, Osborne LR, Scherer SW (2006) Speech and language impairment and oromotor dyspraxia due to deletion of 7q31 that involves FOXP2. Am J Med Genet A 140:509–514 [DOI] [PubMed] [Google Scholar]
- 12.Hannula K, Kere J, Pirinen S, Holmberg C, Lipsanen-Nyman M (2001) Do patients with maternal uniparental disomy for chromosome 7 have a distinct mild Silver-Russell phenotype? J Med Genet 38:273–278 10.1136/jmg.38.4.273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hannula K, Lipsanen-Nyman M, Kristo P, Kaitila I, Simola KO, Lenko HL, Tapanainen P, Holmberg C, Kere J (2002) Genetic screening for maternal uniparental disomy of chromosome 7 in prenatal and postnatal growth retardation of unknown cause. Pediatrics 109:441–448 10.1542/peds.109.3.441 [DOI] [PubMed] [Google Scholar]
- 14.Hannula K, Lipsanen-Nyman M, Kontiokari T, Kere J (2001) A narrow segment of maternal uniparental disomy of chromosome 7q31-qter in Silver-Russell syndrome delimits a candidate gene region. Am J Hum Genet 68:247–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoglund P, Holmberg C, de la Chapelle A, Kere J (1994) Paternal isodisomy for chromosome 7 is compatible with normal growth and development in a patient with congenital chloride diarrhea. Am J Hum Genet 55:747–752 [PMC free article] [PubMed] [Google Scholar]
- 16.Kotzot D, Schmitt S, Bernasconi F, Robinson WP, Lurie IW, Ilyina H, Mehes K, Hamel BC, Otten BJ, Hergersberg M, Werder E, Schoenle E, Schinzel A (1995) Uniparental disomy 7 in Silver-Russell syndrome and primordial growth retardation. Hum Mol Genet 4:583–587 [DOI] [PubMed] [Google Scholar]
- 17.Nakabayashi K, Fernandez BA, Teshima I, Shuman C, Proud VK, Curry CJ, Chitayat D, Grebe T, Ming J, Oshimura M, Meguro M, Mitsuya K, Deb-Rinker P, Herbrick JA, Weksberg R, Scherer SW (2002) Molecular genetic studies of human chromosome 7 in Russell-Silver syndrome. Genomics 79:186–196 10.1006/geno.2002.6695 [DOI] [PubMed] [Google Scholar]
- 18.Hitchins MP, Stanier P, Preece MA, Moore GE (2001) Silver-Russell syndrome: a dissection of the genetic aetiology and candidate chromosomal regions. J Med Genet 38:810–819 10.1136/jmg.38.12.810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shriberg LD, Ballard KJ, Tomblin JB, Duffy JR, Odell KH, Williams CA (2006) Speech, prosody, and voice characteristics of a mother and daughter with a 7;13 translocation affecting FOXP2. J Speech Lang Hear Res 49:500–525 [DOI] [PubMed] [Google Scholar]
- 20.Osborne LR, Li M, Pober B, Chitayat D, Bodurtha J, Mandel A, Costa T, Grebe T, Cox S, Tsui LC, Scherer SW (2001) A 1.5 million-base pair inversion polymorphism in families with Williams-Beuren syndrome. Nat Genet 29:321–325 10.1038/ng753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Newbury DF, Bonora E, Lamb JA, Fisher SE, Lai CS, Baird G, Jannoun L, Slonims V, Stott CM, Merricks MJ, Bolton PF, Bailey AJ, Monaco AP (2002) FOXP2 is not a major susceptibility gene for autism or specific language impairment. Am J Hum Genet 70:1318–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O’Brien EK, Zhang X, Nishimura C, Tomblin JB, Murray JC (2003) Association of specific language impairment (SLI) to the region of 7q31. Am J Hum Genet 72:1536–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ashley-Koch A, Wolpert CM, Menold MM, Zaeem L, Basu S, Donnelly SL, Ravan SA, Powell CM, Qumsiyeh MB, Aylsworth AS, Vance JM, Gilbert JR, Wright HH, Abramson RK, DeLong GR, Cuccaro ML, Pericak-Vance MA (1999) Genetic studies of autistic disorder and chromosome 7. Genomics 61:227–236 10.1006/geno.1999.5968 [DOI] [PubMed] [Google Scholar]
- 24.International Molecular Genetic Study of Autism Consortium (IMGSAC) (2001) A genomewide screen for autism: strong evidence for linkage to chromosomes 2q, 7q, and 16p. Am J Hum Genet 69:570–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Warburton P, Baird G, Chen W, Morris K, Jacobs BW, Hodgson S, Docherty Z (2000) Support for linkage of autism and specific language impairment to 7q3 from two chromosome rearrangements involving band 7q31. Am J Med Genet 96:228–234 [DOI] [PubMed] [Google Scholar]
- 26.Xu J, Zwaigenbaum L, Szatmari P, Scherer SW (2004) Molecular cytogenetics of autism. Curr Genomics 5:347–364 10.2174/1389202043349246 [DOI] [Google Scholar]
- 27.Lamb JA, Barnby G, Bonora E, Sykes N, Bacchelli E, Blasi F, Maestrini E, Broxholme J, Tzenova J, Weeks D, Bailey AJ, Monaco AP (2005) Analysis of IMGSAC autism susceptibility loci: evidence for sex limited and parent of origin specific effects. J Med Genet 42:132–137 10.1136/jmg.2004.025668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wassink TH, Piven J, Vieland VJ, Pietila J, Goedken RJ, Folstein SE, Sheffield VC (2002) Evaluation of FOXP2 as an autism susceptibility gene. Am J Med Genet 114:566–569 10.1002/ajmg.10415 [DOI] [PubMed] [Google Scholar]
- 29.Lehmann OJ, Sowden JC, Carlsson P, Jordan T, Bhattacharya SS (2003) Fox’s in development and disease. Trends Genet 19:339–344 10.1016/S0168-9525(03)00111-2 [DOI] [PubMed] [Google Scholar]
- 30.Li S, Weidenfeld J, Morrisey EE (2004) Transcriptional and DNA binding activity of the Foxp1/2/4 family is modulated by heterotypic and homotypic protein interactions. Mol Cell Biol 24:809–822 10.1128/MCB.24.2.809-822.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Varela MC, Kok F, Otto PA, Koiffmann CP (2004) Phenotypic variability in Angelman syndrome: comparison among different deletion classes and between deletion and UPD subjects. Eur J Hum Genet 12:987–992 10.1038/sj.ejhg.5201264 [DOI] [PubMed] [Google Scholar]
- 32.Chamberlin J, Magenis RE (1980) Parental origin of de novo chromosome rearrangements. Hum Genet 53:343–347 [DOI] [PubMed] [Google Scholar]
- 33.Pan Y, McCaskill CD, Thompson KH, Hicks J, Casey B, Shaffer LG, Craigen WJ (1998) Paternal isodisomy of chromosome 7 associated with complete situs inversus and immotile cilia. Am J Hum Genet 62:1551–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Enard W, Przeworski M, Fisher SE, Lai CS, Wiebe V, Kitano T, Monaco AP, Paabo S (2002) Molecular evolution of FOXP2, a gene involved in speech and language. Nature 418:869–872 10.1038/nature01025 [DOI] [PubMed] [Google Scholar]