Abstract
Claudins are major components of tight junctions and contribute to the epithelial-barrier function by restricting free diffusion of solutes through the paracellular pathway. We have mapped a new locus for recessive renal magnesium loss on chromosome 1p34.2 and have identified mutations in CLDN19, a member of the claudin multigene family, in patients affected by hypomagnesemia, renal failure, and severe ocular abnormalities. CLDN19 encodes the tight-junction protein claudin-19, and we demonstrate high expression of CLDN19 in renal tubules and the retina. The identified mutations interfere severely with either cell-membrane trafficking or the assembly of the claudin-19 protein. The identification of CLDN19 mutations in patients with chronic renal failure and severe visual impairment supports the fundamental role of claudin-19 for normal renal tubular function and undisturbed organization and development of the retina.
Tight junctions constitute cell-cell contacts by forming circumferential belts around epithelial or endothelial cells in many organs. These dynamic microstructures are involved in several major cellular functions1: first, they construct a “fence” that generates cell polarity by separating apical and basolateral cell domains; second, they form trafficking and signaling platforms involved in the regulation of growth, proliferation, and differentiation of cells; third, they serve as a regulatory barrier (“gate”) that separates fluid compartments of different compositions (e.g. in the kidney, intestine, and brain). The gate properties of tight junctions are derived primarily from claudin proteins that form porelike structures that participate in the regulation of paracellular movements of ions such as magnesium. Claudins are thought to form heterodimers or -oligomers, and barrier properties of tight junctions seem to depend on the expression profiles of participating claudins.2,3 In the kidney, claudins confer ion selectivity to the paracellular pathway that, in particular, is important for ion reabsorption in tubular epithelia.4,5
Mutations in CLDN16 (which encodes claudin-16) have been identified in familial hypomagnesemia with hypercalciuria and nephrocalcinosis (FHHNC [MIM *248250]), a recessive renal disorder of paracellular magnesium and calcium reabsorption in the thick ascending limb of Henle’s loop (TAL).6–8 In rare cases, CLDN16 mutations lead to transient hypercalciuria with nephrocalcinosis without significant disturbance of magnesium reabsorption.9
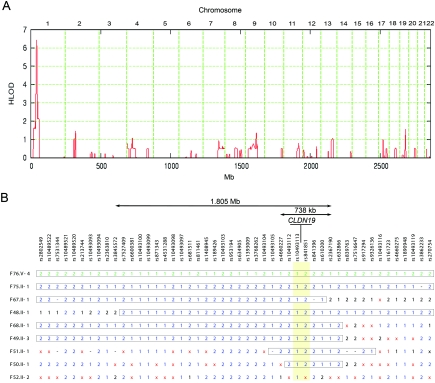
We have clinically characterized one Swiss and eight Spanish/Hispanic families affected with severe hypomagnesemia due to renal wasting, nephrocalcinosis, and progressive renal failure but did not have any mutations in CLDN16 (table 1). Detailed clinical data about families F48–F51 and F76 were reported elsewhere.10,11 The renal phenotype was virtually undistinguishable from that of patients with FHHNC with proven CLDN16 mutations. However, the affected individuals in these families also have severe visual impairment, characterized by macular colobomata, significant myopia, and horizontal nystagmus. With the assumption of the involvement of a second locus in the non-CLDN16 hypomagnesemic patients, we performed a genome scan for shared homozygous regions in these families of known and suspected consanguinity. Informed consent was obtained from all participating individuals. We genotyped five members of the Swiss family and all Spanish/Hispanic patients, using a 50,000-SNP array (Affymetrix GeneChip Human Mapping 50K Array Xba240). Genomewide linkage analysis was performed, with the assumption of heterogeneity and second-degree cousin marriages for the parents of the Spanish/Hispanic families. A critical region on chromosome 1p34.2, with a highly significant heterogeneity LOD score (HLOD) of 6.4, was identified using Genehunter-Modscore12,13 (fig. 1A). On the basis of the haplotype data from patients with extended homozygosity, this interval is delimited by SNP markers rs3845572 and rs2367190 and comprises ∼2 Mb, with CLDN19 as the most promising positional candidate (fig. 1B) (Ensembl). CLDN19 was still in the critical interval of ∼740 kb, even when a less precise definition was used and additional patients with shorter homozygous segments in that region were considered. Only one Spanish/Hispanic patient (F52) did not show a homozygous segment in this interval.
Table 1. .
Phenotypic Characteristics of the Patient Cohort
| Findings for Patient |
||||||||||||
| Characteristic | F76.V-4a | F76.V-1 | F75.II-1 | F48.II-1 | F68.II-1 | F49.II-3 | F51-II-1 | F50.II-1 | F67.II-1 | F52.II-2 | F90.IV-2 | F90.IV-3 |
| Ethnic Origin | Swiss | Swiss | Spanish | Spanish | Hispanic | Spanish | Spanish | Spanish | Hispanic | Spanish | Turkish | Turkish |
| Sex | M | F | M | M | F | F | M | M | M | M | F | M |
| Age (in years) at symptoms onset | 0.8 | 6.0 | 3.0 | 3.6 | 0.5 | 7.7 | 0.2 | 0.8 | 0.1 | 0.8 | 0.9 | 0.2 |
| Present age (in years) | 39 | 36 | 49 | 26 | 3.0 | 18 | 10 | 21 | 14 | 10 | 2.5 | 1.0 |
| Renal phenotypeb: | ||||||||||||
| Nephrolithiasis | Yes | Yes | Yes | No | No | No | No | No | No | Yes | No | Yes |
| Urinary tract infections | Yes | Yes | No | No | Yes | Yes | Yes | No | No | Yes | Yes | Yes |
| Actual renal functionc | TPL | TPL | CRF | CRF | Normal | CRF | CRF | CRF | TPL | TPL | Normal | Normal |
| Ocular phenotype: | ||||||||||||
| Macular colobomata | Yes | Yes | No | Yes | No | No | No | Yes | Unknown | No | Yes | Unknown |
| Nystagmus | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Unknown | Yes | Yes | Yes |
| Myopia | Yes | Yes | Yes | Yes | Yes | Yes | Nod | Yes | Yes | No | Yes | Yes |
Deceased.
All patients are or were affected with hypomagnesemia, hypercalciuria, and nephrocalcinosis.
TPL indicates that the patient underwent kidney transplantation; CRF indicates that the patient experienced chronic renal failure.
Unconfirmed.
Figure 1. .
Mapping of a new gene locus for recessive renal hypomagnesemia on 1p34.2. A, Application of a reduced subset of 7,802 informative autosomal SNPs. The highest (by far) HLOD score was identified on chromosome 1p34.2, with a peak value of 6.4. Physical positions of the SNP markers are depicted on the abscissa. B, A critical interval of <2 Mb, revealed by comparison of the patients’ haplotypes. The position of the candidate gene CLDN19 and the flanking SNP markers (highlighted in yellow) are depicted. Homozygous genotypes are indicated by “1” or “2,” heterozygous genotypes are indicated by a times sign (×), and SNPs without a genotype are indicated by a minus sign (−). The two different haplotypes are drawn in green (for the Swiss sample) or blue (for the Spanish/Hispanic samples). Boxed haplotype segments are identical by descent.
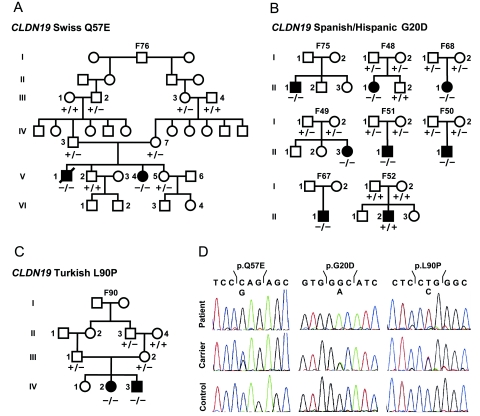
CLDN19 encodes for claudin-19, a recently described member of the claudin multigene family. Claudin-19 is expressed in few organs, and the highest levels are found in the kidney and the eye (UniGene profile). By sequence analysis of the coding region and the adjacent intron/exon boundaries, we identified two different homozygous missense mutations, a Spanish/Hispanic mutation (G20D) located in the first transmembrane domain that was identified in all Spanish/Hispanic patients except F52, and a Swiss mutation (Q57E) located in the first extracellular loop of claudin-19 (figs. 2A, 2B, and 3) (primer sequences are available on request). The G20 and Q57 residues of the claudin-19 protein are highly conserved throughout evolution, and both mutations replace a neutral with a negatively charged amino acid. SIFT (sorting intolerant from tolerant) analysis supports the pathogenicity of both mutations. In addition, the two mutations cosegregate with the disease phenotype and were not found in 196 ethnically matched control chromosomes for the Spanish/Hispanic mutation or 150 Swiss individuals (300 chromosomes) for the Swiss mutation (number of controls allows detection of a 1% polymorphism with a power of 0.8). Obviously, a recent founder effect is responsible for the concordance of the mutation in all Spanish/Hispanic patients, since it is embedded into the same haplotype background that extends over >450 kb (fig. 1B).
Figure 2. .
A, Pedigree of the extended Swiss family with two affected siblings (blackened symbols). Wild-type and mutant sequences for the CLDN19 Q57E mutation analysis are marked by a minus sign (−) and a plus sign (+), respectively. B, Pedigrees of eight Spanish/Hispanic families with results of the CLDN19 G20D mutation. In one family (F52), CLDN19-mutation analysis was negative. C, Turkish family with two affected individuals, with results of the CLDN19 L90P mutation analysis. D, CLDN19 sequence analysis. Chromatograms of homozygous mutations (top), compared with heterozygous carriers (middle) and wild-type sequences (bottom).
Figure 3. .
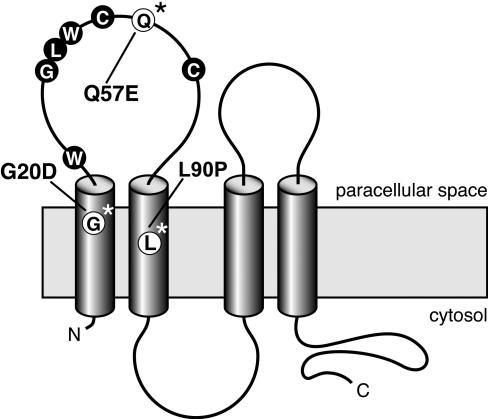
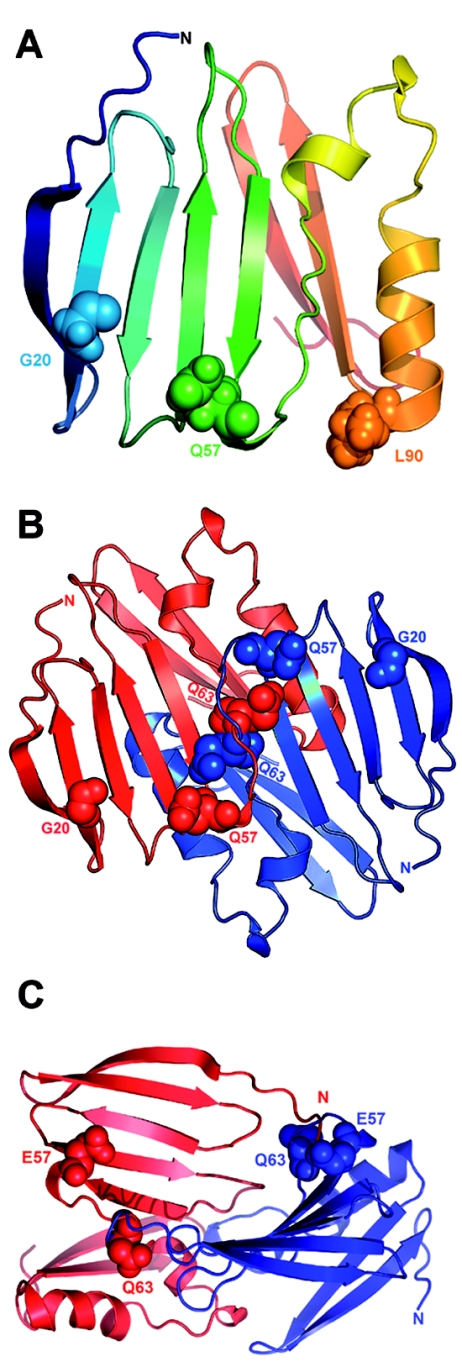
Predicted model of claudin-19, with four transmembrane domains, two extracellular loops, and intracellular termini. The first loop is characterized by the claudin-specific amino acid motif W-GLW-C-C. The Spanish/Hispanic mutation G20D is localized in the first transmembrane domain, the Swiss mutation Q57E affects a residue located between the two cysteines of the W-GLW-C-C motif, and the Turkish mutation L90P resides in the second transmembrane domain.
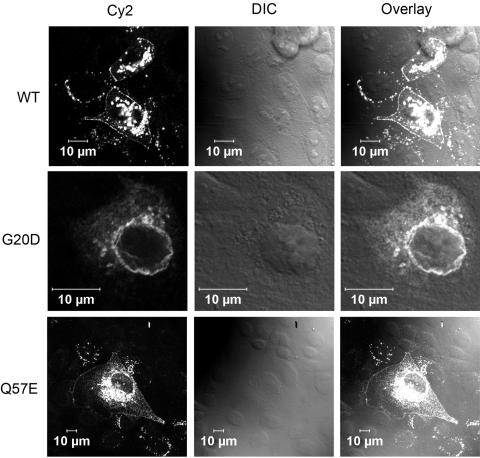
Functional analysis of G20D and Q57E claudin-19 was performed by heterologous expression of the mutants in Madin-Darby canine kidney (MDCK) cells. For immunodetection, a V5-epitope sequence was added to the carboxy terminus of the claudin-19 constructs. These were then transiently expressed in MDCK cells, were stained with a mouse monoclonal anti-V5 antibody (Invitrogen) and a Cy2-coupled goat–anti-mouse polyclonal antibody (Jackson Immuno Research), and were visualized by laser scanning microscopy. Expression analysis revealed perinuclear retention of the G20D mutant, whereas wild-type claudin-19 is targeted to the cell membrane, which points to a trafficking defect (fig. 4). This trafficking defect of the mutant is explained by a disturbance of the signal-peptide sequence, corresponding to the first 20 aa of the claudin-19 protein, as predicted by in silico analysis (SignalP 3.0). The Q57E mutant, however, inserts correctly into the cell membrane. This missense mutation is located in the region of the W-GLW-C-C signature sequence that characterizes all members of the claudin family3 (fig. 3). A 3D model of the first 115 aa of claudin-19 was constructed using ROSETTA de novo methods, with use of a structure library of protein fragments.14 Quality of the model was checked by WHATCHECK15 and Ramachandran plot analysis.16 The final model was energetically stable and had the features characteristic of transmembrane proteins (fig. 5A). Since the first extracellular loop of claudins is thought to bridge the paracellular space and claudin-5 has been said to form homodimers,17 we subsequently tested, by applying the PATCHDOCK software,18 whether claudin-19 can form a homodimer and whether Q57 has any role in this complex formation. Of the best 10 solutions obtained, 8 showed involvement of the first extracellular loop in the dimer formation, with Q57 of the first molecule interacting with Q61 and Q63 of the second molecule (fig. 5B). This computational model cannot predict whether the claudin-19 dimer is located in one cell or in opposing cells bridging the intercellular space; however, it is well known that members of the claudin family form homo- and heterodimers, both interacting laterally and bridging the intercellular space with their extracellular domains.3,19 Docking calculations were repeated after in silico introduction of the mutation Q57E in the monomeric model. The dimer formation was disrupted with the mutant protein, which is possibly due to strong electrostatic repulsion from opposing E residues (fig. 5C).
Figure 4. .
Functional analysis of the CLDN19 mutations. Analysis of the subcellular localization of transiently expressed claudin-19 in MDCK cells demonstrates correct insertion of the wild-type (WT) claudin-19 protein (upper panels) and the Q57E mutant (lower panels) into the cell membrane. The G20D mutant, however, is retained inside the cell (middle panels). Immunofluorescence images, the corresponding differential interference contrast images (DIC), and overlays are depicted.
Figure 5. .
Three-dimensional structural models of claudin-19. A, The first 115 aa of claudin-19 are shown as a ribbon diagram in a gradient of rainbow colors, with the N terminus depicted in blue. The G20D mutation is located in the first transmembrane region that is buried in the membrane. The strong change in the charge pattern by the replacement of glycine with aspartic acid presumably influences intramolecular interactions that will alter the signal sequence. The Q57E mutation is located in the first extracellular loop and is likely to affect electrostatic interactions that result in altered protein-protein interactions. The L90P mutation is located in the second transmembrane domain. B, Potential model of claudin-19 homodimer formation. A representative docking solution—which shows the involvement of the first extracellular loop in the dimer formation with Q57 of the first molecule interacting with Q61 and Q63 of the second molecule—is depicted. The two claudin-19 molecules are colored in red and blue. The claudin-19 homodimer seems to be stabilized by interaction of Q57 of one molecule with Q63 of the partner molecule. C, Effect of the Q57E mutation on claudin-19 homodimer formation. Docking calculations were repeated after performing in silico mutation of Q57E in the monomeric model. The dimer formation was disrupted with the mutant protein, possibly because of strong electrostatic repulsion from opposing E residues.
Subsequently, we identified another consanguineous family with two children with FHHNC and severe ocular involvement (family F90 [table 1 and fig. 2C]). This family is of Turkish origin. Direct sequencing of CLDN19 revealed a homozygous L90P mutation in both affected siblings and was detected at a heterozygous state in the obligate carriers (fig. 2C and 2D). The possibility of a polymorphism was ruled out by testing 220 Turkish control chromosomes. Moreover, SIFT and PolyPhen analyses (score 2.43) indicate pathogenicity of this mutation, and L90 is highly conserved among claudin genes across all species. L90 is located in the second transmembrane domain, and the replacement by a proline disrupts the α-helices, as predicted by the GOR IV secondary structure–prediction method. This might be explained by the rigid structure of the proline residue, where the side chain is covalently bound to its amino-terminal end. Together with the two mutation described above, these data clearly indicate that the pathogenesis of FHHNC with severe ocular abnormalities in humans is explained by mutations in CLDN19.
A recently generated Cldn19 knockout mouse model demonstrates disorganized tight junctions in the Schwann cells of the peripheral nervous system, and homozygous animals show abnormal behavior and peripheral neuropathy.20 Visual impairment, electrolyte imbalances, and renal abnormalities are not reported in that work.20 This might be because these parameters have not been studied. It is also possible that phenotypic disparities actually exist between the human and the murine model, which could be explained by the fact that different members of the large claudin family seem to be able to compensate for specific claudin defects to a variable extent, dependent on the species and the specific genetic background of the model.
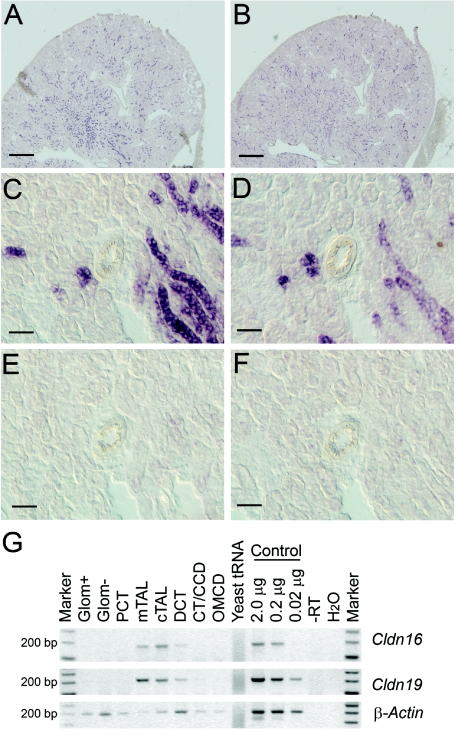
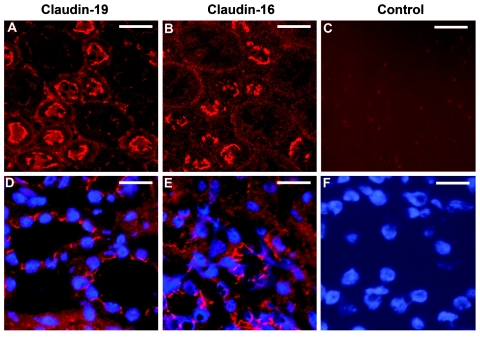
However, previous studies of mice demonstrated a strong expression of claudin-19 in the distal tubules of the kidney cortex,21 which supports a functional role of claudin-19 for paracellular electrolyte-transport mechanisms. We therefore studied the expression of claudin-19 in the kidney in more detail. In situ hybridization analysis was performed using probes that recognize Cldn16 (GenBank accession number NM_053241 [nucleotides 154–1074]) and Cldn19 (GenBank accession number BC115827 [nucleotides 63–908]). These experiments revealed a highly similar expression pattern of claudin-19 and -16 mRNAs in mouse cortical kidney tubules (fig. 6A and 6B). Higher magnification of series kidney sections suggests a colocalization of both claudins (fig. 6C–6F). To identify the nephron segments expressing Cldn19, we performed RT-PCR analysis in microdissected mouse nephron segments (primer sequences are available on request). Cldn19 is expressed in exactly the same nephron segments as is Cldn16—namely, the medullary and cortical TAL, with a lower level of expression in the distal convoluted tubule (fig. 6G). These results were confirmed by immunohistochemistry of series kidney sections with use of polyclonal rabbit anti–claudin-19 (as described by Lee et al.21) and anti–claudin-16 (Zymed) antibodies at a dilution of 1:200. Anti-rabbit Cy3 (Rockland) was used as secondary antibody, at a dilution of 1:1,600. Bisbenzimide H33258 was used for nuclear staining (Serva). Consistent with the results of RT-PCR analysis, immunohistochemistry revealed a marked apical and slighter basolateral expression of both claudins in the TAL and distal tubules (fig. 7A and 7B). Negative controls were performed by omission of the primary antibodies (fig. 7C and 7F). At higher magnification, the expression of both proteins could be detected at cell-cell contacts (fig. 7D and 7E). Thus, it seems possible that both claudins interact at the tight-junction barrier of the renal tubules involved in paracellular magnesium and calcium reabsorption.
Figure 6. .
Localization of claudin-19 and claudin-16 in mouse kidney. A–D, In situ hybridization analysis of kidney sections, which demonstrates highly similar expression patterns of Cldn19 (A and C) and Cldn16 (B and D) in the renal tubules. Scale bars represent 500 μm in panels A and B and represent 50 μm in panels C and D. E and F, Negative controls, with Cldn19 (E) and Cldn16 (F) sense probes, which demonstrate the specificity of the antisense probes. G, RT-PCR analysis of microdissected nephron segments, which revealed a similar expression pattern for both genes in the medullary (mTAL) and cortical (cTAL) TAL and in the distal convoluted tubules (DCT). Marker = 100-bp ladder; Glom = glomeruli with (Glom+) and without (Glom−) blood vessels; PCT = proximal convoluted tubule; CT/CCD = connecting tubule/cortical collecting duct; OMCD = outer medullary collecting duct. Negative and positive control experiments were performed using yeast tRNA/H2O and kidney RNA, respectively. Obtained PCR products were of expected size (176 bp for Cldn16; 198 bp for Cldn19). In parallel with Cldn19 and Cldn16 amplification, a set of marker genes was amplified (SLC12A1, SLC12A3, AQP1, AQP4, PTHR1, and SLC5A2) as controls for segment specificity of the preparation (data not shown).
Figure 7. .
Immunofluorescence analysis of claudin-19 and claudin-16 in mouse kidney. A and B, Series kidney sections showing similar apical staining patterns of claudin-19 and claudin-16 in the TAL and distal tubules. Scale bars represent 25 μm. D and E, Higher magnification, which demonstrates protein expression at cell-cell contacts (nuclear staining in blue). Scale bars represent 15 μm. C and F, Negative control with omission of the primary anti–claudin-19 and anti–claudin-16-antibodies (C) and nuclear staining of the same section (F).
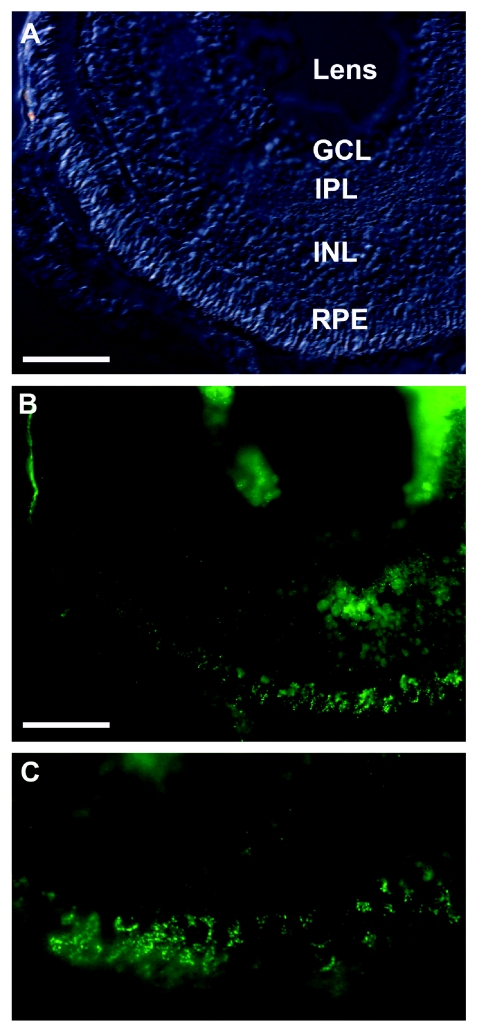
Immunohistochemistry was subsequently performed on ocular sections of zebrafish (claudin-19 antibody dilution 1:100). In the eye, claudin-19 is expressed specifically in the retina, with a strong expression in the retinal pigment epithelium (RPE). Additional claudin-19 staining was observed in the inner plexiform layer (IPL) and the inner nuclear layer (INL) (fig. 8). Negative controls consisted of omission of the primary antibody and the evaluation of autofluorescence (data not shown). Different claudins are known to be expressed during ocular development at the tight junctions of the RPE—for example, claudins-1, -2, -5, and -12—and proper tight-junction formation is required for apical cell polarity and an intact retinal transfusion barrier.22 As a consequence, loss of tight-junction formation can result in the mistargeting of factors that influence retinal cell division.22 To our knowledge, the present study demonstrates for the first time that claudin-19 is an important component of RPE tight junctions and that patients affected by CLDN19 mutations present with a marked misdevelopment of the optic disc, which is associated with near blindness and the manifestation of a horizontal nystagmus. The eye phenotypes in patients with CLDN19 mutations reported here are much more severe than phenotypes reported elsewhere for patients with FHHNC.7 For a long time, ocular involvement with an FHHNC phenotype has been observed with much higher frequency in Spanish patients than in patients from other parts of the world (reviewed by Benigno et al.23). We have now provided evidence that genetic heterogeneity is responsible for this phenotypic difference and that mutations in CLDN19 are the causative defect in this subset of patients (i.e., those with FHHNC and severe ocular involvement). The identification of CLDN19 mutations in patients with hypomagnesemia, chronic renal failure, and severe visual impairment supports the fundamental role of claudin-19 for magnesium homeostasis, normal tubular structures in the kidney, and undisturbed organization and development of the retina.
Figure 8. .
Localization of claudin-19 in zebrafish retina. A, Bright field image of a section of a larval retina at 5 d post fertilization. Scale bar represents 50 μm. GCL = ganglion-cell layer; INL = inner nuclear layer. B, Punctate staining of claudin-19, detected in the RPE, including the interdigitating microvilli. The inner nuclear and inner synaptic layers show less staining. C, Close-up view of claudin-19 staining in the outer retina.
Acknowledgments
We are indebted to the participating patients and their families for their cooperation. We thank Bärbel Philippin, Ulla Pechmann, and Franziska Rubi, for excellent technical assistance. The work was supported by the German Federal Ministry of Science and Education through the National Genome Research Network (support to D.S., C.B., and P.N.), the Swiss National Science Foundation (to S.C.F.N.), and Eidgenössische Technische Hochschule internal grants (to A.L.).
Web Resources
Accession numbers and URLs for data presented herein are as follows:
- Ensembl, http://www.ensembl.org/Homo_sapiens/index.html
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for Cldn16 [accession number NM_053241] and Cldn19 [accession number BC115827])
- GOR IV, http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_gor4.html
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for FHHNC) [PubMed]
- PolyPhen, http://tux.embl-heidelberg.de/ramensky/polyphen.cgi
- SIFT, http://blocks.fhcrc.org/sift/SIFT.html
- SignalP, http://www.cbs.dtu.dk/services/SignalP/
- UniGene, http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=unigene
References
- 1.Lee DB, Huang E, Ward HJ (2006) Tight junction biology and kidney dysfunction. Am J Physiol Renal Physiol 290:F20–F34 10.1152/ajprenal.00052.2005 [DOI] [PubMed] [Google Scholar]
- 2.Furuse M, Tsukita S (2006) Claudins in occluding junctions of humans and flies. Trends Cell Biol 16:181–188 10.1016/j.tcb.2006.02.006 [DOI] [PubMed] [Google Scholar]
- 3.Van Itallie CM, Anderson JM (2006) Claudins and epithelial paracellular transport. Annu Rev Physiol 68:403–429 10.1146/annurev.physiol.68.040104.131404 [DOI] [PubMed] [Google Scholar]
- 4.Colegio OR, Van Itallie CM, McCrea HJ, Rahner C, Anderson JM (2002) Claudins create charge-selective channels in the paracellular pathway between epithelial cells. Am J Physiol Cell Physiol 283:C142–C147 [DOI] [PubMed] [Google Scholar]
- 5.Van Itallie CM, Fanning AS, Anderson JM (2003) Reversal of charge selectivity in cation or anion-selective epithelial lines by expression of different claudins. Am J Physiol Renal Physiol 285:F1078–F1084 [DOI] [PubMed] [Google Scholar]
- 6.Simon DB, Lu Y, Choate KA, Velazquez H, Al-Sabban E, Praga M, Casari G, Bettinelli A, Colussi G, Rodriguez-Soriano J, McCredie D, Milford D, Sanjad S, Lifton RP (1999) Paracellin-1, a renal tight junction protein required for paracellular Mg2+ resorption. Science 285:103–106 10.1126/science.285.5424.103 [DOI] [PubMed] [Google Scholar]
- 7.Weber S, Schneider L, Peters M, Misselwitz J, Ronnefarth G, Boswald M, Bonzel KE, Seeman T, Sulakova T, Kuwertz-Broking E, Gregoric A, Palcoux JB, Tasic V, Manz F, Scharer K, Seyberth HW, Konrad M (2001) Novel paracellin-1 mutations in 25 families with familial hypomagnesemia with hypercalciuria and nephrocalcinosis. J Am Soc Nephrol 12:1872–1881 [DOI] [PubMed] [Google Scholar]
- 8.Blanchard A, Jeunemaitre X, Coudol P, Dechaux M, Froissart M, May A, Demontis R, Fournier A, Paillard M, Houillier P (2001) Paracellin-1 is critical for magnesium and calcium reabsorption in the human thick ascending limb of Henle. Kidney Int 59:2206–2215 [DOI] [PubMed] [Google Scholar]
- 9.Muller D, Kausalya PJ, Claverie-Martin F, Meij IC, Eggert P, Garcia-Nieto V, Hunziker W (2003) A novel claudin 16 mutation associated with childhood hypercalciuria abolishes binding to ZO-1 and results in lysosomal mistargeting. Am J Hum Genet 73:1293–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meier W, Blumberg A, Imahorn W, De Luca F, Wildberger H, Oetliker O (1979) Idiopathic hypercalciuria with bilateral macular colobomata: a new variant of oculo-renal syndrome. Helv Paediatr Acta 34:257–269 [PubMed] [Google Scholar]
- 11.Rodriguez-Soriano J, Vallo A (1994) Pathophysiology of the renal acidification defect present in the syndrome of familial hypomagnesaemia-hypercalciuria. Pediatr Nephrol 8:431–435 10.1007/BF00856522 [DOI] [PubMed] [Google Scholar]
- 12.Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES (1996) Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet 58:1347–1363 [PMC free article] [PubMed] [Google Scholar]
- 13.Strauch K, Fimmers R, Kurz T, Deichmann KA, Wienker TF, Baur MP (2000) Parametric and nonparametric multipoint linkage analysis with imprinting and two-locus–trait models: application to mite sensitization. Am J Hum Genet 66:1945–1957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schueler-Furman O, Wang C, Bradley P, Misura K, Baker D (2005) Progress in modeling of protein structures and interactions. Science 310:638–642 10.1126/science.1112160 [DOI] [PubMed] [Google Scholar]
- 15.Hooft RW, Vriend G, Sander C, Abola EE (1996) Errors in protein structures. Nature 381:272 10.1038/381272a0 [DOI] [PubMed] [Google Scholar]
- 16.Ramachandra GN, Ramakrishnan C, Sasisekharan V (1963) Stereochemistry of polypeptide chain configurations. J Mol Biol 7:95–99 [DOI] [PubMed] [Google Scholar]
- 17.Blasig IE, Winkler L, Lassowski B, Mueller SL, Zuleger N, Krause E, Krause G, Gast K, Kolbe M, Piontek J (2006) On the self-association potential of transmembrane tight junction proteins. Cell Mol Life Sci 63:505–514 10.1007/s00018-005-5472-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schneidman-Duhovny D, Inbar Y, Polak V, Shatsky M, Halperin I, Benyamini H, Barzilai A, Dror O, Haspel N, Nussinov R, Wolfson HJ (2003) Taking geometry to its edge: fast unbound rigid (and hinge-bent) docking. Proteins 52:107–112 10.1002/prot.10397 [DOI] [PubMed] [Google Scholar]
- 19.Furuse M, Sasaki H, Tsukita S (1999) Manner of interaction of heterogeneous claudin species within and between tight junction strands. J Cell Biol 147:891–903 10.1083/jcb.147.4.891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miyamoto T, Morita K, Takemoto D, Takeuchi K, Kitano Y, Miyakawa T, Nakayama K, Okamura Y, Sasaki H, Miyachi Y, Furuse M, Tsukita S (2005) Tight junctions in Schwann cells of peripheral myelinated axons: a lesson from claudin-19-deficient mice. J Cell Biol 169:527–538 10.1083/jcb.200501154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee NP, Tong MK, Leung PP, Chan VW, Leung S, Tam PC, Chan KW, Lee KF, Yeung WS, Luk JM (2006) Kidney claudin-19: localization in distal tubules and collecting ducts and dysregulation in polycystic renal disease. FEBS Lett 580:923–931 10.1016/j.febslet.2006.01.019 [DOI] [PubMed] [Google Scholar]
- 22.Rahner C, Fukuhara M, Peng S, Kojima S, Rizzolo LJ (2004) The apical and basal environments of the retinal pigment epithelium regulate the maturation of tight junctions during development. J Cell Sci 117:3307–3318 10.1242/jcs.01181 [DOI] [PubMed] [Google Scholar]
- 23.Benigno V, Canonica CS, Bettinelli A, von Vigier RO, Truttmann AC, Bianchetti MG (2000) Hypomagnesaemia-hypercalciuria-nephrocalcinosis: a report of nine cases and a review. Nephrol Dial Transplant 15:605–610 10.1093/ndt/15.5.605 [DOI] [PubMed] [Google Scholar]