Abstract
The 13 polypeptides encoded in mitochondrial DNA (mtDNA) are synthesized in the mitochondrial matrix on a dedicated protein-translation apparatus that resembles that found in prokaryotes. Here, we have investigated the genetic basis for a mitochondrial protein-synthesis defect associated with a combined oxidative phosphorylation enzyme deficiency in two patients, one of whom presented with encephalomyopathy and the other with hypertrophic cardiomyopathy. Sequencing of candidate genes revealed the same homozygous mutation (C997T) in both patients in TSFM, a gene coding for the mitochondrial translation elongation factor EFTs. EFTs functions as a guanine nucleotide exchange factor for EFTu, another translation elongation factor that brings aminoacylated transfer RNAs to the ribosomal A site as a ternary complex with guanosine triphosphate. The mutation predicts an Arg333Trp substitution at an evolutionarily conserved site in a subdomain of EFTs that interacts with EFTu. Molecular modeling showed that the substitution disrupts local subdomain structure and the dimerization interface. The steady-state levels of EFTs and EFTu in patient fibroblasts were reduced by 75% and 60%, respectively, and the amounts of assembled complexes I, IV, and V were reduced by 35%–91% compared with the amounts in controls. These phenotypes and the translation defect were rescued by retroviral expression of either EFTs or EFTu. These data clearly establish mutant EFTs as the cause of disease in these patients. The fact that the same mutation is associated with distinct clinical phenotypes suggests the presence of genetic modifiers of the mitochondrial translation apparatus.
Defects in mitochondrial oxidative phosphorylation (OXPHOS) are estimated to occur in ∼1 in 5,000 live births.1 They can be classified biochemically as either isolated or combined deficiencies of any of the five OXPHOS complexes.2 Genetic defects leading to combined deficiencies of complexes I, III, IV, and V can be caused by mutations in mtDNA or in the components of the mitochondrial translation apparatus encoded by nuclear genes.3
Synthesis of the 13 mitochondrial-encoded proteins occurs on a dedicated mitochondrial translation apparatus similar to that found in prokaryotes and requires, in addition to the tRNAs and ribosomal RNAs (rRNAs) encoded in mtDNA, the concerted action of several translation factors and a large number of mitochondrial ribosomal (mitoribosome) proteins, all of which are encoded by nuclear genes. Two mammalian initiation factors, IF2 and IF3; four elongation factors, EFTu, EFTs, EFG1, and EFG2 (reviewed by Spremulli et al.4); one release factor, RF1; and one ribosomal recycling factor, RRF,5 have been cloned and sequenced. Mitoribosomes are distinct from both prokaryotic and eukaryotic cytosolic ribosomes; they are 55S particles composed of a small (28S) and a large (39S) subunit and contain a much higher protein:RNA ratio than that of the bacterial 70S ribosome.6
Although the vast majority of components of the mitochondrial translation system are nuclear encoded, most mutations associated with mitochondrial translation defects have been reported in mtDNA-encoded tRNAs and rRNAs.7,8 However, we recently reported mutations in the nuclear-encoded translation elongation factor EFG19,10 and in MRPS16, a protein of the small ribosomal subunit,11 in patients with autosomal recessive mitochondrial translation defects. Encouraged by these findings, we investigated a cohort of 16 patients with combined OXPHOS enzyme defects expressed in both muscle and fibroblasts in which mtDNA sequencing and complex II activity–measurement results were normal. We describe the first mutation in the mitochondrial elongation factor EFTs (encoded by TSFM [MIM 604723]) associated with a fatal mitochondrial encephalomyopathy in one pedigree and with fatal hypertrophic cardiomyopathy in another, and we demonstrate the molecular basis for the defect.
Material and Methods
Case Reports
Patient 1, a boy, was born at term as the second child of consanguineous Turkish parents by cesarean section performed on maternal indication. Birth weight (3,140 g), length (50 cm), and head circumference (36 cm) were age appropriate. Muscular hypotonia, sucking weakness, and a severe lactic acidosis (pH 6.8; base excess −26; lactic acid 42 mM [control <2.1 mM]) were the initial clinical signs and symptoms, followed by rhabdomyolysis with creatine kinase values up to 7,252 U/liter (control <248 U/liter). The initial lactate:pyruvate (L:P) ratio was 92 (control 12–15). During life, lactic acid dropped to values between 4 and 5.5 mM, with L:P ratios ∼25. Artificial ventilation was needed because of persistent dyspnea. On the 3rd day of life, generalized convulsions became obvious. The electroencephalogram showed a discontinuous pattern with bifrontal symmetrical sharper spikes. Repeated ultrasound imaging of the brain showed reduced gyri on day 4, plexus bleeding with enlarged ventricles on day 6, and abnormal signal intensity of the thalami on day 9. Echocardiographic examination on day 6 revealed a persistent ductus arteriosus Botalli, with left-to-right shunting, and a persistent foramen ovale. Septum thickness and contractility were age appropriate. On the basis of the suspicion of a mitochondrial disorder, a fibroblast culture was established and a skeletal muscle (m. vastus lateralis dextra) biopsy sample was obtained for measurements of the OXPHOS enzymes. Despite intensive treatment, the patient died of progressive encephalomyopathy and respiratory failure at age 7 wk.
Patient 2, a girl, was the third child of second cousins once removed whose parents are of Kurdish Jewish origin. Two older children were healthy. The mother reported a paucity of fetal movements throughout the pregnancy. Delivery was vaginal, and birth weight was 2,600 g. Initial examination results were normal, and the patient was breast-fed. At age 36 h, apathy, irregular breathing, and severe muscular hypotonia were noted. Laboratory investigation revealed severe metabolic acidosis (pH 6.93; HCO3 −4.6; base excess −26). Serum lactate level was increased (17.6 mM; control <2.4 mM), as were blood ketone levels (3-OH-butyrate 4,803 μM; acetoacetate 257 μM; control values <200 μM and <100 μM, respectively) and serum ammonia levels (268 μM; control <32 μM). Blood count and liver enzyme levels were normal. Echocardiographic examination disclosed severe concentric hypertrophic cardiomyopathy with normal contractility. Apart from generalized muscle hypotonia, neurological examination, including fundoscopy and brain CT scan, were normal. Skin biopsy for fibroblast culture and muscle biopsy from the right quadriceps were performed at age 2 wk; histochemical analysis of the muscle revealed a generally decreased cytochrome c oxidase (COX) stain, whereas the succinate dehydrogenase stain was normal. The patient was given treatment with carnitine, sodium-dichloroacetate, thiamine, idebenone, and riboflavin, but the lactate level remained elevated, and cardiac function gradually deteriorated. Her death at age 7 wk was preceded by severe hyponatremia with a low urinary output for 2 d. An open-liver biopsy was performed 20 min after her death.
Southern-blot analysis of the muscle, liver, and fibroblast DNA revealed normal size and abundance of mtDNA. Northern-blot analysis showed that the steady-state levels of several mitochondrial transcripts (ND6, COXI, COXII, and 12S rRNA) were normal. Sequencing of the 22 mitochondrial tRNA genes failed to identify a pathogenic mutation.
Human Studies
Informed consent was obtained, and research studies were approved by the Helsinki committee of Hadassah–Hebrew University Medical Center, the Radboud University Medical Centre Nijmegen, and the Montreal Neurological Institute Institutional Review Boards.
Cell Culture
Primary human skin fibroblasts were immortalized with a retrovirus expressing the E7 gene of type 16 human papilloma virus and a retroviral vector expressing the protein component (htert) of human telomerase.12 Patient and control skin fibroblasts were cultured in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% fetal calf serum.
Enzyme Measurements
The activities of the OXPHOS enzyme complexes in skeletal muscle and fibroblasts were measured according to described protocols.13,14 COX and citrate synthase activities were measured in fibroblast cell extracts as described elsewhere.15
Sequence Analysis
Total RNA was extracted from cultured skin fibroblasts of the patients by use of RNAzol (Campro Scientific). The TSFM cDNA (GenBank accession number NM_005726) was amplified by superscript II reverse transcriptase/Taq DNA polymerase (Life Technologies) in four overlapping fragments (primer sequences available on request). After treatment with ExoI (Promega) and SAP (Promega), the PCR fragments were used as templates in sequencing reactions with the chemistry of BigDye1.1 (Applied Biosystems) according to the manufacturer’s protocol. The labeled fragments were separated on a 3130xl Genetic Analyzer (Applied Biosystems). Sequence data were evaluated with Sequencher 4.2 sequence-analysis software (Gene Codes) by comparison with the reference sequence of TSFM. Sequence analysis of the last part of exon 7 of the TSFM gene was performed on genomic DNA by use of the primers 5′-CTTGGGCAGCCATGTGG-3′ (forward) and 5′-ACCCATGCATTCTCGGTCTG-3′ (reverse).
Blue-Native PAGE and Immunoblotting
Blue-native PAGE was used for separation of the OXPHOS complexes on 6%–15% polyacrylamide-gradient gels.16 Mitoplasts, which were prepared from fibroblasts by treatment with 0.8 mg of digitonin per mg of protein, were solubilized with 1% lauryl maltoside, and 20 μg of the solubilized protein was used for electrophoresis.17 Assembly of the OXPHOS complexes was established by immunoblot analysis with the use of monoclonal antibodies against subunits of complexes II–V (Molecular Probes) and a polyclonal antibody against the ND1 subunit of complex I (a kind gift of A. Lombes, Paris). For immunoblotting, fibroblasts were solubilized with 1.5% lauryl maltoside in PBS, and 20 μg of protein was separated by TRIS-Glycine SDS-PAGE gel. After transfer to a nitrocellulose membrane, individual proteins were detected using monoclonal antibodies against subunits of complexes I–V (Molecular Probes), porin (Sigma), MnSOD (Stressgen), and polyconal antibodies directed against ND1, EFTu (a kind gift of N. Takeuchi, Tokyo), EFTu/Ts (a kind gift of L. Spremulli, Chapel Hill, NC), and EFG1.10
Pulse Labeling of Mitochondrial Translation Products
In vitro labeling of mitochondrial translation was performed as described elsewhere.18 In brief, cells were labeled for 60 min at 37°C in methionine-free DMEM with 10% dialyzed fetal bovine serum, containing 200 μCi/ml of [35S]methionine and 100 μg/ml of emetine, followed by a 10-min incubation in regular DMEM with 10% fetal bovine serum. Total cellular protein (50 μg) was resuspended in loading buffer containing 93 mM Tris-HCl (pH 6.7), 7.5% glycerol, 1% SDS, 0.25 mg bromophenol blue per ml, and 3% mercaptoethanol, was sonicated for 3–8 s, and was run on 12%–20% polyacrylamide-gradient gels. The gels were scanned on a Storm Phosphorimager (Molecular Dynamics) and were analyzed with ImageQuant software. Under the conditions of the experiment, all signals were in the linear range.
cDNA Constructs, Virus Production, and Infection
Retroviral vectors containing the cDNA sequence of TUFM (GenBank accession number NM_003321) and TSFM were created with the Gateway cloning system (Invitrogen) as described elsewhere.10 The PCR constructs were cloned into a Gateway-modified retroviral-expression vector (pBABE-puro). Retroviral constructs were transiently transfected into Phoenix packaging cell line by use of the HBS/Ca3(PO4)2 method (Helper Dependent Protocol Web site). Patient and control fibroblasts were infected 48 h later by exposure to virus-containing medium in the presence of 4 μg/ml of polybrene, as described elsewhere.12
Computational Modeling
The crystal structure of the bovine EFTu/EFTs complex has been described elsewhere.19 On the basis of this model, the structure of wild-type and mutant EFTs protein were determined with the YASARA–WHAT IF twin set software.20 The R333W mutation was predicted using the method of De Fillippis et al.21 Molecular figures were drawn with the POV-Ray module in YASARA. Further modeling details are available at the TS Modelling Web site.
Results
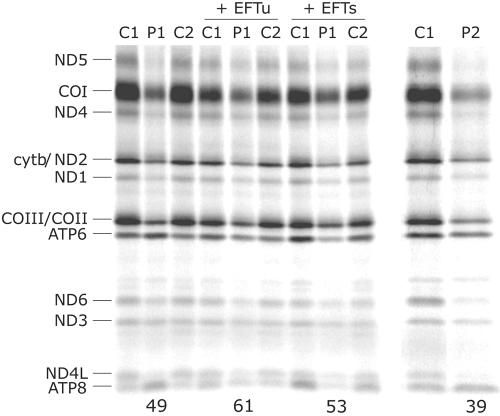
Skeletal muscle and fibroblasts from patient 1 showed decreased enzyme activities of complexes I, III, and IV of the respiratory chain (table 1). The activities of complexes I, III, and IV in the skeletal muscle of patient 2 were similarly reduced to 30%, 14%, and 14% of the control means, respectively, whereas complex II was unaffected (123% of the control mean). In fibroblast mitochondria from patient 2, complexes I, IV, II+III, and II activities were 44%, 25%, 77%, and 107% of the control mean, respectively. Pulse labeling of the mitochondrial translation products in fibroblasts from both patients revealed a generalized mitochondrial protein-synthesis defect; however, different polypeptides were variably affected (fig. 1 and table 2). The overall incorporation of [35S]methionine was 39%–49% of that in controls, and the rate of synthesis of most polypeptides was reduced to 30%–60% of control values. However, the synthesis of the two ATP synthase subunits (ATP6 and ATP8) actually increased relative to control levels in patient 1 and was similar to control levels in patient 2 (table 2).
Table 1. .
OXPHOS Enzyme Activities in Skeletal Muscle and Fibroblasts of Patient 1[Note]
| Activities (mU/U citrate synthase) in |
||||
| Skeletal Muscle |
Fibroblasts |
|||
| Complex | Patient | Reference Range | Patient | Reference Range |
| I | 0 | 70–250 | 53 | 100–310 |
| III | 753 | 2,200–6,610 | 1,250 | 1,320–2,610 |
| II + III | 228 | 300–970 | 163 | 110–470 |
| IV | 186 | 810–3,120 | 397 | 680–1,190 |
| PDH | 60 | 34–122 | ND | ND |
Note.— Samples were 600 g of supernatant of skeletal muscle or mitochondrial enriched fraction from fibroblasts. ND = not determined; PDH= pyruvate dehydrogenase complex.
Figure 1. .
Pulse labeling of mitochondrial translation products in patient fibroblasts. Fibroblasts from the two patients (P1 and P2) and two controls (C1 and C2) were labeled with [35S]methionine in the presence of emetine, a cytosolic translation inhibitor, as outlined in the “Material and Methods” section. Fibroblasts from patient 1 were also analyzed after transduction with retroviral vectors overexpressing the mitochondrial translation factors EFTu or EFTs. The mitochondrial translation products are indicated on the left of the autoradiogram. COI–COIII = subunits of cytochrome c oxidase; cytb = cytochrome b subunit; ND1, ND2, ND3, ND4, ND4L, ND5, and ND6 = subunits of NADH CoQ oxidoreductase. The numbers at the bottom of the gel indicate the total incorporation of [35S]methionine relative to the corresponding control.
Table 2. .
Synthesis of Individual Mitochondrial Polypeptides in Fibroblasts from Patients P1 and P2 after Pulse Labeling with [35S]Methionine[Note]
| Incorporation of [35S]Methionine(% of Control) |
||||
| Polypeptide | P1 | P2 | P1 + EFTu | P1 + EFTs |
| ND5 | 22 | 18 | 59 | 49 |
| COI | 34 | 32 | 59 | 61 |
| ND4 | 42 | 33 | 65 | 52 |
| ND2/cytb | 61 | 51 | 56 | 43 |
| ND1 | 56 | 55 | 58 | 46 |
| COIII/COII | 42 | 34 | 56 | 60 |
| ATP6 | 134 | 64 | 69 | 31 |
| ND6 | 59 | 21 | 60 | 72 |
| ND3 | 103 | 68 | 112 | 91 |
| ND4L | 29 | 30 | 43 | 48 |
| ATP8 | 244 | 113 | 94 | 22 |
Note.— The results are presented as a percentage of the average incorporation of [35S]methionine in two relevant controls. Mitochondrial translation was also analyzed in patient 1 after transduction with retroviral constructs overexpressing EFTu or EFTs. COI–COIII = subunits of cytochrome c oxidase; cytb = cytochrome b subunit; ND1, ND2, ND3, ND4, ND4L, ND5, and ND6 = subunits of NADH CoQ oxidoreductase.
These data suggested that the genetic defect was due to a component of the mitochondrial translation apparatus, and the failure to identify any abnormalities in mtDNA (data not shown) suggested a nuclear gene defect. Two strategies were employed to identify the underlying genetic defect. In patient 1 (and 14 additional patients with a similar combined OXPHOS deficiency), we sequenced the cDNAs for the known mitochondrial translation initiation, elongation, and termination factors. In patient 2, we used the Affymetrix Human Mapping 50K Array Xba240 to genotype 58,495 SNPs to identify regions of homozygosity. The latter analysis identified a long segment of consecutive homozygous SNPs on chromosome 2 spanning 22.1 Mb (213.1–235.2 Mb) and another segment on chromosome 12 spanning 30.79 Mb (26.48–57.27 Mb). With use of the MitoP2 prediction program22 for mitochondrial localization (score >60), two proteins with known function in mitochondrial translation were identified within these regions—the mitochondrial ribosomal protein S35 and the mitochondrial translation elongation factor EFTs (encoded by TSFM).
Sequence analysis of TSFM revealed the same homozygous C997T mutation in exon 7 in both patients (fig. 2). A BLAST search showed that the C997T mutation was not present in any reported human EST, and the mutation was not found in the other 14 patients tested or in 135 controls. The mutation was heterozygous in the parents in both families and in the two healthy sibs of patient 2 (data not shown). A sib of patient 1 was homozygous wild type (data not shown). Analysis of microsatellite markers on chromosome 12 demonstrated that the mutation did not arise on a common haplotype in the two patients (fig. 2D). Human TSFM appears to be alternatively spliced, since exon 5 was not present in any of the patient or control cDNAs analyzed. Exon 5 is present in a single EST sequence in the human database, derived form bone marrow of a patient with acute myelogenous leukemia.
Figure 2. .
Sequence analysis of TSFM. A, DNA sequence chromatogram from a control and patient 1 (homozygous C997T) and a heterozygous chorionic villus sample. B, Domain structure of EFTs (N-terminal in black, subdomain N in white, and subdomain C in gray) showing the predicted Arg333Trp substitution in subdomain. C, Protein sequence alignment for Homo sapiens, Bos taurus, Caenorhabditis elegans, and E. coli. The alignment from human to bacteria shows that the mutated Arg333Trp is highly conserved among different species. D, Analysis of microsatellite markers spanning the TSFM locus on chromosome 12. The markers are arbitrarily assigned to chromosomes in both patients.
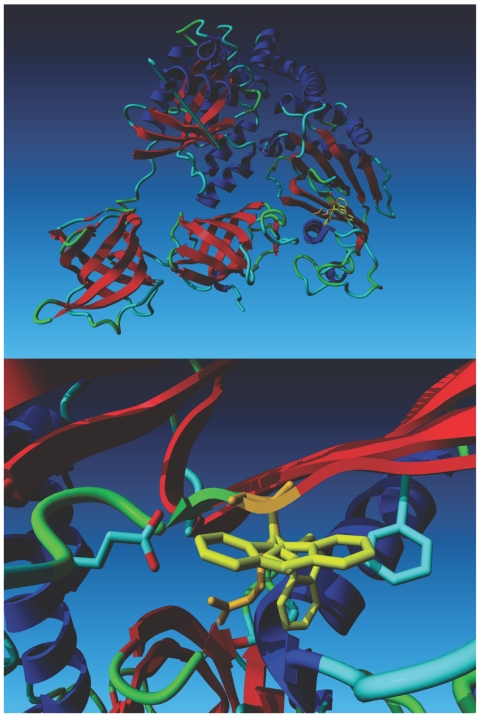
The mutation predicts an Arg333Trp substitution in the C-terminal domain of EFTs, and sequence alignment of this protein from several species shows that the mutated amino acid is highly conserved from human to bacteria (fig. 2B). This Arg residue makes favorable hydrophobic and electrostatic interactions within the helical part of subdomain C (fig. 3). Mutational analysis using the crystal structure of the bovine EFTu•EFTs complex shows that it is not possible to introduce a Trp at this position without serious clashes between the Trp side chain and the backbone atoms in the immediate surroundings. The close-up of the region of Ts that harbors the mutation shows three preferred rotamers of Trp, representing three different energy minima (fig. 3, lower panel). The rotamer that is predicted to be the most favorable in the absence of a surrounding bumps into the helical domain immediately beneath it, and the other two rotamers that normally are energetically favorable bump into other local residues, as indicated in the legend of figure 3. The mutation is predicted to disrupt the interaction between the long helix and two strands in subdomain C, moving the domain that holds the long helix away from domain III of EFTu 19 and preventing it from contributing to EFTu•EFTs binding (fig. 3).
Figure 3. .
Computational modeling based on the crystal structure of the bovine EFTu•EFTs complex. The top panel shows the structure of the Tu•Ts dimer. Tu is on the left side of the model, and Ts is on the right. The wild-type Arg 333 residue (depicted in orange) is superimposed on three Trp rotamers (depicted in yellow). The lower panel is a close-up of the region containing the mutation, which shows that the Arg residue fits well in front of the C-terminal side of the helix beneath it. The most favorable Trp rotamer bumps into this same helix. The second- and third-best local minima for the Trp rotamer bump into Glu 644 and Phe 637, respectively. The second rotamer also bumps into its own backbone.
To directly test whether the lack of a functional EFTs protein was the cause of the defective mitochondrial translation, we transduced patient fibroblasts with a retroviral vector expressing the wild-type TSFM cDNA. Since EFTs and EFTu form a dimer in human cells, we also tested whether overexpression of wild-type EFTu could suppress the translation defect. Overexpression of either of these constructs (two-to-threefold for EFTu and three-to-fourfold for EFTs; fig. 4) had a dominant negative effect on mitochondrial translation in control cells. In independent experiments with different control lines, global mitochondrial translation was 61% ± 18% (five independent transductions in two control lines) of control in cells overexpressing EFTu and 74% ± 14% of control in cells overexpressing EFTs (four independent transductions in two control lines). In the results shown in figure 1, global mitochondrial translation is 68% and 75% of the control in cells overexpressing EFTu and EFTs, respectively. A relatively modest increase in global mitochondrial protein synthesis was observed in patient cells overexpressing EFTs compared with the appropriate control; however, there was a significant increase in the rate of synthesis of several polypeptides of complex I and complex IV, and the rate of synthesis of ATP6 and ATP8 was significantly reduced relative to that observed in untransduced patient fibroblasts (table 2 and fig. 1). The results were generally similar in patient cells overexpressing EFTu. Thus, overexpression of either translation elongation factor could partially suppress the translation defect in the patient cells.
Figure 4. .
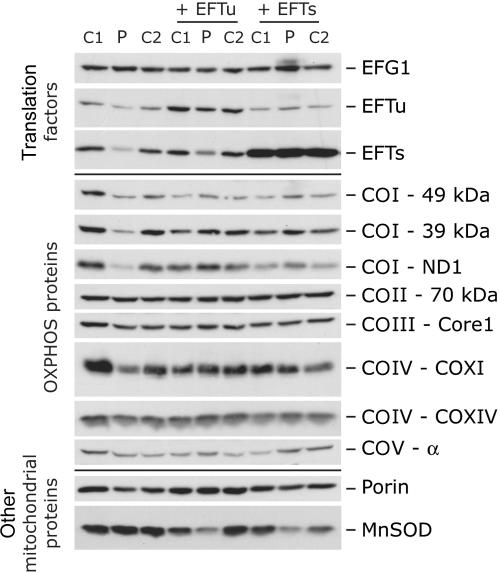
Immunoblot analysis of fibroblasts from patient 1. Immunoblot analysis of the steady-state levels of three translation elongation factors (EFG1, EFTu, and EFTs) and eight OXPHOS system subunits (COI–49 kDa, COI–39 kDa, COI-ND1, COII–70 kDa, COIII-Core1, COIV-COXI, COIV-COXIV, and COV-α) in control and patient fibroblasts before and after transduction with retroviral constructs expressing the mitochondrial translation elongation factors EFTu or EFTs. Porin and manganese superoxide dismutase (MnSOD) were used as loading controls.
To investigate the mechanism of suppression of the translation defect by EFTu, we analyzed the steady-state levels of the translation elongation factors by immunoblot analysis. EFTs and EFTu in patient fibroblasts were reduced to 25% and 40% of control levels, respectively (fig. 4). Overexpression of EFTu partially rescued EFTs levels, and EFTu levels were completely rescued by overexpression of EFTs. Overexpression of EFTs also slightly reduced EFTu levels in control cells. The levels of another mitochondrial translation elongation factor, EFG1, were unaltered in patient cells and were not changed in any of the cells overexpressing EFTs or EFTu.
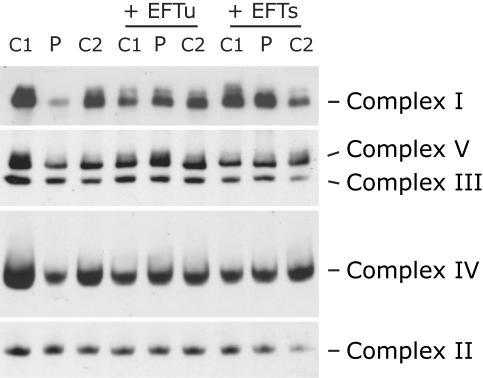
To investigate whether the increased mitochondrial protein synthesis in patient cells overexpressing EFTu or EFTs resulted in increased OXPHOS function, we examined the assembly of the OXPHOS complexes by blue-native PAGE analysis (fig. 5). Patient fibroblasts showed reduced levels of fully assembled complexes I, IV, and V but normal levels of complex II and III, consistent with the deficiencies demonstrated by enzyme activity assays (table 1). The assembly of all three affected complexes was restored to control levels (in comparison with control fibroblasts overexpressing the same construct) by overexpression of either elongation factor (fig. 5). Functional complementation of the biochemical defect in patient cells overexpressing the elongation factors was also demonstrated by immunoblot analysis, which showed similar steady-state levels of both nuclear- and mitochondrial-encoded subunits of the OXPHOS complexes in patient cells and controls (fig. 4). Interestingly, overexpression of both factors led to slightly higher steady-state levels of some complex I subunits (39 kDa, 49 kDa, and ND1) in the patient cell line compared with controls and a markedly decreased expression of MnSOD, the mitochondrial superoxide dismutase.
Figure 5. .
Blue-native PAGE analysis of the assembly of the OXPHOS complexes in patient fibroblasts. Fibroblasts from patient 1 and two controls (C1 and C2) were analyzed by blue-native PAGE before and after transduction with retroviral constructs expressing the mitochondrial translation elongation factors EFTu or EFTs. The gels were immunoblotted with antibodies directed against specific individual subunits to assess the amount of each of the fully assembled complexes.
Discussion
This study firmly establishes mutations in the mitochondrial translation elongation factor EFTs as the cause of disease in two unrelated patients with mitochondrial dysfunction due to a combined deficiency of OXPHOS enzymes. Several pieces of evidence support this conclusion. First, pulse labeling of the mitochondrial translation products in patient fibroblasts identified a global translation defect in both patients, leading to a failure to assemble adequate amounts of three of the OXPHOS complexes containing subunits encoded in mtDNA. This phenotype could be rescued by retroviral expression of the wild-type cDNAs for either EFTs or EFTu. Second, a homozygous missense mutation, predicting an amino acid substitution at an evolutionarily conserved site, was identified in TSFM in both patients. Molecular modeling predicts that this mutation would disrupt the Tu•Ts dimerization interface. Finally, the steady-state levels of mutant EFTs and wild-type EFTu were reduced in patient fibroblasts, and this phenotype was rescued by overexpression of either EFTu or EFTs.
Much of our knowledge of the mechanism of mitochondrial translation in mammals comes from studies of prokaryotic organisms, which share most of the same initiation, elongation, and termination factors.4 EFTs is a guanine nucleotide exchange factor that binds to the EFTu•guanosine diphosphate (GDP) complex, promoting the release of GDP and the formation of a stable EFTu•EFTs heterodimer. Guanosine triphosphate (GTP) then promotes dissociation of EFTs from this complex, regenerating EFTu•GTP, which can then bind another aminoacyl tRNA. This ternary complex associates with the ribosomal A site, and, when correct codon-anticodon recognition is established, GTP is hydrolyzed, releasing EFTu•GDP, and the cycle repeats itself. Interestingly, not all organisms require EFTs. For instance, the budding yeast Saccaromyces cerevisiae contains only EFTu, which can function both as a guanosine triphosphatase and its own guanine nucleotide exchange factor.23
Human TSFM maps to chromosome 12q13-q14, has an ORF of 1,040 bp, and comprises seven exons, although it appears that exon 5 is spliced out of the coding sequence in the human protein. EFTs from Escherichia coli is organized into four recognizable modules: N-terminus, core domain (consisting of subdomains N and C), coiled-coil domain, and C-terminal domain.24 The crystal structure of the bovine mitochondrial elongation EFTu•EFTs complex has been determined to a 0.22-nm resolution,19 and it appears to be broadly similar to that in E. coli, with a notable exception of the virtual absence of the coiled-coil and C-terminal domains in the mammalian protein.19 Human EFTu is 95% identical25 and human EFTs is 91% identical26 to the bovine sequence, and this permitted us to model the effects of the homozygous Arg333Trp mutation onto the bovine sequence without altering the backbone structure.
Molecular modeling predicts that the mutation will alter the local structure of subdomain C, abolishing most of the important interactions of this portion of EFTs with domain III of EFTu (fig. 3). This is likely to lead to the destabilization of the EFTu•EFTs complex and an increased turnover of its components, explaining the decreased steady-state levels of both factors we observed in the patient cells (fig. 4). Expression of the wild-type version of either factor would stabilize the complex, thus increasing the total EFTu•EFTs pool. This can explain why overexpression of EFTu acts as a suppressor of the translation defect in patient cells. However, the observation that overexpression of either EFTu or EFTs in control cells acts as a dominant negative—decreasing the rate of mitochondrial protein synthesis by ∼35%–40% and ∼20%–25%, respectively10 (fig. 1)—shows that the relative ratio of these elongation factors is a critical determinant of mitochondrial translation efficiency. Indeed, we observed that overexpression of EFTs slightly reduced the levels of EFTu in control fibroblasts, again suggesting that the relative expression of both factors is finely tuned. Consistent with this interpretation, co-overexpression of both EFTu and EFTs partially rescued the dominant negative effect of EFTu in control fibroblasts.10
It is not clear why the synthesis of the different mtDNA-encoded polypeptides is variably affected as a result of the TSFM mutation, but a similar phenomenon has been observed in fibroblasts from patients with mutations in the elongation factor EFG1.9,10 The most striking difference in this study, and in patients with EFG1 mutations, is in the synthesis of the two complex V subunits (ATP6 and ATP8), for which the rate of synthesis is similar to the control or even increased in the presence of the mutant elongation factor. The most parsimonious interpretation of this observation is that the mRNA coding for these polypeptides, a bicistronic mRNA with overlapping reading frames, can be translated more efficiently than can mRNAs coding for the other polypeptides, most of which (except ND4/4L) are translated from monocistronic mRNAs. Whether this difference reflects an increased efficiency of translation of the ATP6 and ATP8 mRNA per se, or simply reduced competition from other transcripts for a limited number of ribosomes, remains uncertain, since the factors that regulate the efficiency of translation initiation and elongation in mitochondria are largely unknown.
The Arg333Trp TSFM mutation in patient 1 led to a fatal mitochondrial encephalomyopathy, with muscle weakness, hypotonia, rhabdomyolysis, and epilepsy as the most prominent clinical signs and symptoms. In striking contrast, the identical mutation in patient 2 was associated with a hypertrophic cardiomyopathy, but the results of neurological examination and CT scan were normal. Although tissue specificity in the clinical phenotype is common in mitochondrial disease, as far as we are aware, this is the first example of a nuclear-encoded mitochondrial accessory factor in which the identical mutation is associated with such a remarkable difference in clinical presentation. In addition, in the patients with EFG1 mutations, cardiac function was normal, and the only apparent OXPHOS defect in this tissue was a modest reduction in the assembly of complex IV.9,10 Moreover, none of the other study patients with combined OXPHOS enzyme deficiencies presented with early-onset cardiomyopathy as the major clinical symptom. This suggests the presence of genetic modifiers, but these await identification. Combined enzyme deficiencies of the OXPHOS system are a frequent cause of mitochondrial disorders. The traditional genetic diagnostic approach in affected patients often includes the study of mitochondrial tRNAs and, more recently, sequencing of the entire mitochondrial genome. This study suggests that it will be useful to include a genetic screen of nuclear-encoded translation elongation factors in the diagnostic program for these patients or to perform SNP genotyping in all patients originating from consanguineous families.
Acknowledgments
We thank the support staff of the Nijmegen Centre for Mitochondrial Disorders for biochemical analysis of the patient skeletal muscle and fibroblast OXPHOS-system enzyme activities. J.S. and B.v.d.H. are currently supported by grants from the Radboud University Medical Centre Nijmegen, the Prinses Beatrix Fonds, and the European Union (EUMITOCOMBAT [LSHM-CT-2004-503116] and MitoCircle). This research was supported by the Canadian Institutes of Health Research (CIHR) (to E.A.S.), by the Israel Science Foundation (grant 1354-2005 [to O.E. and A. Saada]), and the Israeli Ministry of Science and Technology (grant 3-904 [to O.E.]). G.V. acknowledges BioSapiens, which is funded by the European Commission within its FP6 Programme, under the thematic area “Life sciences, genomics and biotechnology for health,” contract LSHG-CT-2003-503265. E.A.S. is an international scholar of the Howard Hughes Medical Institute and a senior scientist of the CIHR.
Web Resources
Accession numbers and URLs for data presented herein are as follows:
- BLAST, http://www.ncbi.nlm.nih.gov/BLAST/
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for TSFM [accession number NM_005726] and TUFM [accession number NM_003321])
- Helper Dependent Protocol, http://www.stanford.edu/group/nolan/protocols/pro_helper_dep.html
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for TSFM) [PubMed]
- POV-Ray, http://www.povray.org/
- TS Modelling, http://swift.cmbi.ru.nl/gv/service/ts/
- YASARA, http://www.yasara.org/
References
- 1.Thorburn DR (2004) Mitochondrial disorders: prevalence, myths and advances. J Inherit Metab Dis 27:349–362 10.1023/B:BOLI.0000031098.41409.55 [DOI] [PubMed] [Google Scholar]
- 2.Smeitink J, van den Heuvel L, DiMauro S (2001) The genetics and pathology of oxidative phosphorylation. Nat Rev Genet 2:342–352 10.1038/35072063 [DOI] [PubMed] [Google Scholar]
- 3.Jacobs HT (2003) Disorders of mitochondrial protein synthesis. Hum Mol Genet Spec No 2 12:R293–R301 10.1093/hmg/ddg285 [DOI] [PubMed] [Google Scholar]
- 4.Spremulli LL, Coursey A, Navratil T, Hunter SE (2004) Initiation and elongation factors in mammalian mitochondrial protein biosynthesis. Prog Nucleic Acid Res Mol Biol 77:211–261 [DOI] [PubMed] [Google Scholar]
- 5.Zhang Y, Spremulli LL (1998) Identification and cloning of human mitochondrial translational release factor 1 and the ribosome recycling factor. Biochim Biophys Acta 1443:245–250 [DOI] [PubMed] [Google Scholar]
- 6.O’Brien TW (2002) Evolution of a protein-rich mitochondrial ribosome: implications for human genetic disease. Gene 286:73–79 10.1016/S0378-1119(01)00808-3 [DOI] [PubMed] [Google Scholar]
- 7.DiMauro S, Schon EA (2003) Mitochondrial respiratory-chain diseases. N Engl J Med 348:2656–2668 10.1056/NEJMra022567 [DOI] [PubMed] [Google Scholar]
- 8.Taylor RW, Turnbull DM (2005) Mitochondrial DNA mutations in human disease. Nat Rev Genet 6:389–402 10.1038/nrg1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coenen MJ, Antonicka H, Ugalde C, Sasarman F, Rossi R, Heister JG, Newbold RF, Trijbels FJ, van den Heuvel LP, Shoubridge EA, Smeitink JA (2004) Mutant mitochondrial elongation factor G1 and combined oxidative phosphorylation deficiency. N Engl J Med 351:2080–2086 10.1056/NEJMoa041878 [DOI] [PubMed] [Google Scholar]
- 10.Antonicka H, Sasarman F, Kennaway NG, Shoubridge EA (2006) The molecular basis for tissue specificity of the oxidative phosphorylation deficiencies in patients with mutations in the mitochondrial translation factor EFG1. Hum Mol Genet 15:1835–1846 10.1093/hmg/ddl106 [DOI] [PubMed] [Google Scholar]
- 11.Miller C, Saada A, Shaul N, Shabtai N, Ben-Shalom E, Shaag A, Hershkovitz E, Elpeleg O (2004) Defective mitochondrial translation caused by a ribosomal protein (MRPS16) mutation. Ann Neurol 56:734–738 10.1002/ana.20282 [DOI] [PubMed] [Google Scholar]
- 12.Lochmuller H, Johns T, Shoubridge EA (1999) Expression of the E6 and E7 genes of human papillomavirus (HPV16) extends the life span of human myoblasts. Exp Cell Res 248:186–193 10.1006/excr.1999.4407 [DOI] [PubMed] [Google Scholar]
- 13.Smeitink J, Sengers R, Trijbels F, van den Heuvel L (2001) Human NADH:ubiquinone oxidoreductase. J Bioenerg Biomembr 33:259–266 10.1023/A:1010743321800 [DOI] [PubMed] [Google Scholar]
- 14.Janssen AJ, Smeitink JA, van den Heuvel LP (2003) Some practical aspects of providing a diagnostic service for respiratory chain defects. Ann Clin Biochem 40:3–8 10.1258/000456303321016114 [DOI] [PubMed] [Google Scholar]
- 15.Zhu Z, Yao J, Johns T, Fu K, De Bie I, Macmillan C, Cuthbert AP, Newbold RF, Wang J, Chevrette M, Brown GK, Brown RM, Shoubridge EA (1998) SURF1, encoding a factor involved in the biogenesis of cytochrome c oxidase, is mutated in Leigh syndrome. Nat Genet 20:337–343 10.1038/3804 [DOI] [PubMed] [Google Scholar]
- 16.Schagger H, von Jagow G (1991) Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal Biochem 199:223–231 10.1016/0003-2697(91)90094-A [DOI] [PubMed] [Google Scholar]
- 17.Klement P, Nijtmans LG, Van den Bogert C, Houstek J (1995) Analysis of oxidative phosphorylation complexes in cultured human fibroblasts and amniocytes by blue-native-electrophoresis using mitoplasts isolated with the help of digitonin. Anal Biochem 231:218–224 10.1006/abio.1995.1523 [DOI] [PubMed] [Google Scholar]
- 18.Boulet L, Karpati G, Shoubridge EA (1992) Distribution and threshold expression of the tRNA(Lys) mutation in skeletal muscle of patients with myoclonic epilepsy and ragged-red fibers (MERRF). Am J Hum Genet 51:1187–1200 [PMC free article] [PubMed] [Google Scholar]
- 19.Jeppesen MG, Navratil T, Spremulli LL, Nyborg J (2005) Crystal structure of the bovine mitochondrial elongation factor Tu.Ts complex. J Biol Chem 280:5071–5081 10.1074/jbc.M411782200 [DOI] [PubMed] [Google Scholar]
- 20.Vriend G (1990) WHAT IF: a molecular modeling and drug design program. J Mol Graph 8:52–56, 29 10.1016/0263-7855(90)80070-V [DOI] [PubMed] [Google Scholar]
- 21.De Filippis V, Sander C, Vriend G (1994) Predicting local structural changes that result from point mutations. Protein Eng 7:1203–1208 [DOI] [PubMed] [Google Scholar]
- 22.Andreoli C, Prokisch H, Hortnagel K, Mueller JC, Munsterkotter M, Scharfe C, Meitinger T (2004) MitoP2, an integrated database on mitochondrial proteins in yeast and man. Nucleic Acids Res 32:D459–D462 10.1093/nar/gkh137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chiron S, Suleau A, Bonnefoy N (2005) Mitochondrial translation: elongation factor Tu is essential in fission yeast and depends on an exchange factor conserved in humans but not in budding yeast. Genetics 169:1891–1901 10.1534/genetics.104.037473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kawashima T, Berthet-Colominas C, Wulff M, Cusack S, Leberman R (1996) The structure of the Escherichia coli EF-Tu.EF-Ts complex at 2.5 Å resolution. Nature 379:511–518 10.1038/379511a0 [DOI] [PubMed] [Google Scholar]
- 25.Woriax VL, Burkhart W, Spremulli LL (1995) Cloning, sequence analysis and expression of mammalian mitochondrial protein synthesis elongation factor Tu. Biochim Biophys Acta 1264:347–356 [DOI] [PubMed] [Google Scholar]
- 26.Xin H, Woriax V, Burkhart W, Spremulli LL (1995) Cloning and expression of mitochondrial translational elongation factor Ts from bovine and human liver. J Biol Chem 270:17243–17249 10.1074/jbc.270.29.17243 [DOI] [PubMed] [Google Scholar]